Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Exposure Conditions
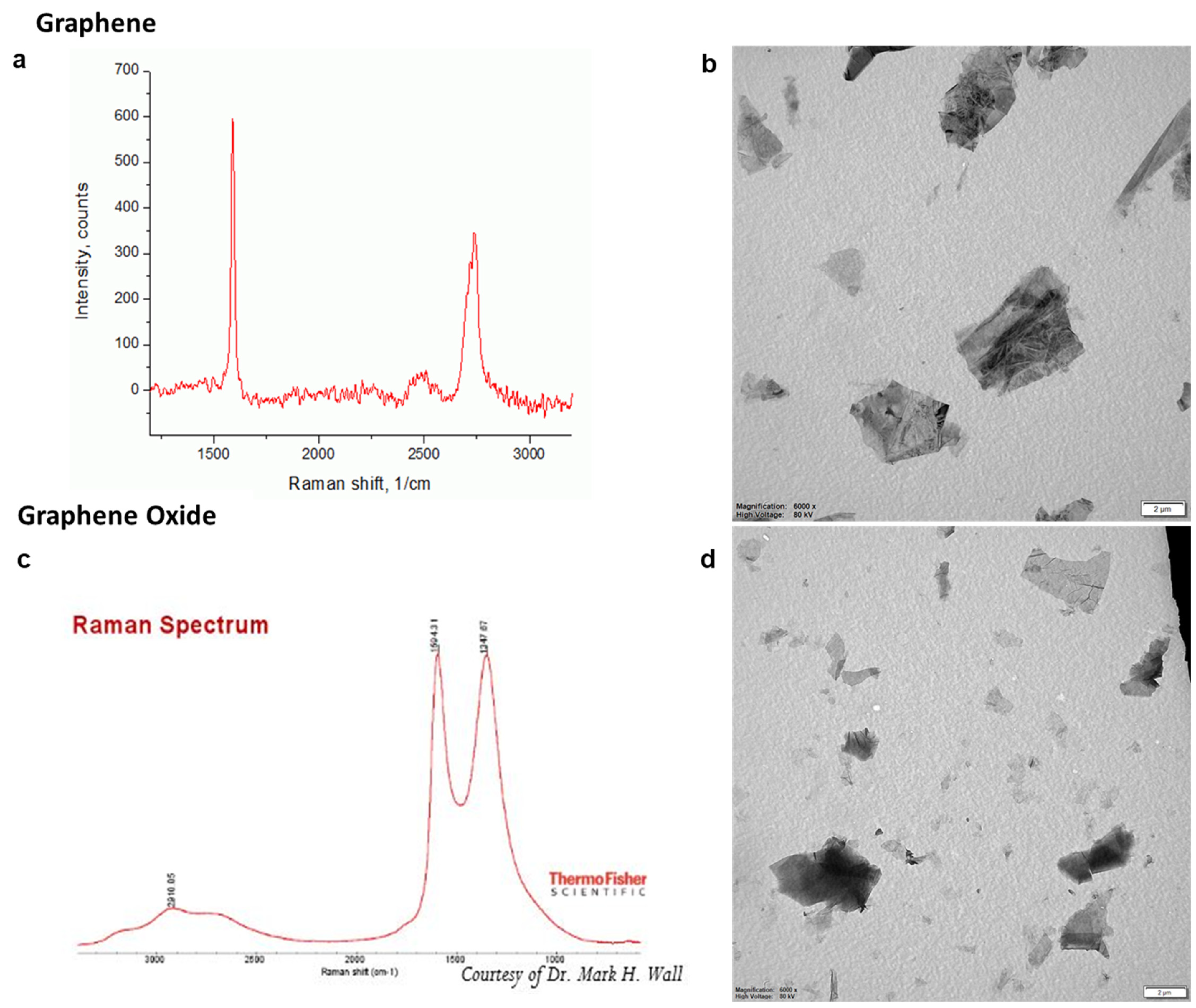
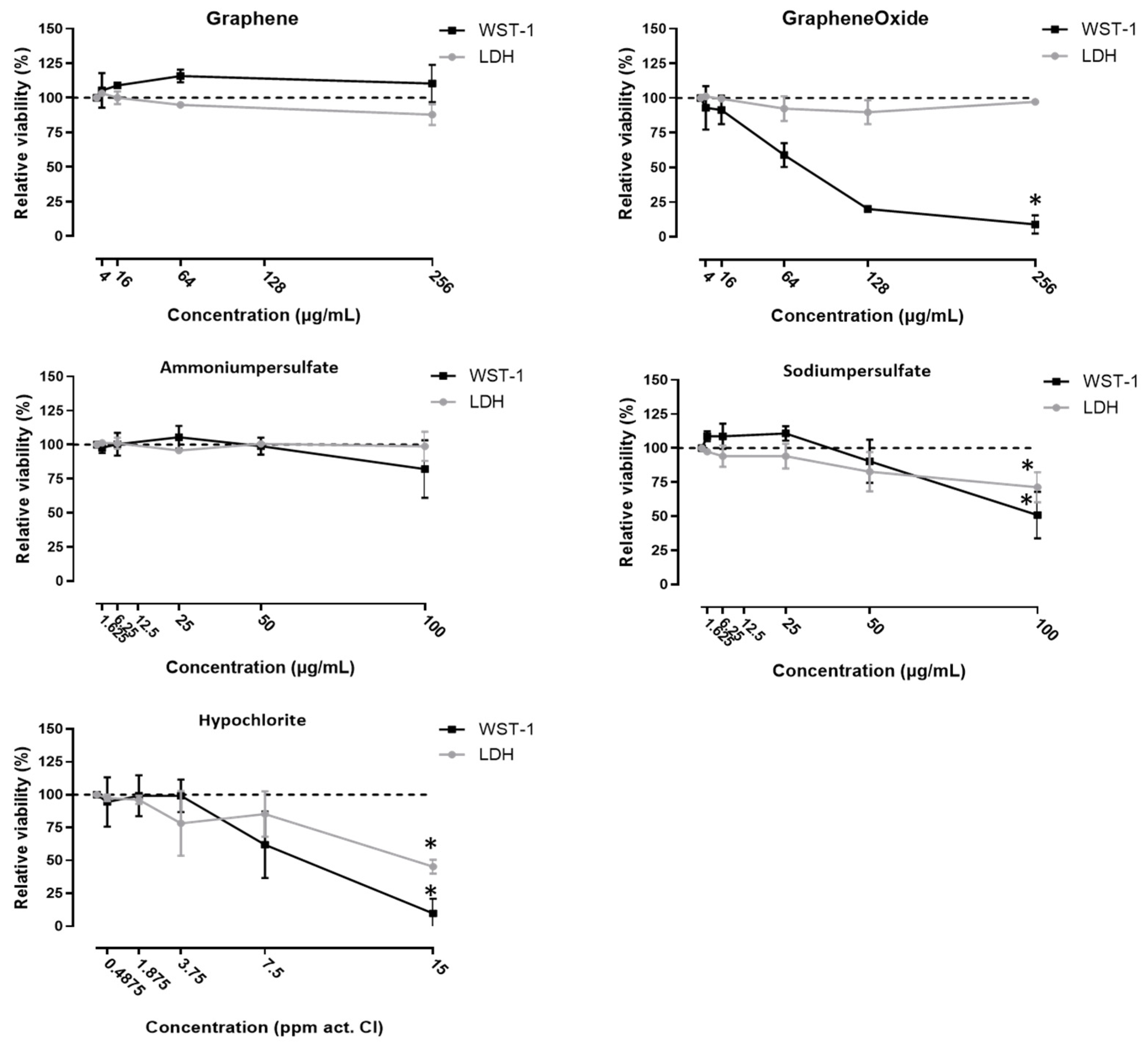
2.3. Cytotoxicity Measurements
2.4. Coculture
2.4.1. Setup
2.4.2. Exposure
2.4.3. Measurement of the Transepithelial Electrical Resistance (TEER)
2.4.4. Paracellular Flux of Fluorescent Labelled Dextran
2.4.5. Staining of the Intercellular Junctional Proteins
2.5. Phosphoproteomics
2.6. Statistical Analysis
3. Results
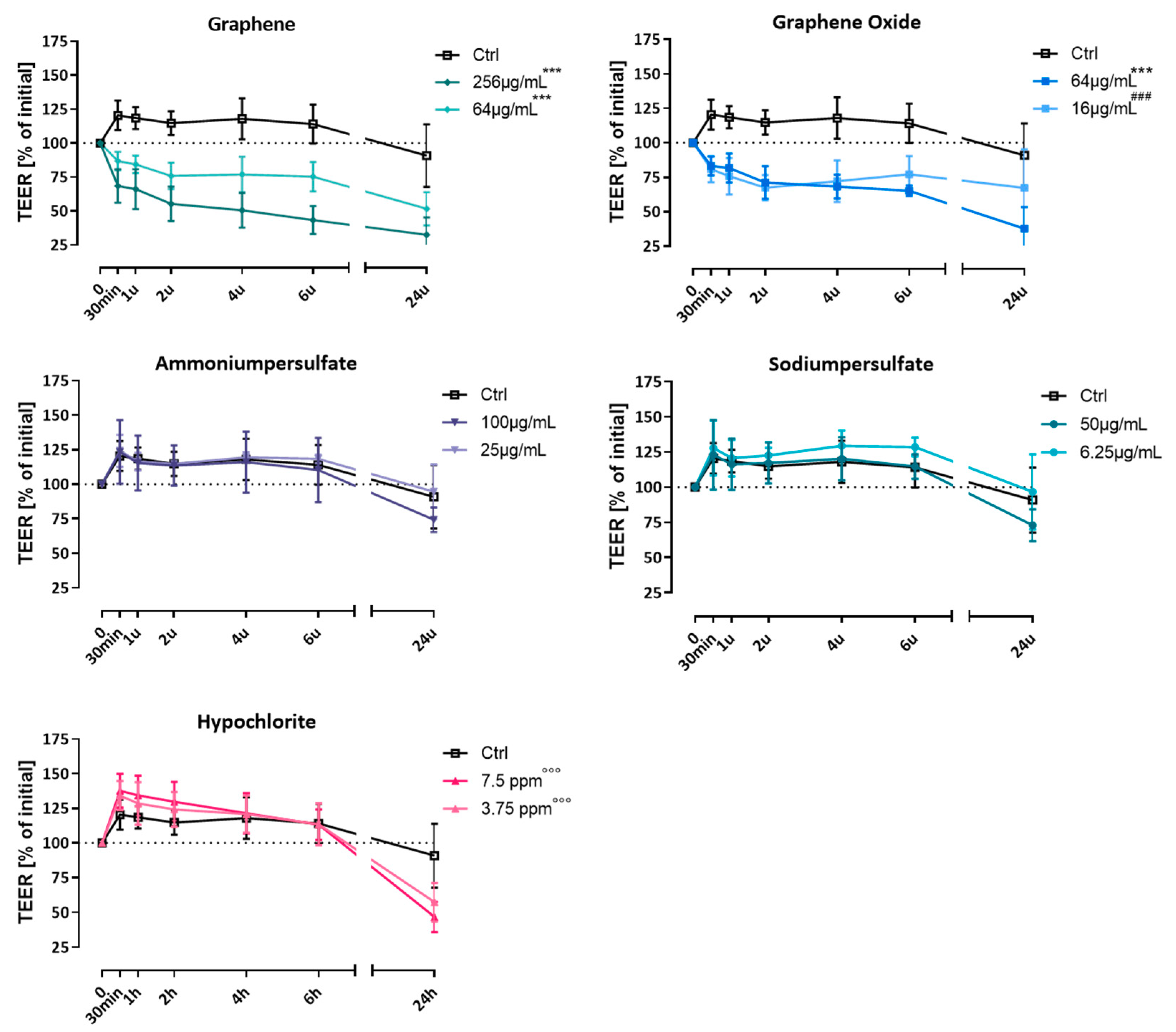
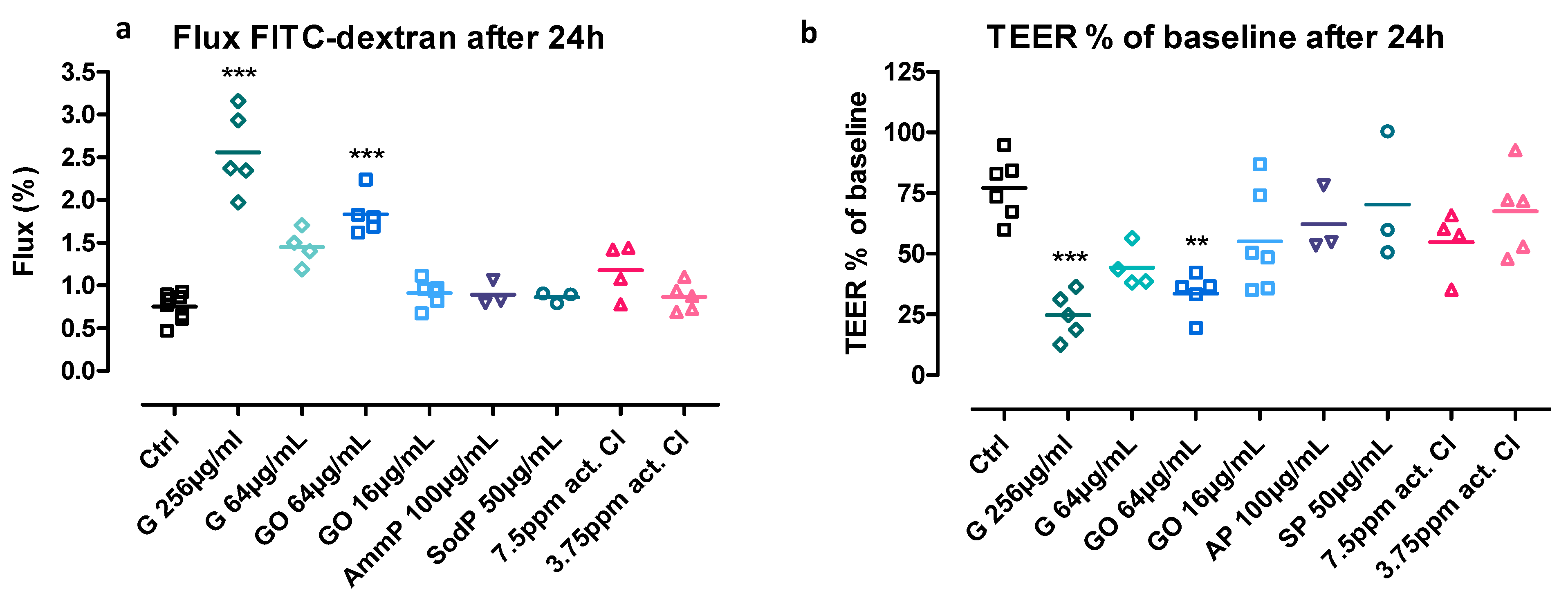
3.1. Effect on Transepithelial Electrical Resistance (TEER)
3.2. Flux of Fluorescent-Labelled Dextran
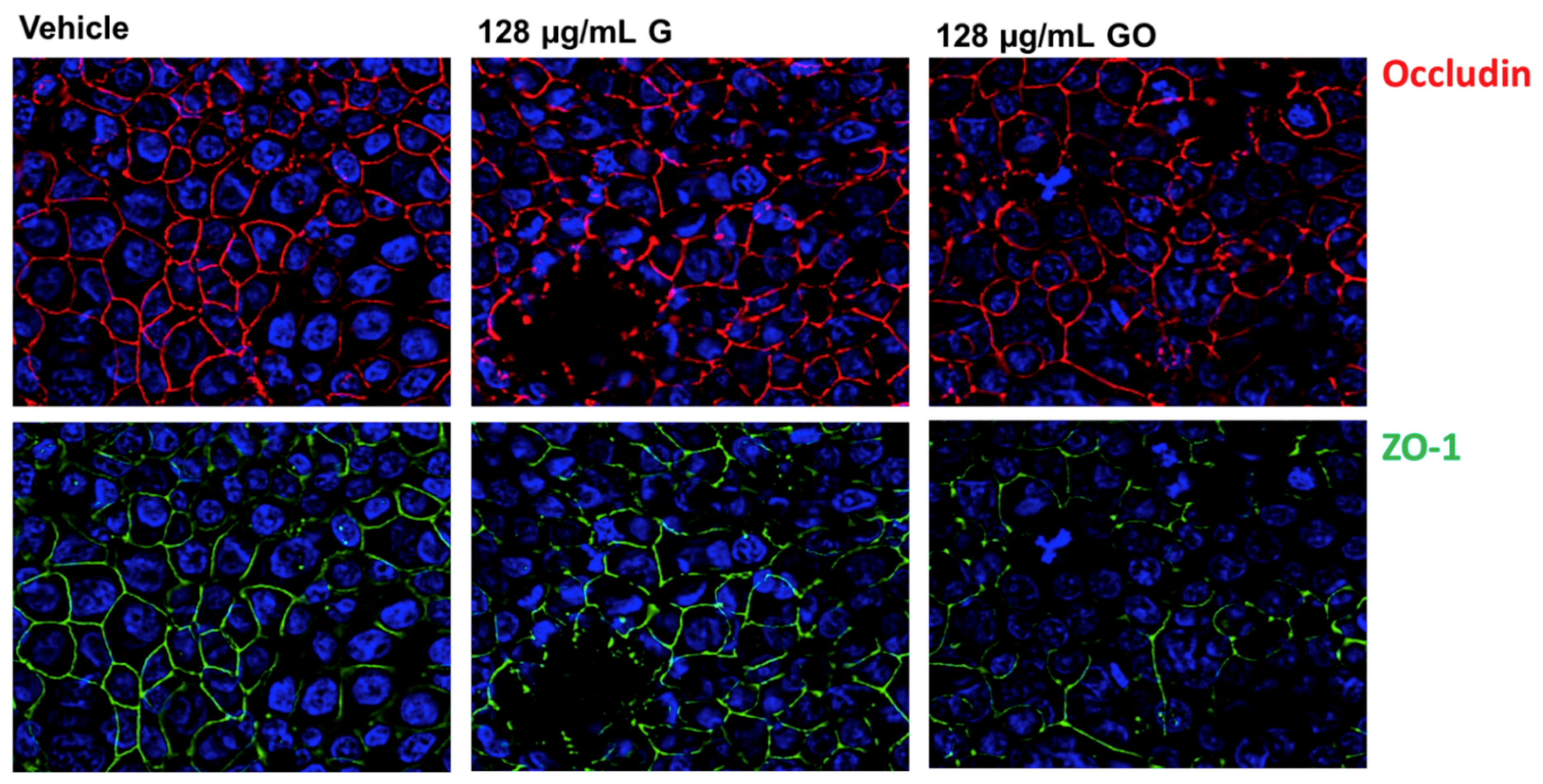
3.3. Effect of GNPs on the Airway Epithelium-Staining of Tight Junction (TJ) Proteins
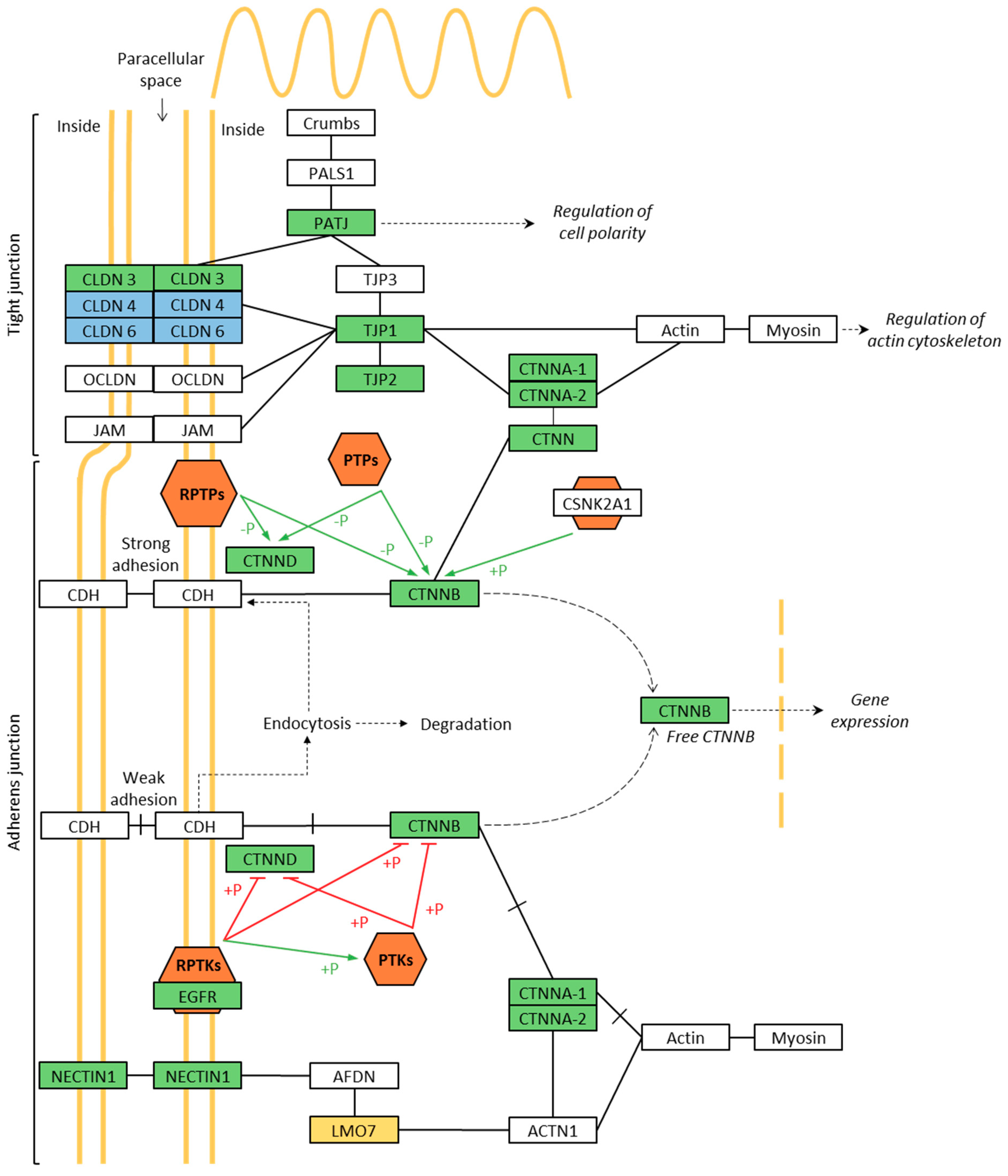
3.4. Phosphoproteomics of Membrane-Associated Proteins
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ganesan, S.; Comstock, A.T.; Sajjan, U.S. Barrier function of airway tract epithelium. Tissue Barriers 2013, 1, e24997. [Google Scholar] [CrossRef] [PubMed]
- Nava, P.; Kamekura, R.; Nusrat, A. Cleavage of transmembrane junction proteins and their role in regulating epithelial homeostasis. Tissue Barriers 2013, 1, e24783. [Google Scholar] [CrossRef]
- Wittekindt, O.H. Tight junctions in pulmonary epithelia during lung inflammation. Pflügers Arch. 2017, 469, 135–147. [Google Scholar] [CrossRef]
- Rezaee, F.; Georas, S.N. Breaking barriers. new insights into airway epithelial barrier function in health and disease. Am. J. Respir. Cell Mol. Biol. 2014, 50, 857–869. [Google Scholar] [CrossRef]
- Georas, S.N.; Rezaee, F. Epithelial barrier function: At the frontline of asthma immunology and allergic airway inflammation. J. Allergy Clin. Immunol. 2014, 134, 509–520. [Google Scholar] [CrossRef]
- Xiao, C.; Puddicombe, S.M.; Field, S.; Haywood, J.; Broughton-Head, V.; Puxeddu, I.; Haitchi, H.M.; Vernon-Wilson, E.; Sammut, D.; Bedke, N.; et al. Defective epithelial barrier function in asthma. J. Allergy Clin. Immunol. 2011, 128, 549. [Google Scholar] [CrossRef] [PubMed]
- Soyka, M.B.; Wawrzyniak, P.; Eiwegger, T.; Holzmann, D.; Treis, A.; Wanke, K.; Kast, J.I.; Akdis, C.A. Defective epithelial barrier in chronic rhinosinusitis: The regulation of tight junctions by IFN-γ and IL-4. J. Allergy Clin. Immunol. 2012, 130, 1087–1096.e10. [Google Scholar] [CrossRef]
