Incorporation of Antibiotics into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence
Abstract
:1. Introduction
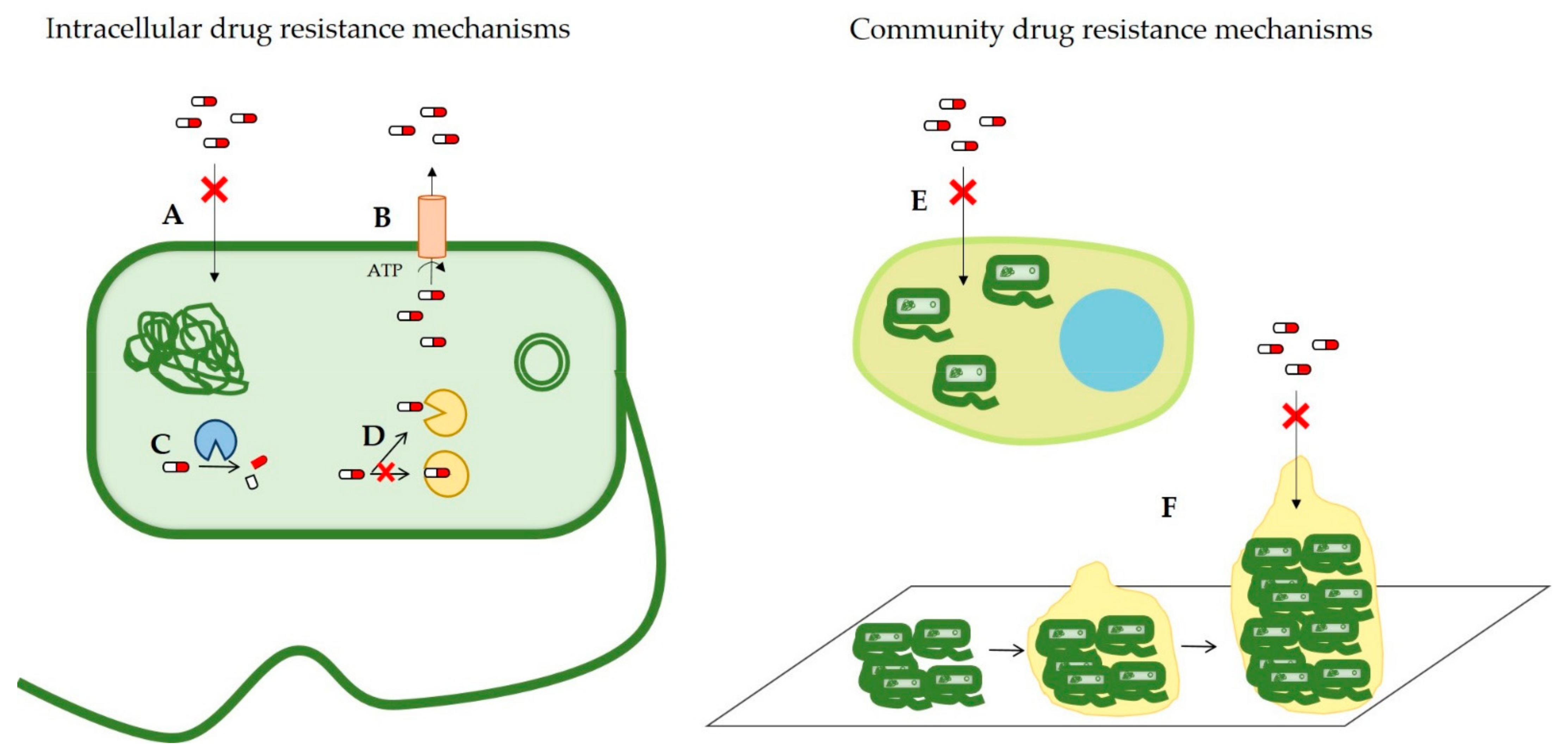
2. Antimicrobial Resistance Mechanisms
2.1. Reduced Drug Permeability
2.2. Overexpression of Efflux Pumps
2.3. Antibiotic-Modifying Enzymes
2.4. Modification of the Drug Target
2.5. Intracellular Infection
2.6. Biofilm Formation
3. New Strategies to Overcome Antimicrobial Resistance Mechanisms
4. Relevant SLNs Characteristics for an Efficient Drug Delivery
4.1. Size, Polydispersity and Zeta-Potential
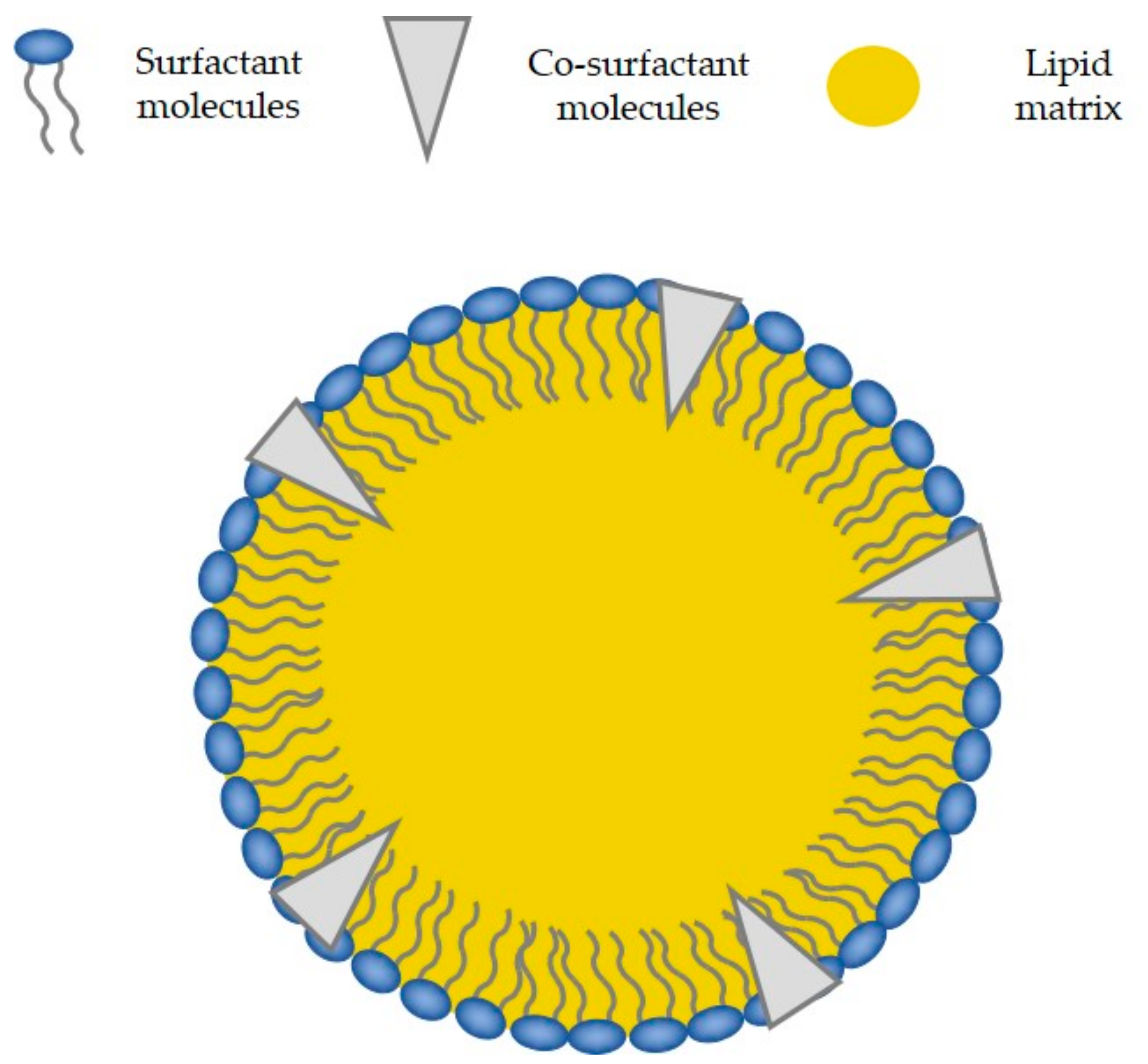
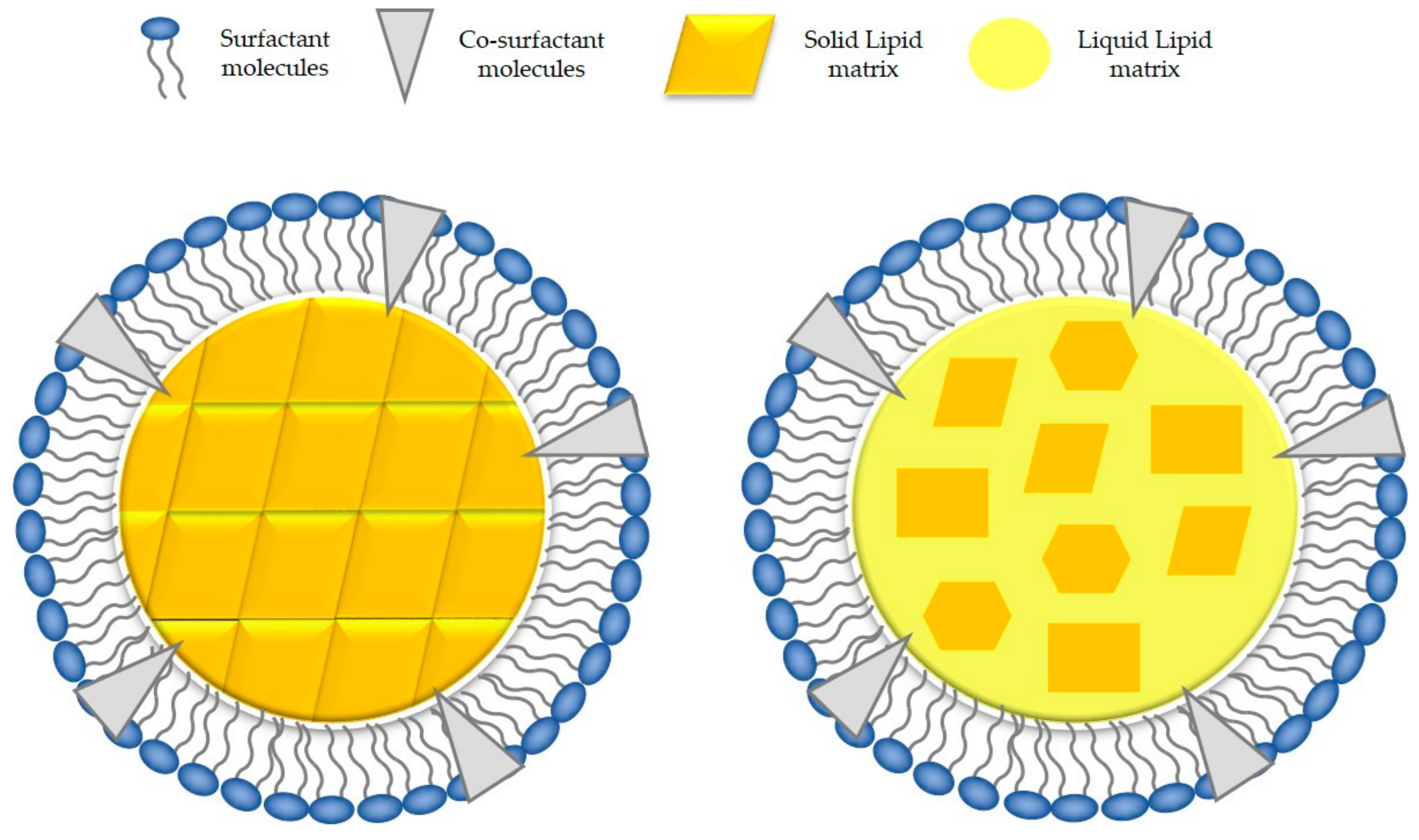
4.2. Solid State and Crystalline Structure of the Lipid Core
4.3. Entrapment Efficiency and Loading Capacity of the Drug
5. Solid Lipid Nanoparticle to Improve Drug Delivery
5.1. Improved Permeation and Bioavailability
5.2. Improved Selectivity
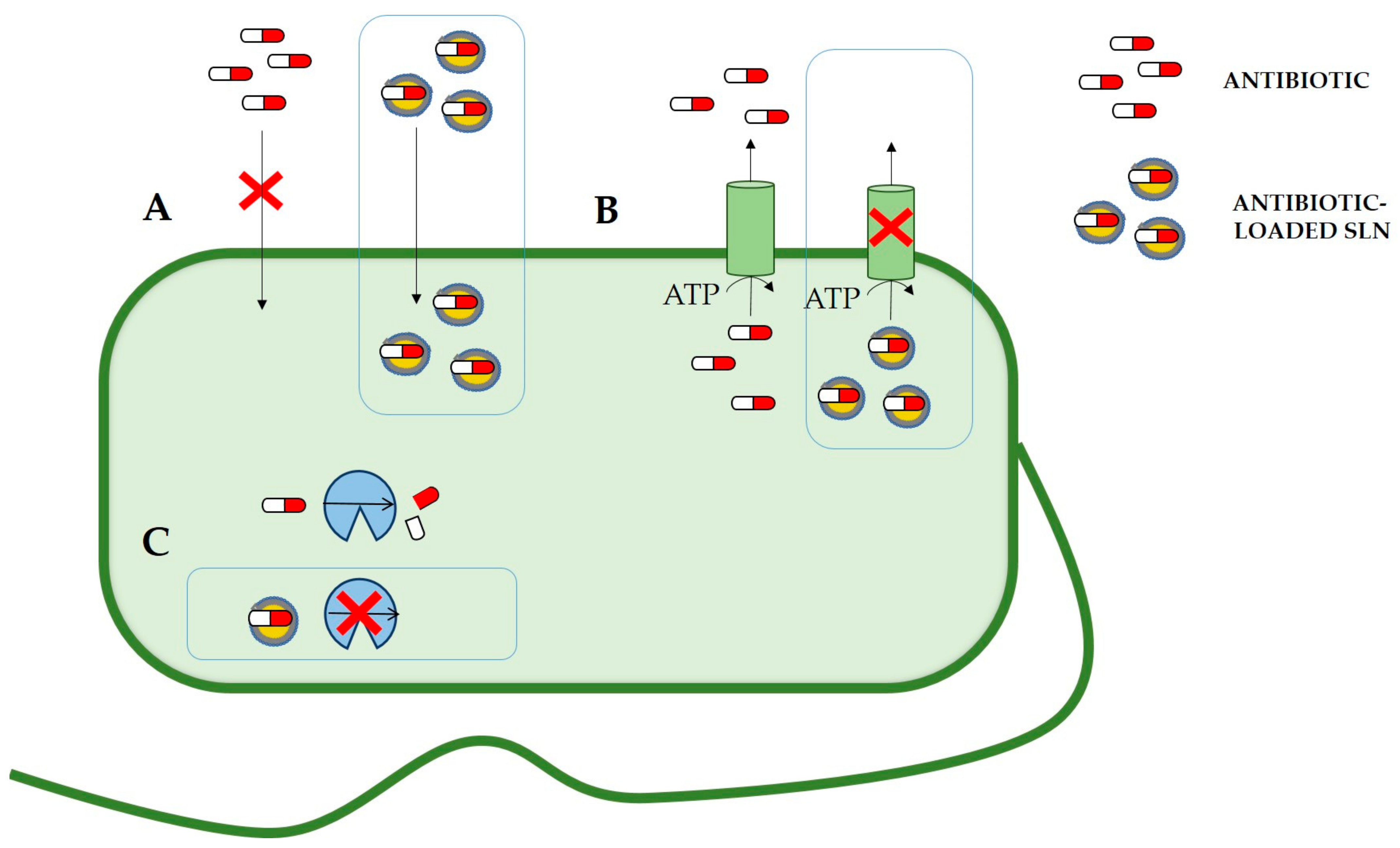
6. Solid Lipid Nanoparticles Can Reduce Antibiotic Resistance Mechanisms
6.1. Drug Efflux Pumps
6.2. Enzymatic Degradation
6.3. Infections by Intracellular Pathogens
6.4. Biofilm Formation and Quorum Sensing
7. Nanoparticles for Drug Combination Strategy
8. Solid Lipid Nanoparticles for the Delivery of New Antibiotic Agents
8.1. Oligonucleotides
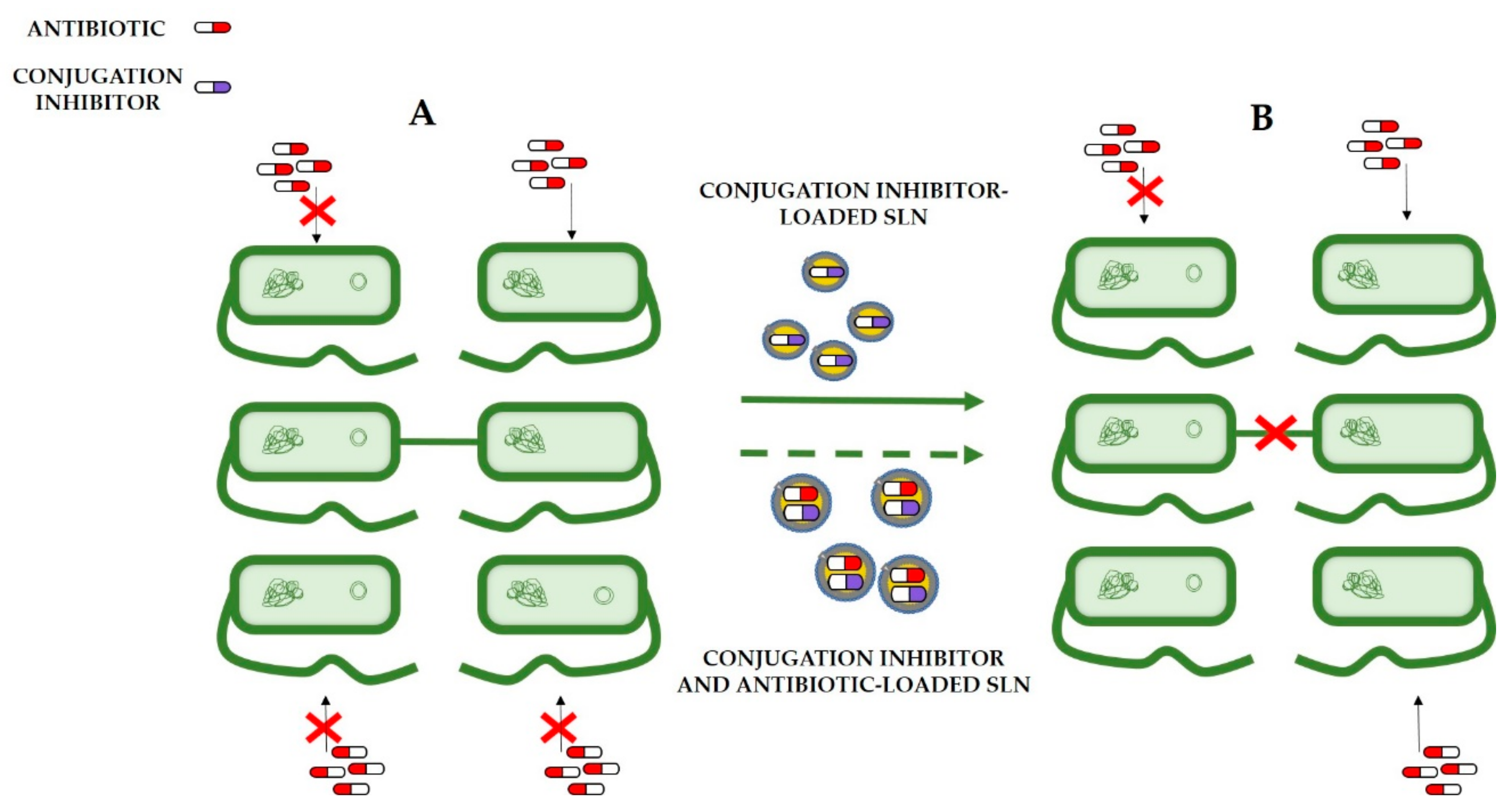
8.2. Conjugation Inhibitors
9. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Organization, W.H. Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 21 April 2021).
- Ling, Z.; Yonghong, L.; Junfeng, L.; Li, Z.; Xianqiang, L. Tilmicosin- and florfenicol-loaded hydrogenated castor oil-solid lipid nanoparticles to pigs: Combined antibacterial activities and pharmacokinetics. J. Vet. Pharmacol. Ther. 2018, 41, 307–313. [Google Scholar] [CrossRef]
- Nations, U. No Time to Wait: Securing the Future from Drug-Resistant Infections; Report to the Secretary-General of the United Nations; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- European Centre for Disease Prevention and Control. Strategies and Action Plans on Antimicrobial Resistance. Available online: https://www.ecdc.europa.eu/en/publications-data/directory-guidance-prevention-and-control/antimicrobial-resistance-strategies (accessed on 21 April 2021).
- McKenna, M. The antibiotic paradox: Why companies can’t afford to create life-saving drugs. Nature 2020, 584, 338–341. [Google Scholar] [CrossRef]
- Qin, S.Y.; Zhang, A.Q.; Cheng, S.X.; Rong, L.; Zhang, X.Z. Drug self-delivery systems for cancer therapy. Biomaterials 2017, 112, 234–247. [Google Scholar] [CrossRef]
- Thakur, S.; Riyaz, B.; Patil, A.; Kaur, A.; Kapoor, B.; Mishra, V. Novel drug delivery systems for NSAIDs in management of rheumatoid arthritis: An overview. Biomed. Pharmacother. 2018, 106, 1011–1023. [Google Scholar] [CrossRef]
- Akhtar, A.; Andleeb, A.; Waris, T.S.; Bazzar, M.; Moradi, A.R.; Awan, N.R.; Yar, M. Neurodegenerative diseases and effective drug delivery: A review of challenges and novel therapeutics. J. Control. Release 2021, 330, 1152–1167. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.; King, C.; Kalan, L.; Morar, M.; Sung, W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Aminov, R. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.; Webber, M.; Baylay, A.; Ogbolu, D.; Piddock, L. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Álvarez-Rodríguez, I.; Arana, L.; Ugarte-Uribe, B.; Gómez-Rubio, E.; Martín-Santamaría, S.; Garbisu, C.; Alkorta, I. Type IV Coupling Proteins as Potential Targets to Control the Dissemination of Antibiotic Resistance. Front. Mol. Biosci 2020, 7, 201. [Google Scholar] [CrossRef]
- Baquero, F.; Baquero-Artigao, G.; Cantón, R.; García-Rey, C. Antibiotic consumption and resistance selection in Streptococcus pneumoniae. J. Antimicrob Chemother 2002, 50 (Suppl. S2), 27–37. [Google Scholar] [CrossRef] [Green Version]
- Kamaruzzaman, N.F.; Kendall, S.; Good, L. Targeting the hard to reach: Challenges and novel strategies in the treatment of intracellular bacterial infections. Br. J. Pharmacol. 2017, 174, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Diab, R.; Khameneh, B.; Joubert, O.; Duval, R. Insights in Nanoparticle-Bacterium Interactions: New Frontiers to Bypass Bacterial Resistance to Antibiotics. Curr. Pharm. Des. 2015, 21, 4095–4105. [Google Scholar] [CrossRef]
- Lambert, P.A. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 2002, 92, 46S–54S. [Google Scholar] [CrossRef]
- Li, X.Z.; Nikaido, H. Efflux-mediated drug resistance in bacteria: An update. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Cerezales, M.; Ocampo-Sosa, A.A.; Álvarez Montes, L.; Díaz Ríos, C.; Bustamante, Z.; Santos, J.; Martínez-Martínez, L.; Higgins, P.G.; Gallego, L. High Prevalence of Extensively Drug-resistant Acinetobacter baumannii at a Children Hospital in Bolivia. Pediatr. Infect. Dis. J. 2018, 37, 1118–1123. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerezales, M.; Xanthopoulou, K.; Wille, J.; Krut, O.; Seifert, H.; Gallego, L.; Higgins, P.G. Mobile Genetic Elements Harboring Antibiotic Resistance Determinants in. Front. Microbiol 2020, 11, 919. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol 2004, 28, 47–62. [Google Scholar] [CrossRef]
- Águila-Arcos, S.; Ding, S.; Aloria, K.; Arizmendi, J.M.; Fearnley, I.M.; Walker, J.E.; Goñi, F.M.; Alkorta, I. A Commensal Strain of Staphylococcus epidermidis Overexpresses Membrane Proteins Associated with Pathogenesis When Grown in Biofilms. J. Membr. Biol. 2015, 248, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirtane, A.R.; Verma, M.; Karandikar, P.; Furin, J.; Langer, R.; Traverso, G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, D.; Kiselev, M.; Caccamo, M. Smart Nanoparticles for Drug Delivery Application: Development of Versatile Nanocarrier Platforms in Biotechnology and Nanomedicine. J. Nanomater. 2019, 2019. [Google Scholar] [CrossRef]
- Thorn, C.R.; Thomas, N.; Boyd, B.J.; Prestidge, C.A. Nano-fats for bugs: The benefits of lipid nanoparticles for antimicrobial therapy. Drug Deliv. Transl. Res. 2021. [Google Scholar] [CrossRef]
- Bayón-Cordero, L.; Alkorta, I.; Arana, L. Application of Solid Lipid Nanoparticles to Improve the Efficiency of Anticancer Drugs. Nanomaterials 2019, 9, 474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thi, T.T.H.; Suys, E.J.A.; Lee, J.S.; Nguyen, D.H.; Park, K.D.; Truong, N.P. Lipid-Based Nanoparticles in the Clinic and Clinical Trials: From Cancer Nanomedicine to COVID-19 Vaccines. Vaccines 2021, 9, 359. [Google Scholar] [CrossRef]
- Geszke-Moritz, M.; Moritz, M. Solid lipid nanoparticles as attractive drug vehicles: Composition, properties and therapeutic strategies. Mater. Sci Eng. C Mater. Biol Appl 2016, 68, 982–994. [Google Scholar] [CrossRef]
- Gordillo-Galeano, A.; Mora-Huertas, C.E. Solid lipid nanoparticles and nanostructured lipid carriers: A review emphasizing on particle structure and drug release. Eur. J. Pharm. Biopharm. 2018, 133, 285–308. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, W.; Mäder, K. Solid lipid nanoparticles: Production, characterization and applications. Adv. Drug Deliv. Rev. 2001, 47, 165–196. [Google Scholar] [CrossRef]
- Uner, M.; Yener, G. Importance of solid lipid nanoparticles (SLN) in various administration routes and future perspectives. Int. J. Nanomed. 2007, 2, 289–300. [Google Scholar]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale 2019, 11, 8760–8775. [Google Scholar] [CrossRef]
- Nishihira, V.S.K.; Rubim, A.M.; Brondani, M.; Dos Santos, J.T.; Pohl, A.R.; Friedrich, J.F.; de Lara, J.D.; Nunes, C.M.; Feksa, L.R.; Simão, E.; et al. In vitro and in silico protein corona formation evaluation of curcumin and capsaicin loaded-solid lipid nanoparticles. Toxicol. In Vitro 2019, 61, 104598. [Google Scholar] [CrossRef]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. Plasma protein adsorption and biological identity of systemically administered nanoparticles. Nanomedicine 2017, 12, 2113–2135. [Google Scholar] [CrossRef]
- Partikel, K.; Korte, R.; Stein, N.C.; Mulac, D.; Herrmann, F.C.; Humpf, H.U.; Langer, K. Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles. Eur. J. Pharm. Biopharm. 2019, 141, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S. DLS and zeta potential-What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Jo, D.H.; Kim, J.H.; Lee, T.G. Size, surface charge, and shape determine therapeutic effects of nanoparticles on brain and retinal diseases. Nanomedicine 2015, 11, 1603–1611. [Google Scholar] [CrossRef]
- Gastaldi, L.; Battaglia, L.; Peira, E.; Chirio, D.; Muntoni, E.; Solazzi, I.; Gallarate, M.; Dosio, F. Solid lipid nanoparticles as vehicles of drugs to the brain: Current state of the art. Eur. J. Pharm. Biopharm. 2014, 87, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Jang, J.H.; Cho, H.Y.; Lee, Y.B. Soft- and hard-lipid nanoparticles: A novel approach to lymphatic drug delivery. Arch. Pharm. Res. 2018, 41, 797–814. [Google Scholar] [CrossRef] [PubMed]
- Vranic, S.; Boggetto, N.; Contremoulins, V.; Mornet, S.; Reinhardt, N.; Marano, F.; Baeza-Squiban, A.; Boland, S. Deciphering the mechanisms of cellular uptake of engineered nanoparticles by accurate evaluation of internalization using imaging flow cytometry. Part. Fibre Toxicol. 2013, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Bragonzi, A.; Conese, M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol. Ther. 2005, 12, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Bhanjana, G.; Kumar, A.; Taneja, K.; Dilbaghi, N.; Kim, K. Synthesis and optimization of ceftriaxone-loaded solid lipid nanocarriers. Chem. Phys. Lipids 2016, 200, 126–132. [Google Scholar] [CrossRef]
- Pignatello, R.; Leonardi, A.; Fuochi, V.; Petronio Petronio, G.; Greco, A.S.; Furneri, P.M. A Method for Efficient Loading of Ciprofloxacin Hydrochloride in Cationic Solid Lipid Nanoparticles: Formulation and Microbiological Evaluation. Nanomaterials 2018, 8, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Paredes, A.; Sitia, L.; Ruyra, A.; Morris, C.J.; Wheeler, G.N.; McArthur, M.; Gasco, P. Solid lipid nanoparticles for the delivery of anti-microbial oligonucleotides. Eur. J. Pharm. Biopharm. 2019, 134, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.F.; Zhang, S.L.; Zhu, L.Y.; Xie, S.Y.; Dong, Z.; Wang, Y.; Zhou, W.Z. Enhancement of antibacterial activity of tilmicosin against Staphylococcus aureus by solid lipid nanoparticles in vitro and in vivo. Vet. J. 2012, 191, 115–120. [Google Scholar] [CrossRef]