- Heijink, I.H.; Noordhoek, J.A.; Timens, W.; Van Oosterhout, A.J.M.; Postma, D.S. Abnormalities in airway epithelial junction formation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2014, 189, 1439–1442. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.M.; Weiss, M.A.; Bernstein, I.L. Reactive airways dysfunction syndrome (RADS). Persistent asthma syndrome after high level irritant exposures. Chest 1985, 88, 376–384. [Google Scholar] [CrossRef]
- Brooks, S.M.; Malo, J.-L.; Gautrin, D. Irritant-induced asthma and reactive airways dysfunction syndrome. In Asthma in the Workplace; CRC Press: Boca Raton, FL, USA, 2013; Volume 21, pp. 305–324. [Google Scholar]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 1–24. [Google Scholar] [CrossRef]
- Arvidsson, R.; Molander, S.; Sandén, B.A. Review of potential environmental and health risks of the nanomaterial graphene. Hum. Ecol. Risk Assess. 2013, 19, 873–887. [Google Scholar] [CrossRef]
- Sanchez, V.C.; Jachak, A.; Hurt, R.H.; Kane, A.B. Biological interactions of graphene-family nanomaterials: An interdisciplinary review. Chem. Res. Toxicol. 2012, 25, 15–34. [Google Scholar] [CrossRef]
- Su, W.-C.; Ku, B.-K.; Kulkarni, P.; Cheng, Y.S. Deposition of graphene nanoparticles in human upper airways. J. Occup. Environ. Hyg. 2016, 13, 48–59. [Google Scholar] [CrossRef]
- Bianco, A. Graphene: Safe or toxic? The two faces of the medal. Angew. Chem. Int. Ed. 2013, 52, 4986–4997. [Google Scholar] [CrossRef]
- Horváth, L.; Magrez, A.; Burghard, M.; Kern, K.; Forró, L.; Schwaller, B. Evaluation of the toxicity of graphene derivatives on cells of the lung luminal surface. Carbon 2013, 64, 45–60. [Google Scholar] [CrossRef]
- Chang, Y.; Yang, S.-T.; Liu, J.-H.; Dong, E.; Wang, Y.; Cao, A.; Liu, Y.; Wang, H. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 2011, 200, 201–210. [Google Scholar] [CrossRef]
- Park, E.-J.; Lee, G.-H.; Han, B.S.; Lee, B.-S.; Lee, S.; Cho, M.-H.; Kim, J.-H.; Kim, D.-W. Toxic response of graphene nanoplatelets in vivo and in vitro. Arch. Toxicol. 2015, 89, 1557–1568. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, G.W.; Svendsen, E.R. Persistent effects of chlorine inhalation on respiratory health. Ann. N. Y. Acad. Sci. 2016, 1378, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Wigenstam, E.; Elfsmark, L.; Koch, B.; Bucht, A.; Jonasson, S. Acute respiratory changes and pulmonary inflammation involving a pathway of TGF-β1 induction in a rat model of chlorine-induced lung injury. Toxicol. Appl. Pharmacol. 2016, 309, 44–54. [Google Scholar] [CrossRef] [PubMed]
- White, C.W.; Martin, J.G. Chlorine gas inhalation: Human clinical evidence of toxicity and experience in animal models. Proc. Am. Thorac. Soc. 2010, 7, 257–263. [Google Scholar] [CrossRef]
- Kamath, Y.K.; Ruetsch, S.B. Reduction-induced surface modification of human hair. J. Cosmet. Sci. 2010, 61, 1–12. [Google Scholar] [PubMed]
- De Vooght, V.; Cruz, M.-J.; Haenen, S.; Wijnhoven, K.; Muñoz, X.; Hoet, P.H.; Morell, F.; Nemery, B.; Vanoirbeek, J.A. Ammonium persulfate can initiate an asthmatic response in mice. Thorax 2010, 65, 252–257. [Google Scholar] [CrossRef]
- Cruz, M.-J.; De Vooght, V.; Muñoz, X.; Hoet, P.H.M.; Morell, F.; Nemery, B.; Vanoirbeek, J.A.J. Assessment of the sensitization potential of persulfate salts used for bleaching hair. Contact Dermat. 2009, 60, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.J.; Olle-Monge, M.; Vanoirbeek, J.A.; Assialioui, A.; Gómez-Ollés, S.; Muñoz, X. Persistence of respiratory and inflammatory responses after dermal sensitization to persulfate salts in a mouse model of non-atopic asthma. Allergy Asthma Clin. Immunol. 2016, 12, 26. [Google Scholar] [CrossRef]
- Tarlo, S.M.; Lemiere, C. Occupational asthma. N. Engl. J. Med. 2014, 370, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Luyts, K.; Napierska, R.; Dinsdale, D.; Klein, S.G.; Serchi, T.; Hoet, P.H. A coculture model of the lung–blood barrier: The role of activated phagocytic cells. Toxicol. Vitro 2015, 29, 234–241. [Google Scholar] [CrossRef]
- Broucke, S.V.D.; Vanoirbeek, J.; Alfaro-Moreno, E.; Hoet, P. Contribution of mast cells in irritant-induced airway epithelial barrier impairment in vitro. Toxicol. Ind. Health 2020, 36, 823–834. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Wu, D.; De Wever, V.; Derua, R.; Winkler, C.; Beullens, M.; Van Eynde, A.; Bollen, M. A substrate-trapping strategy for protein phosphatase PP1 holoenzymes using hypoactive subunit fusions. J. Biol. Chem. 2018, 293, 15152–15162. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Matter, K.; Balda, M.S. Functional analysis of tight junctions. Methods 2003, 30, 228–234. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Phosphorylation of tight junction transmembrane proteins: Many sites, much to do. Tissue Barriers 2017, 6, e1382671. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Zhang, L.; Roy-Engel, A.M.; Saito, S.; A Lasky, J.; Wang, G.; Wang, H. Carcinogenic effects of oil dispersants: A KEGG pathway-based RNA-seq study of human airway epithelial cells. Gene 2017, 602, 16–23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, H.D.; Kim, T.-S.; Kim, J. Aberrant ribosome biogenesis activates c-Myc and ASK1 pathways resulting in p53-dependent G1 arrest. Oncogene 2011, 30, 3317–3327. [Google Scholar] [CrossRef][Green Version]
- Dai, M.-S.; Lu, H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J. Cell. Biochem. 2008, 105, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kwon, A.T.; Ramilowski, J.A.; Silberberg, G.; Söderhäll, C.; Orsmark-Pietras, C.; Nordlund, B.; Konradsen, J.R.; De Hoon, M.J.; Melén, E.; et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J. Allergy Clin. Immunol. 2015, 136, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mao, Z.D.; Shi, Y.J.; Qian, Y.; Liu, Z.G.; Yin, X.W.; Zhang, Q. Comprehensive analysis of miRNA-mRNA-lncRNA networks in severe asthma. Epigenomics 2019, 11, 115–131. [Google Scholar] [CrossRef]
- Huang, Y.; Pan, X.-W.; Li, L.; Chen, L.; Liu, X.; Lu, J.-L.; Zhu, X.-M.; Huang, H.; Yang, Q.-W.; Ye, J.-Q.; et al. Overexpression of USP39 predicts poor prognosis and promotes tumorigenesis of prostate cancer via promoting EGFR mRNA maturation and transcription elongation. Oncotarget 2016, 7, 22016–22030. [Google Scholar] [CrossRef]
- Fraile, J.M.; Manchado, E.; Lujambio, A.; Quesada, V.; Campos-Iglesias, D.; Webb, T.R.; Lowe, S.W.; López-Otín, C.; Freije, J.M.P. USP39 deubiquitinase is essential for KRAS oncogene-driven cancer. J. Biol. Chem. 2017, 292, 4164–4175. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, H.; Mei, M.; Du, C.; Huang, X.; Li, J.; Wang, Y.; Bao, S.; Zheng, H. Phosphorylation of serine/arginine-rich splicing factor 1 at tyrosine 19 promotes cell proliferation in pediatric acute lymphoblastic leukemia. Cancer Sci. 2018, 109, 3805–3815. [Google Scholar] [CrossRef]