- Severino, P.; Chaud, M.; Shimojo, A.; Antonini, D.; Lancelloti, M.; Santana, M.; Souto, E. Sodium alginate-cross-linked polymyxin B sulphate-loaded solid lipid nanoparticles: Antibiotic resistance tests and HaCat and NIH/3T3 cell viability studies. Colloids Surf. B Biointerfaces 2015, 129, 191–197. [Google Scholar] [CrossRef]
- Severino, P.; Silveira, E.F.; Loureiro, K.; Chaud, M.V.; Antonini, D.; Lancellotti, M.; Sarmento, V.H.; da Silva, C.F.; Santana, M.H.A.; Souto, E.B. Antimicrobial activity of polymyxin-loaded solid lipid nanoparticles (PLX-SLN): Characterization of physicochemical properties and in vitro efficacy. Eur. J. Pharm. Sci. 2017, 106, 177–184. [Google Scholar] [CrossRef]
- Fazly Bazzaz, B.S.; Khameneh, B.; Namazi, N.; Iranshahi, M.; Davoodi, D.; Golmohammadzadeh, S. Solid lipid nanoparticles carrying Eugenia caryophyllata essential oil: The novel nanoparticulate systems with broad-spectrum antimicrobial activity. Lett. Appl. Microbiol. 2018, 66, 506–513. [Google Scholar] [CrossRef]
- Maretti, E.; Costantino, L.; Buttini, F.; Rustichelli, C.; Leo, E.; Truzzi, E.; Iannuccelli, V. Newly synthesized surfactants for surface mannosylation of respirable SLN assemblies to target macrophages in tuberculosis therapy. Drug Deliv. Transl. Res. 2019, 9, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Yang, F.; Tao, Y.; Chen, D.; Qu, W.; Huang, L.; Liu, Z.; Pan, Y.; Yuan, Z. Enhanced intracellular delivery and antibacterial efficacy of enrofloxacin-loaded docosanoic acid solid lipid nanoparticles against intracellular Salmonella. Sci. Rep. 2017, 7, 41104. [Google Scholar] [CrossRef] [Green Version]
- Müller, R.H.; Shegokar, R.; Keck, C.M. 20 years of lipid nanoparticles (SLN and NLC): Present state of development and industrial applications. Curr. Drug Discov. Technol. 2011, 8, 207–227. [Google Scholar] [CrossRef] [PubMed]
- Daneshmand, S.; Golmohammadzadeh, S.; Jaafari, M.R.; Movaffagh, J.; Rezaee, M.; Sahebkar, A.; Malaekeh-Nikouei, B. Encapsulation challenges, the substantial issue in solid lipid nanoparticles characterization. J. Cell. Biochem. 2018, 119, 4251–4264. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.; Khatik, R.; Khandelwal, K.; Taneja, I.; Raju, K.S.; Wahajuddin; Paliwal, S.K.; Dwivedi, A.K.; Mishra, P.R. Pharmacokinetics study of arteether loaded solid lipid nanoparticles: An improved oral bioavailability in rats. Int. J. Pharm. 2014, 466, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yang, F.; Meng, K.; Chen, D.; Yang, Y.; Zhou, K.; Luo, W.; Qu, W.; Pan, Y.; Yuan, Z.; et al. Exploitation of enrofloxacin-loaded docosanoic acid solid lipid nanoparticle suspension as oral and intramuscular sustained release formulations for pig. Drug Deliv. 2019, 26, 273–280. [Google Scholar] [CrossRef]
- Pandey, R.; Sharma, S.; Khuller, G.K. Oral solid lipid nanoparticle-based antitubercular chemotherapy. Tuberculosis 2005, 85, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhu, L.; Dong, Z.; Wang, Y.; Wang, X.; Zhou, W. Preparation and evaluation of ofloxacin-loaded palmitic acid solid lipid nanoparticles. Int. J. Nanomed. 2011, 6, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Xie, S.; Zhu, L.; Wang, Y.; Wang, X.; Zhou, W. Preparation and in vitro, in vivo evaluations of norfloxacin-loaded solid lipid nanopartices for oral delivery. Drug Deliv. 2011, 18, 441–450. [Google Scholar] [CrossRef]
- Nirbhavane, P.; Vemuri, N.; Kumar, N.; Khuller, G.K. Lipid Nanocarrier-Mediated Drug Delivery System to Enhance the Oral Bioavailability of Rifabutin. AAPS PharmSciTech 2017, 18, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Yan, Y.; Chen, D.; Huang, L.; Li, C.; Meng, K.; Wang, S.; Algharib, S.; Yuan, Z.; Xie, S. Solid Lipid Nanoparticles for Duodenum Targeted Oral Delivery of Tilmicosin. Pharmaceutics 2020, 12, 731. [Google Scholar] [CrossRef]
- Singh, H.; Jindal, S.; Singh, M.; Sharma, G.; Kaur, I. Nano-formulation of rifampicin with enhanced bioavailability: Development, characterization and in-vivo safety. Int. J. Pharm. 2015, 485, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Zhu, L.; Dong, Z.; Wang, X.; Wang, Y.; Li, X.; Zhou, W. Preparation, characterization and pharmacokinetics of enrofloxacin-loaded solid lipid nanoparticles: Influences of fatty acids. Colloids Surf. B Biointerfaces 2011, 83, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Pastor, M.; Moreno-Sastre, M.; Esquisabel, A.; Sans, E.; Viñas, M.; Bachiller, D.; Asensio, V.J.; Pozo, A.D.; Gainza, E.; Pedraz, J.L. Sodium colistimethate loaded lipid nanocarriers for the treatment of Pseudomonas aeruginosa infections associated with cystic fibrosis. Int. J. Pharm. 2014, 477, 485–494. [Google Scholar] [CrossRef]
- Gaspar, D.P.; Gaspar, M.M.; Eleutério, C.V.; Grenha, A.; Blanco, M.; Gonçalves, L.M.D.; Taboada, P.; Almeida, A.J.; Remuñán-López, C. Microencapsulated Solid Lipid Nanoparticles as a Hybrid Platform for Pulmonary Antibiotic Delivery. Mol. Pharm. 2017, 14, 2977–2990. [Google Scholar] [CrossRef] [PubMed]
- Wosicka-Frąckowiak, H.; Cal, K.; Stefanowska, J.; Główka, E.; Nowacka, M.; Struck-Lewicka, W.; Govedarica, B.; Pasikowska, M.; Dębowska, R.; Jesionowski, T.; et al. Roxithromycin-loaded lipid nanoparticles for follicular targeting. Int. J. Pharm. 2015, 495, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Butani, D.; Yewale, C.; Misra, A. Topical Amphotericin B solid lipid nanoparticles: Design and development. Colloids Surf. B Biointerfaces 2016, 139, 17–24. [Google Scholar] [CrossRef]
- Raza, K.; Singh, B.; Singla, S.; Wadhwa, S.; Garg, B.; Chhibber, S.; Katare, O. Nanocolloidal Carriers of Isotretinoin: Antimicrobial Activity against Propionibacterium acnes and Dermatokinetic Modeling. Mol. Pharm. 2013, 10, 1958–1963. [Google Scholar] [CrossRef]
- Han, C.; Qi, C.M.; Zhao, B.K.; Cao, J.; Xie, S.Y.; Wang, S.L.; Zhou, W.Z. Hydrogenated castor oil nanoparticles as carriers for the subcutaneous administration of tilmicosin: In vitro and in vivo studies. J. Vet. Pharmacol. Ther. 2009, 32, 116–123. [Google Scholar] [CrossRef]
- Xu, X.M.; Wang, Y.S.; Chen, R.Y.; Feng, C.L.; Yao, F.; Tong, S.S.; Wang, L.; Yamashita, F.; Yu, J.N. Formulation and pharmacokinetic evaluation of tetracycline-loaded solid lipid nanoparticles for subcutaneous injection in mice. Chem. Pharm. Bull. 2011, 59, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Chetoni, P.; Burgalassi, S.; Monti, D.; Tampucci, S.; Tullio, V.; Cuffini, A.M.; Muntoni, E.; Spagnolo, R.; Zara, G.P.; Cavalli, R. Solid lipid nanoparticles as promising tool for intraocular tobramycin delivery: Pharmacokinetic studies on rabbits. Eur. J. Pharm. Biopharm. 2016, 109, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Eid, H.M.; Elkomy, M.H.; El Menshawe, S.F.; Salem, H.F. Development, Optimization, and In Vitro/In Vivo Characterization of Enhanced Lipid Nanoparticles for Ocular Delivery of Ofloxacin: The Influence of Pegylation and Chitosan Coating. AAPS PharmSciTech 2019, 20, 183. [Google Scholar] [CrossRef]
- Cavalli, R.; Gasco, M.R.; Chetoni, P.; Burgalassi, S.; Saettone, M.F. Solid lipid nanoparticles (SLN) as ocular delivery system for tobramycin. Int. J. Pharm. 2002, 238, 241–245. [Google Scholar] [CrossRef]
- Kalam, M.A.; Sultana, Y.; Ali, A.; Aqil, M.; Mishra, A.K.; Chuttani, K. Preparation, characterization, and evaluation of gatifloxacin loaded solid lipid nanoparticles as colloidal ocular drug delivery system. J. Drug Target. 2010, 18, 191–204. [Google Scholar] [CrossRef]
- Abul Kalam, M.; Sultana, Y.; Ali, A.; Aqil, M.; Mishra, A.K.; Chuttani, K.; Aljuffali, I.A.; Alshamsan, A. Part II: Enhancement of transcorneal delivery of gatifloxacin by solid lipid nanoparticles in comparison to commercial aqueous eye drops. J. Biomed. Mater. Res. A 2013, 101, 1828–1836. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.S.; Ahad, A.; Aslam, M.; Imam, S.S.; Aqil, M.; Ali, A. Application of Box-Behnken design for preparation of levofloxacin-loaded stearic acid solid lipid nanoparticles for ocular delivery: Optimization, in vitro release, ocular tolerance, and antibacterial activity. Int. J. Biol. Macromol. 2016, 85, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, R.; Zara, G.P.; Caputo, O.; Bargoni, A.; Fundarò, A.; Gasco, M.R. Transmucosal transport of tobramycin incorporated in SLN after duodenal administration to rats. Part I--a pharmacokinetic study. Pharmacol. Res. 2000, 42, 541–545. [Google Scholar] [CrossRef]
- Lopes-de-Campos, D.; Pinto, R.M.; Lima, S.A.C.; Santos, T.; Sarmento, B.; Nunes, C.; Reis, S. Delivering amoxicillin at the infection site-a rational design through lipid nanoparticles. Int. J. Nanomed. 2019, 14, 2781–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargoni, A.; Cavalli, R.; Zara, G.P.; Fundarò, A.; Caputo, O.; Gasco, M.R. Transmucosal transport of tobramycin incorporated in solid lipid nanoparticles (SLN) after duodenal administration to rats. Part II--tissue distribution. Pharmacol. Res. 2001, 43, 497–502. [Google Scholar] [CrossRef]
- Ghanbar, S.; Fumakia, M.; Ho, E.A.; Liu, S. A new strategy for battling bacterial resistance: Turning potent, non-selective and potentially non-resistance-inducing biocides into selective ones. Nanomedicine 2018, 14, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Sandri, G.; Bonferoni, M.C.; D’Autilia, F.; Rossi, S.; Ferrari, F.; Grisoli, P.; Sorrenti, M.; Catenacci, L.; Del Fante, C.; Perotti, C.; et al. Wound dressings based on silver sulfadiazine solid lipid nanoparticles for tissue repairing. Eur. J. Pharm. Biopharm. 2013, 84, 84–90. [Google Scholar] [CrossRef]
- Xie, S.; Wang, F.; Wang, Y.; Zhu, L.; Dong, Z.; Wang, X.; Li, X.; Zhou, W. Acute toxicity study of tilmicosin-loaded hydrogenated castor oil-solid lipid nanoparticles. Part. Fibre Toxicol. 2011, 8, 33. [Google Scholar] [CrossRef] [Green Version]
- Kalhapure, R.S.; Sikwal, D.R.; Rambharose, S.; Mocktar, C.; Singh, S.; Bester, L.; Oh, J.K.; Renukuntla, J.; Govender, T. Enhancing targeted antibiotic therapy via pH responsive solid lipid nanoparticles from an acid cleavable lipid. Nanomedicine 2017, 13, 2067–2077. [Google Scholar] [CrossRef]
- Mhule, D.; Kalhapure, R.S.; Jadhav, M.; Omolo, C.A.; Rambharose, S.; Mocktar, C.; Singh, S.; Waddad, A.Y.; Ndesendo, V.M.K.; Govender, T. Synthesis of an oleic acid based pH-responsive lipid and its application in nanodelivery of vancomycin. Int. J. Pharm. 2018, 550, 149–159. [Google Scholar] [CrossRef] [Green Version]