- Issouf, M.; Vargas, A.; Boivin, R.; Lavoie, J. SRSF6 is upregulated in asthmatic horses and involved in the MYH11 SMB expression. Physiol. Rep. 2018, 6, e13896. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-T.T.; Hagner, S.; Ruchti, F.; Radzikowska, U.; Tan, G.; Altunbulakli, C.; Eljaszewicz, A.; Moniuszko, M.; Akdis, M.; Akdis, C.A.; et al. Tight junction, mucin, and inflammasome-related molecules are differentially expressed in eosinophilic, mixed, and neutrophilic experimental asthma in mice. Allergy 2019, 74, 294–307. [Google Scholar] [CrossRef]
- Olivera, D.; Knall, C.; Boggs, S.; Seagrave, J. Cytoskeletal modulation and tyrosine phosphorylation of tight junction proteins are associated with mainstream cigarette smoke-induced permeability of airway epithelium. Exp. Toxicol. Pathol. 2010, 62, 133–143. [Google Scholar] [CrossRef]
- Rao, R.K.; Basuroy, S.; Rao, V.U.; Karnaky, K.J., Jr.; Gupta, A. Tyrosine phosphorylation and dissociation of occludin-ZO-1 and E-cadherin-β-catenin complexes from the cytoskeleton by oxidative stress. Biochem. J. 2002, 368, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Blagoev, B.; Ong, S.-E.; Kratchmarova, I.; Mann, M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat. Biotechnol. 2004, 22, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, J.; Nika, H.; Hawke, D.; Keezer, S.; Ge, Q.; Fang, B.; Fang, X.; Fang, D.; Litchfield, D.W.; et al. EGF-induced ERK activation promotes CK2-mediated disassociation of α-Catenin from β-Catenin and transactivation of β-Catenin. Mol. Cell 2009, 36, 547–559. [Google Scholar] [CrossRef]
- Fang, D.; Hawke, D.; Zheng, Y.; Xia, Y.; Meisenhelder, J.; Nika, H.; Mills, G.B.; Kobayashi, R.; Hunter, T.; Lu, Z. Phosphorylation of β-Catenin by AKT Promotes β-Catenin Transcriptional Activity. J. Biol. Chem. 2007, 282, 11221–11229. [Google Scholar] [CrossRef]
- Wu, X.; Tu, X.; Joeng, K.S.; Hilton, M.J.; Williams, D.A.; Long, F. Rac1 activation and subsequent β-catenin phosphorylation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell 2008, 133, 340–353. [Google Scholar] [CrossRef]
- Le Bras, G.F.; Taubenslag, K.J.; Andl, C.D. The regulation of cell-cell adhesion during epithelial-mesenchymal transition, motility and tumor progression. Cell Adhes. Migr. 2012, 6, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Warzecha, C.C.; Carstens, R.P. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 2012, 22, 417–427. [Google Scholar] [CrossRef]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Silvestrini, V.C.; Lanfredi, G.P.; Masson, A.P.; Poersch, A.; Ferreira, G.A.; Thomé, C.H.; Faça, V.M. A proteomics outlook towards the elucidation of epithelial–mesenchymal transition molecular events. Mol. Omics 2019, 15, 316–330. [Google Scholar] [CrossRef] [PubMed]