- Lei, Z.; Karim, A. The challenges and applications of nanotechnology against bacterial resistance. J. Vet. Pharmacol. Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Meng, K.; Chen, D.; Yang, F.; Zhang, A.; Tao, Y.; Qu, W.; Pan, Y.; Hao, H.; Xie, S. Intracellular delivery, accumulation, and discrepancy in antibacterial activity of four enrofloxacin-loaded fatty acid solid lipid nanoparticles. Colloids Surf. B Biointerfaces 2020, 194. [Google Scholar] [CrossRef]
- Moazeni, M.; Kelidari, H.R.; Saeedi, M.; Morteza-Semnani, K.; Nabili, M.; Gohar, A.A.; Akbari, J.; Lotfali, E.; Nokhodchi, A. Time to overcome fluconazole resistant Candida isolates: Solid lipid nanoparticles as a novel antifungal drug delivery system. Colloids Surf. B Biointerfaces 2016, 142, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Aboutaleb, E.; Noori, M.; Gandomi, N.; Atyabi, F.; Fazeli, M.R.; Jamalifar, H.; Dinarvand, R. Improved antimycobacterial activity of rifampin using solid lipid nanoparticles. Int. Nano Lett. 2012, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Shaaban, M.I.; Shaker, M.A.; Mady, F.M. Imipenem/cilastatin encapsulated polymeric nanoparticles for destroying carbapenem-resistant bacterial isolates. J. Nanobiotechnol. 2017, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Nacucchio, M.C.; Bellora, M.J.; Sordelli, D.O.; D’Aquino, M. Enhanced liposome-mediated activity of piperacillin against staphylococci. Antimicrob. Agents Chemother. 1985, 27, 137–139. [Google Scholar] [CrossRef] [Green Version]
- Groo, A.C.; Matougui, N.; Umerska, A.; Saulnier, P. Reverse micelle-lipid nanocapsules: A novel strategy for drug delivery of the plectasin derivate AP138 antimicrobial peptide. Int. J. Nanomed. 2018, 13, 7565–7574. [Google Scholar] [CrossRef] [Green Version]
- Battaglia, L.; Trotta, M.; Gallarate, M.; Carlotti, M.E.; Zara, G.P.; Bargoni, A. Solid lipid nanoparticles formed by solvent-in-water emulsion-diffusion technique: Development and influence on insulin stability. J. Microencapsul. 2007, 24, 660–672. [Google Scholar] [CrossRef]
- Dumont, C.; Bourgeois, S.; Fessi, H.; Dugas, P.Y.; Jannin, V. In-vitro evaluation of solid lipid nanoparticles: Ability to encapsulate, release and ensure effective protection of peptides in the gastrointestinal tract. Int. J. Pharm. 2019, 565, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Arana, L.; Bayón-Cordero, L.; Sarasola, L.I.; Berasategi, M.; Ruiz, S.; Alkorta, I. Solid Lipid Nanoparticles Surface Modification Modulates Cell Internalization and Improves Chemotoxic Treatment in an Oral Carcinoma Cell Line. Nanomaterials 2019, 9, 464. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.M.; Abbasalipourkabir, R.; Jalilian, F.A.; Asl, S.S.; Farmany, A.; Roshanaei, G.; Arabestani, M.R. Doxycycline-encapsulated solid lipid nanoparticles as promising tool against. Antimicrob. Resist. Infect. Control 2019, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wu, M.; Ye, W.; Huang, Z.; Ma, X.; Wang, W.; Huang, Y.; Pan, X.; Wu, C. Inhalable solid lipid nanoparticles for intracellular tuberculosis infection therapy: Macrophage-targeting and pH-sensitive properties. Drug Deliv. Transl. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Farmany, A.; Abbasalipourkabir, R.; Soleimani Asl, S.; Nourian, A.; Arabestani, M.R. Doxycycline-encapsulated solid lipid nanoparticles for the enhanced antibacterial potential to treat the chronic brucellosis and preventing its relapse: In vivo study. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.E. Recognition of complex carbohydrates by the macrophage mannose receptor. Biochem. Soc. Trans. 1993, 21, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Nimje, N.; Agarwal, A.; Saraogi, G.K.; Lariya, N.; Rai, G.; Agrawal, H.; Agrawal, G.P. Mannosylated nanoparticulate carriers of rifabutin for alveolar targeting. J. Drug Target. 2009, 17, 777–787. [Google Scholar] [CrossRef]
- Águila-Arcos, S.; Álvarez-Rodríguez, I.; Garaiyurrebaso, O.; Garbisu, C.; Grohmann, E.; Alkorta, I. Biofilm-Forming Clinical. Front. Microbiol 2017, 8, 2018. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Vuddanda, P.R.; Vijayakumar, M.R.; Kumar, V.; Saxena, P.S.; Singh, S. Cefuroxime axetil loaded solid lipid nanoparticles for enhanced activity against S. aureus biofilm. Colloids Surf. B Biointerfaces 2014, 121, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Fazly Bazzaz, B.S.; Khameneh, B.; Zarei, H.; Golmohammadzadeh, S. Antibacterial efficacy of rifampin loaded solid lipid nanoparticles against Staphylococcus epidermidis biofilm. Microb. Pathog. 2016, 93, 137–144. [Google Scholar] [CrossRef]
- Sharma, M.; Gupta, N.; Gupta, S. Implications of designing clarithromycin loaded solid lipid nanoparticles on their pharmacokinetics, antibacterial activity and safety. RSC Adv. 2016. [Google Scholar] [CrossRef]
- Luan, L.; Chi, Z.; Liu, C. Chinese White Wax Solid Lipid Nanoparticles as a Novel Nanocarrier of Curcumin for Inhibiting the Formation of Staphylococcus aureus Biofilms. Nanomaterials 2019, 9, 763. [Google Scholar] [CrossRef] [Green Version]
- Sans-Serramitjana, E.; Fusté, E.; Martínez-Garriga, B.; Merlos, A.; Pastor, M.; Pedraz, J.L.; Esquisabel, A.; Bachiller, D.; Vinuesa, T.; Viñas, M. Killing effect of nanoencapsulated colistin sulfate on Pseudomonas aeruginosa from cystic fibrosis patients. J. Cyst. Fibros. 2016, 15, 611–618. [Google Scholar] [CrossRef] [Green Version]
- Sans-Serramitjana, E.; Jorba, M.; Fusté, E.; Pedraz, J.L.; Vinuesa, T.; Viñas, M. Free and Nanoencapsulated Tobramycin: Effects on Planktonic and Biofilm Forms of Pseudomonas. Microorganisms 2017, 5, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reading, N.C.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nafee, N.; Husari, A.; Maurer, C.K.; Lu, C.; de Rossi, C.; Steinbach, A.; Hartmann, R.W.; Lehr, C.M.; Schneider, M. Antibiotic-free nanotherapeutics: Ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J. Control. Release 2014, 192, 131–140. [Google Scholar] [CrossRef]
- Zhang, R.X.; Wong, H.L.; Xue, H.Y.; Eoh, J.Y.; Wu, X.Y. Nanomedicine of synergistic drug combinations for cancer therapy-Strategies and perspectives. J. Control. Release 2016, 240, 489–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Han, L.; Liu, J.; Han, S.; Chen, Z.; Jiang, L. Co-delivery of paclitaxel and TOS-cisplatin via TAT-targeted solid lipid nanoparticles with synergistic antitumor activity against cervical cancer. Int. J. Nanomed. 2017, 12, 955–968. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Ji, H.; Ren, J.; Li, M.; Zheng, N.; Wu, L. Solid lipid nanoparticles with TPGS and Brij 78: A co-delivery vehicle of curcumin and piperine for reversing P-glycoprotein-mediated multidrug resistance. Oncol. Lett. 2017, 13, 389–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Han, L.; Deng, L.; Zhang, Y.; Shen, H.; Gong, T.; Zhang, Z.; Sun, X. Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J. Control. Release 2014, 194, 228–237. [Google Scholar] [CrossRef]
- Yu, Y.H.; Kim, E.; Park, D.E.; Shim, G.; Lee, S.; Kim, Y.B.; Kim, C.W.; Oh, Y.K. Cationic solid lipid nanoparticles for co-delivery of paclitaxel and siRNA. Eur. J. Pharm. Biopharm. 2012, 80, 268–273. [Google Scholar] [CrossRef]
- Ling, Z.; Yonghong, L.; Changqing, S.; Junfeng, L.; Li, Z.; Chunyu, J.; Xianqiang, L. Preparation, characterization, and pharmacokinetics of tilmicosin- and florfenicol-loaded hydrogenated castor oil-solid lipid nanoparticles. J. Vet. Pharmacol. Ther. 2017, 40, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Fuochi, V.; Zielińska, A.; Musumeci, T.; Souto, E.B.; Bonaccorso, A.; Puglia, C.; Petronio Petronio, G.; Furneri, P.M. Dual-drugs delivery in solid lipid nanoparticles for the treatment of Candida albicans mycosis. Colloids Surf. B Biointerfaces 2020, 186, 110705. [Google Scholar] [CrossRef]
- Kali, A.; Bhuvaneshwar, D.; Charles, P.M.; Seetha, K.S. Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates. J. Basic Clin. Pharm. 2016, 7, 93–96. [Google Scholar] [CrossRef] [Green Version]
- Alihosseini, F.; Azarmi, S.; Ghaffari, S.; Haghighat, S.; Rezayat Sorkhabadi, S.M. Synergic Antibacterial Effect of Curcumin with Ampicillin; Free Drug Solutions in Comparison with SLN Dispersions. Adv. Pharm. Bull. 2016, 6, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Rodenak-Kladniew, B.; Scioli Montoto, S.; Sbaraglini, M.L.; Di Ianni, M.; Ruiz, M.E.; Talevi, A.; Alvarez, V.A.; Durán, N.; Castro, G.R.; Islan, G.A. Hybrid Ofloxacin/eugenol co-loaded solid lipid nanoparticles with enhanced and targetable antimicrobial properties. Int. J. Pharm. 2019, 569, 118575. [Google Scholar] [CrossRef]
- Akhtari, H.; Bazzaz, B.; Golmohammadzadeh, S.; Movaffagh, J.; Soheili, V.; Khameneh, B. Rifampin and Cis-2-Decenoic Acid Co-entrapment in Solid Lipid Nanoparticles as an Efficient Nano-system with Potent Anti-biofilm Activities. J. Pharm. Innov. 2020. [Google Scholar] [CrossRef]
- Lewies, A.; Wentzel, J.F.; Jordaan, A.; Bezuidenhout, C.; Du Plessis, L.H. Interactions of the antimicrobial peptide nisin Z with conventional antibiotics and the use of nanostructured lipid carriers to enhance antimicrobial activity. Int. J. Pharm. 2017, 526, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Kalhapure, R.S.; Sonawane, S.J.; Sikwal, D.R.; Jadhav, M.; Rambharose, S.; Mocktar, C.; Govender, T. Solid lipid nanoparticles of clotrimazole silver complex: An efficient nano antibacterial against Staphylococcus aureus and MRSA. Colloids Surf. B Biointerfaces 2015, 136, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Meng, J.; Cao, L.; Li, Y.; Deng, P.; Pan, P.; Hu, C.; Yang, H. Synthesis and investigations of ciprofloxacin loaded engineered selenium lipid nanocarriers for effective drug delivery system for preventing lung infections of interstitial lung disease. J. Photochem. Photobiol. B 2019, 197, 111510. [Google Scholar] [CrossRef]
- Fumakia, M.; Ho, E.A. Nanoparticles Encapsulated with LL37 and Serpin A1 Promotes Wound Healing and Synergistically Enhances Antibacterial Activity. Mol. Pharm. 2016, 13, 2318–2331. [Google Scholar] [CrossRef]