| No. | Full Gene Name | Gene ID | p-Value | Enrichment | FDR |
|---|---|---|---|---|---|
| Spliceosome | 6.92 × 10−13 | 10.28 | 8.15 × 10−10 | ||
| 1 | Apoptotic chromatin condensation inducer 1 | ACIN1 | |||
| 2 | Calcium homeostasis endoplasmic reticulum protein | CHERP | |||
| 3 | DEAH-box helicase 16 | DHX16 | |||
| 4 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | HNRNPC | |||
| 5 | Heterogeneous nuclear ribonucleoprotein K | HNRNPK | |||
| 6 | Pre-mRNA processing factor 38A | PRPF38A | |||
| 7 | RNA binding motif protein 17 | RBM17 | |||
| 8 | RNA binding motif protein 8A | RBM8A | |||
| 9 | X-linked RNA binding motif protein X-linked | RBMX | |||
| 10 | Squamous cell carcinoma antigen recognized by T-cells 1 | SART1 | |||
| 11 | Small nuclear ribonucleoprotein U5 subunit 200 | SNRNP200 | |||
| 12 | Serine and arginine rich splicing factor 4 | SRSF4 | |||
| 13 | Serine and arginine rich splicing factor 6 | SRSF6 | |||
| 14 | Serine and arginine rich splicing factor 9 | SRSF9 | |||
| 15 | THO complex 2 | THOC2 | |||
| 16 | Transformer 2 alpha homolog | TRA2A | |||
| 17 | Transformer 2 beta homolog | TRA2B | |||
| 18 | Ubiquitin specific peptidase 39 | USP39 | |||
| Adherens Junction | 3.40 × 10−5 | 8.56 | 0.04 | ||
| 1 | Catenin alpha 1 | CTNNA1 | |||
| 2 | Catenin alpha 2 | CTNNA2 | |||
| 3 | Catenin beta 1 | CTNNB1 | |||
| 4 | Catenin delta 1 | CTNND1 | |||
| 5 | Epidermal growth factor receptor | EGFR | |||
| 6 | Lim domain 7 | LMO7 | |||
| 7 | Nectin cell adhesion molecule 1 | NECTIN1 | |||
| 8 | Tight junction protein 1 | TJP1 | |||
| RNA Transport | 3.72 × 10−4 | 4.41 | 0.44 | ||
| 1 | Apoptotic chromatin condensation inducer 1 | ACIN1 | |||
| 2 | FMR1 autosomal homolog 2 | FXR2 | |||
| 3 | Nucleoporin 88 | NUP88 | |||
| 4 | Nucleoporin 93 | NUP93 | |||
| 5 | Nucleoporin 98 | NUP98 | |||
| 6 | Pinin desmosome associated protein | PNN | |||
| 7 | RNA binding motif protein 8A | RBM8A | |||
| 8 | Serine and arginine repetitive matrix 1 | SRRM1 | |||
| 9 | THO complex 2 | THOC2 | |||
| 10 | THO complex 5 | THOC5 | |||