- Islan, G.A.; Tornello, P.C.; Abraham, G.A.; Duran, N.; Castro, G.R. Smart lipid nanoparticles containing levofloxacin and DNase for lung delivery. Design and characterization. Colloids Surf. B Biointerfaces 2016, 143, 168–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjum, M.M.; Patel, K.K.; Dehari, D.; Pandey, N.; Tilak, R.; Agrawal, A.K.; Singh, S. Anacardic acid encapsulated solid lipid nanoparticles for Staphylococcus aureus biofilm therapy: Chitosan and DNase coating improves antimicrobial activity. Drug Deliv. Transl. Res. 2021, 11, 305–317. [Google Scholar] [CrossRef]
- Xue, X.Y.; Mao, X.G.; Zhou, Y.; Chen, Z.; Hu, Y.; Hou, Z.; Li, M.K.; Meng, J.R.; Luo, X.X. Advances in the delivery of antisense oligonucleotides for combating bacterial infectious diseases. Nanomedicine 2018, 14, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 2021, 11, 614–648. [Google Scholar] [CrossRef]
- Küçüktürkmen, B.; Bozkır, A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and miR-21 antisense oligonucleotide to glioblastoma cells. Drug Dev. Ind. Pharm. 2018, 44, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, X.; Xu, X.; Shen, L.; Yao, Y.; Yang, Z.; Liu, P. STAT3 Decoy Oligodeoxynucleotides-Loaded Solid Lipid Nanoparticles Induce Cell Death and Inhibit Invasion in Ovarian Cancer Cells. PLoS ONE 2015, 10, e0124924. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.E.; Kim, C.K. Long-term stable cationic solid lipid nanoparticles for the enhanced intracellular delivery of SMAD3 antisense oligonucleotides in activated murine macrophages. J. Pharm. Sci. 2012, 15, 467–482. [Google Scholar] [CrossRef] [Green Version]
- Bikard, D.; Euler, C.; Jiang, W.; Nussenzweig, P.; Goldberg, G.; Duportet, X.; Fischetti, V.; Marraffini, L. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Gomaa, A.; Klumpe, H.; Luo, M.; Selle, K.; Barrangou, R.; Beisel, C. Programmable Removal of Bacterial Strains by Use of Genome-Targeting CRISPR-Cas Systems. Mbio 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014, 32, 1141–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, A. CRISPR-Based Antibacterials: Transforming Bacterial Defense into Offense. Trends Biotechnol. 2018, 36, 127–130. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graf, F.E.; Palm, M.; Warringer, J.; Farewell, A. Inhibiting conjugation as a tool in the fight against antibiotic resistance. Drug Dev. Res. 2019, 80, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Getino, M.; Fernández-López, R.; Palencia-Gándara, C.; Campos-Gómez, J.; Sánchez-López, J.M.; Martínez, M.; Fernández, A.; de la Cruz, F. Tanzawaic Acids, a Chemically Novel Set of Bacterial Conjugation Inhibitors. PLoS ONE 2016, 11, e0148098. [Google Scholar] [CrossRef]
- Fernandez-Lopez, R.; Machón, C.; Longshaw, C.M.; Martin, S.; Molin, S.; Zechner, E.L.; Espinosa, M.; Lanka, E.; de la Cruz, F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 2005, 151, 3517–3526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Getino, M.; Sanabria-Ríos, D.J.; Fernández-López, R.; Campos-Gómez, J.; Sánchez-López, J.M.; Fernández, A.; Carballeira, N.M.; de la Cruz, F. Synthetic Fatty Acids Prevent Plasmid-Mediated Horizontal Gene Transfer. mBio 2015, 6, e01015–e01032. [Google Scholar] [CrossRef] [Green Version]

| Drug | Size | pdi | Zeta-Potential (mV) | EE (%) | Efficiency Enhancement | Organism | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Efflux pumps | Fluconazole | 84.8 ± 4.2 | 0.291 ± 0.012 | −25 ± 4.1 | 89.6 ± 3.97 | Avoidance of drug recognition by efflux pump proteins | Candida glabrata | [91] |
| Rifampin | 108.7 ± 5.5 | 0.18 | −10.7 ± 0.5 | 82 | Reduction of drug expulsion | Mycobacterium fortuitum (ATCC 2701P) | [92] | |
| Infections by intracellular pathogens | Rifampicin | 440 ± 40 | 0.37 ± 0.01 | − 49.73 ± 0.50 | 52.45 | Relevant and significant increase in drug content within the macrophage; Uptake by macrophages involving mannose receptors | J774A.1 | [55] |
| Enrofloxacin; docosanoic acid; 2% PVA; dimethyldioctadecyl ammonium chloride (0.5–4%) | 414.5 ± 3.8; 617.5 ± 7.1; 532.1 ± 10.0; 501.3 ± 16.6; 345.2 ± 9.6 | 0.265 ± 0.019; 0.458 ± 0.010; 0.461 ± 0.058; 0.417 ± 0.016; 0.393 ± 0.011 | −22.1 ± 0.1; −17.5 ± 0.6; −8.1 ± 0.4; 7.1 ± 0.5; 18.8 ± 0.2 | 86.6 ± 1.7; 42.8 ± 2.3; 41.2 ± 0.8; 46.7 ± 2.4; 45.6 ± 1.8 | Enhanced cellular uptake; Slower elimination of enrofloxacin after removing extracellular drug; Stronger inhibition effect against intracellular Salmonella CVCC541 | Intracellular Salmonella CVCC541 | [56] | |
| Doxycycline | 299 ± 34 | 0.29 ± 0.027 | −28.7 ± 3.2 | 94.9 ± 3.2 | Reduced number of bacteria inside J444A.1 macrophages | Intracellular Brucella melitensis | [99] | |
| isoniazid | 236 ± 9 | 0.240 ± 0.012 | − 19 ± 2 | 75.13 ± 0.97 | Increased intracellular antibiotic efficiency for the in vitro latent tuberculosis infection model; Superior antibiotic efficacy in the in vivo antibiotic tests compared to the INH solution | Mycobacterium tuberculosis; Wistar rats | [100] | |
| Enrofloxacin | 341.4 ± 4.9; 348.8 ± 3.5; 408.5 ± 6.3; 414.5±3.8 | 0.241 ± 0.014; 0.264 ± 0.013; 0.352 ± 0.015; 0.265±0.019 | −19.9 ± 0.4; −20.6 ± 0.9; −21.3 ± 0.6; −22.1 ± 0.1 | 65.2 ± 1.76; 67.53 ± 2.25; 72.57 ± 2.90; 86.56±1.60 | Enhanced cellular uptake; Slower elimination of enrofloxacin after removing extracellular drug; Stronger inhibition effect against intracellular Salmonella CVCC541 | Intracellular Salmonella CVCC541 | [90] | |