| Ribosome Biogenesis in Eukaryotes | 8.95 × 10−4 | 6.11 | 1.05 | ||
| 1 | Dyskerin pseudouridine synthase 1 | DKC1 | |||
| 2 | NOP56 ribonucleoprotein | NOP56 | |||
| 3 | NOP58 ribonucleoprotein | NOP58 | |||
| 4 | Treacle ribosome biogenesis factor 1(TCOF1) | TCOF1 | |||
| 5 | UTP 14 small subunit processome component | UTP14A | |||
| 6 | UTP 18 small subunit processome component 18 | UTP18 | |||
| 7 | 5′-3′ exoribonuclease 2 | XRN2 | |||
| Tight Junction | 1.96 × 10−3 | 4.43 | 2.29 | ||
| 1 | Claudin 3 | CLDN3 | |||
| 2 | Catenin alpha 1 | CTNNA1 | |||
| 3 | Catenin alpha 2 | CTNNA2 | |||
| 4 | Catenin beta 1 | CTNNB1 | |||
| 5 | Cortactin | CTTN | |||
| 6 | Crumbs cell polarity complex component | PATJ | |||
| 7 | Tight junction protein 1 | TJP1 | |||
| 8 | Tight junction protein 2 | TJP2 |
| No. | Full Gene Name | Gene ID | p-Value | Enrichment | FDR |
|---|---|---|---|---|---|
| Spliceosome | 1.75 × 10−12 | 9.74 | 2.08 × 10−9 | ||
| 1 | Apoptotic chromatin condensation inducer 1 | ACIN1 | |||
| 2 | Calcium homeostasis endoplasmic reticulum protein | CHERP | |||
| 3 | DEAH-box helicase 16 | DHX16 | |||
| 4 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | HNRNPC | |||
| 5 | Heterogeneous nuclear ribonucleoprotein K | HNRNPK | |||
| 6 | Pre-mRNA processing factor 38A | PRPF38A | |||
| 7 | RNA binding motif protein 8A | RBM8A | |||
| 8 | X-linked RNA binding motif protein X-linked | RBMX | |||
| 9 | Squamous cell carcinoma antigen recognized by T-cells 1 | SART1 | |||
| 10 | Small nuclear ribonucleoprotein U5 subunit 200 | SNRNP200 | |||
| 11 | Serine and arginine rich splicing factor 10 | SRSF10 | |||
| 12 | Serine and arginine rich splicing factor 4 | SRSF4 | |||
| 13 | Serine and arginine rich splicing factor 6 | SRSF6 | |||
| 14 | Serine and arginine rich splicing factor 9 | SRSF9 | |||
| 15 | THO complex 2 | THOC2 | |||
| 16 | Transformer 2 alpha homolog | TRA2A | |||
| 17 | Transformer 2 beta homolog | TRA2B | |||
| 18 | Ubiquitin specific peptidase 39 | USP39 | |||
| Tight Junction | 1.01 × 10−4 | 5.25 | 0.12 | ||
| 1 | Claudin 3 | CLDN3 | |||
| 2 | Claudin 4 | CLDN4 | |||
| 3 | Claudin 6 | CLDN6 | |||
| 4 | Catenin alpha 1 | CTNNA1 | |||
| 5 | Catenin alpha 2 | CTNNA2 | |||
| 6 | Catenin beta 1 | CTNNB1 | |||
| 7 | Cortactin | CTTN | |||
| 8 | Crumbs cell polarity complex component | PATJ | |||
| 9 | Tight junction protein 1 | TJP1 | |||
| 10 | Tight junction protein 2 | TJP2 | |||
| Adherens Junction | 4.02 × 10−4 | 7.10 | 4.75 × 10−1 | ||
| 1 | Catenin alpha 1 | CTNNA1 | |||
| 2 | Catenin alpha 2 | CTNNA2 | |||
| 3 | Catenin beta 1 | CTNNB1 | |||
| 4 | Catenin delta 1 | CTNND1 | |||
| 5 | Epidermal growth factor receptor | EGFR | |||
| 6 | Nectin cell adhesion molecule 1 | NECTIN1 | |||
| 7 | Tight junction protein 1 | TJP1 | |||
| RNA Transport | 5.57 × 10−4 | 4.18 | 0.66 | ||
| 1 | Apoptotic chromatin condensation inducer 1 | ACIN1 | |||
| 2 | Eukaryotic translation initiation factor 5B | EIF5B | |||
| 3 | FMR1 autosomal homolog 2 | FXR2 | |||
| 4 | Nucleoporin 93 | NUP93 | |||
| 5 | Nucleoporin 98 | NUP98 | |||
| 6 | Pinin desmosome associated protein | PNN | |||
| 7 | RNA binding motif protein 8A | RBM8A | |||
| 8 | Serine and arginine repetitive matrix 1 | SRRM1 | |||
| 9 | THO complex 2 | THOC2 | |||
| 10 | THO complex 5 | THOC5 | |||
| Cell Adhesion Molecules (CAM) | 7.04 × 10−4 | 4.56 | 0.83 | ||
| 1 | Claudin 3 | CLDN3 | |||
| 2 | Claudin 4 | CLDN4 | |||
| 3 | Claudin 6 | CLDN6 | |||
| 4 | Major histocompatibility complex class I A | HLA-A | |||
| 5 | Major histocompatibility complex class I B | HLA-B | |||
| 6 | Major histocompatibility complex class I C | HLA-C | |||
| 7 | L1 cell adhesion molecule | L1CAM | |||
| 8 | Myelin protein zero like 1 | MPZL1 | |||
| 9 | Nectin cell adhesion molecule 1 | NECTIN1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Den Broucke, S.; Vanoirbeek, J.A.J.; Derua, R.; Hoet, P.H.M.; Ghosh, M. Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways. Nanomaterials 2021, 11, 1283. https://doi.org/10.3390/nano11051283
Van Den Broucke S, Vanoirbeek JAJ, Derua R, Hoet PHM, Ghosh M. Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways. Nanomaterials. 2021; 11(5):1283. https://doi.org/10.3390/nano11051283
Chicago/Turabian StyleVan Den Broucke, Sofie, Jeroen A. J. Vanoirbeek, Rita Derua, Peter H. M. Hoet, and Manosij Ghosh. 2021. "Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways" Nanomaterials 11, no. 5: 1283. https://doi.org/10.3390/nano11051283
APA StyleVan Den Broucke, S., Vanoirbeek, J. A. J., Derua, R., Hoet, P. H. M., & Ghosh, M. (2021). Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways. Nanomaterials, 11(5), 1283. https://doi.org/10.3390/nano11051283