| Rifabutin-uncoatedrifabutin–mannose | 389 ± 2.3; 251 ± 5.1 | 0.357; 0.439 | 3.38 ± 0.3; −11.7 ± 0.8 | 87.8 ± 1.2; 82.6 ± 1.2 | Mannosylation enhances macrophage uptake Mannosylation promotes; selective uptake by lung tissues | J774 macrophages; Healthy albino rats | [103] | |
| Biofilm formation and quorum sensing | Cefuroxime axetil | 279.2 ± 28.5 | 0.107 ± 0.07 | −23.58 | 70.62 ± 0.82 | Drug minimum biofilm inhibitory concentration is 50% lower in SLNs | Staphylococcus aureus (ATCC-25923 | [105] |
| Rifampin | 101.7 ± 4.7 | 0.284 ± 0.024 | +17.1 ± 0.7 | 69% ± 2.1 | Significant reduction of the viability of bacteria embedded in biofilms | Biofilm-producing Staphylococcus epidermidis | [106] | |
| Clarithromycin | 307 ± 23 | 0.21 ± 0.04 | −29.0 | 84 ± 9 | Enhanced in vitro antibacterial activity; Higher potential in biofilm eradication compared to free drugs; Almost 5-fold improvement in relative oral bioavailability | Staphylococcus aureus; (MTCC86)Wistar rats | [107] | |
| Curcumin | 423.7 ± 23.2 | 0.310 ± 0.076 | −25.9 ± 6.7 | 85 | Satisfactory inhibition of biofilms | Staphylococcus aureus; (ATCC-12600) | [108] | |
| Colistin sulfate | 300–427 | 0.3–0.4 | n.d. | 80–95 | Efficient eradication of biofilms | Pseudomonas aeruginosa | [109] | |
| Tobramycin | 302 ± 20.5 | 0.361 ± 0.02 | −20.5 ± 6.09 | n.d. | Increased biofilms eradication | Pseudomonas aeruginosa | [110] | |
| Quorum sensing inhibitor (2-heptyl-6-nitro-4-oxo-1,4-dihydroquinoline-3-carboxamide) | <100 nm | <0.2 | −(15–35) | Reduction in pyocyanin 73.4 | (virulence factor) formation; High deposition in the bronchial area, the target site | Pseudomonas aeruginosa; Calu-3 cells | [112] |
| Drug | Size | pdi | Zeta-Potential (mV) | EE (%) | Efficiency Enhancement | Organism | Ref. |
|---|---|---|---|---|---|---|---|
| Ampicillin and; curcumin | 163 nm | <0.5 | n.d. | n.d. | Overcome resistance to free antibiotic; Overcome resistance to free antibiotic; Minimum bactericidal concentration decreased 4 times comparing to free drugs; Resistance of bacteria to free drugs is broken | Bacillus subtilis; Pseudomonas aeruginosa; Corynebacterium diphtheriae; Methicillin-resistant Staphylococcus aureus | [121] |
| Chitosan + ofloxacin/eugenol | 210.1 ± 5.9 | 0.418 ± 0.033 | 15.47 ± 0.21 | Ofloxacin 33.5 ± 1.9 | Minimum inhibitory concentration six-fold lower concerning the free antibiotic; MIC 16-times lower than that of free drug | Pseudomonas aeruginosa; Staphylococcus aureus | [122] |
| Rifampin + cis-2-decenoic acid | 127.2 ± 2.8 | 0.263 ± 0.017 | 19.0 ± 7.64 | Rifampin 69 ± 5.10C2DA 46 ± 4.23 | In vitro anti-biofilm activities at both formation and eradication stages | Staphylococcus aureus; Staphylococcus epidermidis | [123] |
| Ampicillin + nisin Z | 175.457± 17.885 | 0.279 ± 0.057 | −42.078 ± 0.903 | Ampicillin 43.826 ± 4.596 | Selective toxicity toward bacterial cells; Enhanced antibacterial activity of nisin ZNo improvement (electrostatic problems) | Staphylococcus aureus; Staphylococcus epidermidis Escherichia coli | [124] |
| Clotrimazole–Ag | 124.1± 2.5 | 0.235 ± 0.02 | −30.3 ± 5.9 | CTM 96.94 ± 0.42 | Enhanced and sustained antibacterial activity | Methicillin-resistant Staphylococcus aureus Methicillin-susceptible Staphylococcus aureus | [125] |
| Ciprofloxacin–selenium | 153.6 ±1.8 | 0.134 ± 0.03 | −1.74 ± 0.27 | CPF 40.4±4.4 | Greater antibacterial activity; Prevented the liver tissue damage | Pseudomonas aeruginosa; Mice | [126] |
| LL37 + serpin A1 | 214.9 ± 2.2; 261.7 ± 4.4 | n.d.; n.d. | −20 ± 1.8; −21 ± 2.1 | LL37:84.8 ± 2.7 A1:87 ± 3.5; LL37:81.6 ± 3.2A1:83.3 ± 4.1 | Synergistically enhance the antibacterial activity; In vitro accelerated wound healing | Staphylococcus aureus, Escherichia coli; Fibroblast and keratinocytes | [127] |
| Levofloxacin + DNase | 162.9 ± 5.3 | 0.340 ± 0.014 | −10.3 ± 0.3 | Levo 55.9 ± 1.6% | A strong antibacterial activity (less than free drug); Destroy biofilms after 24 h (Pseudomonas aeruginosa) | Pseudomonas aeruginosa Staphylococcus aureus | [128] |
| Anacardic acid + chitosan + DNAse | 212.8 ± 4.21 | 0.285 ± 0.04 | +13.5 ± 1.92 | Ana 73.8 ± 1.23% | Higher biofilm eradication activity | Staphylococcus aureus | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arana, L.; Gallego, L.; Alkorta, I. Incorporation of Antibiotics into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence. Nanomaterials 2021, 11, 1251. https://doi.org/10.3390/nano11051251
Arana L, Gallego L, Alkorta I. Incorporation of Antibiotics into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence. Nanomaterials. 2021; 11(5):1251. https://doi.org/10.3390/nano11051251
Chicago/Turabian StyleArana, Lide, Lucia Gallego, and Itziar Alkorta. 2021. "Incorporation of Antibiotics into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence" Nanomaterials 11, no. 5: 1251. https://doi.org/10.3390/nano11051251
APA StyleArana, L., Gallego, L., & Alkorta, I. (2021). Incorporation of Antibiotics into Solid Lipid Nanoparticles: A Promising Approach to Reduce Antibiotic Resistance Emergence. Nanomaterials, 11(5), 1251. https://doi.org/10.3390/nano11051251






