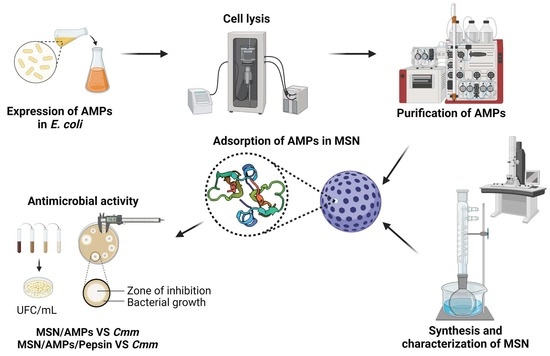
Adsorption of Recombinant Human β-Defensin 2 and Two Mutants on Mesoporous Silica Nanoparticles and Its Effect against Clavibacter michiganensis subsp. michiganensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
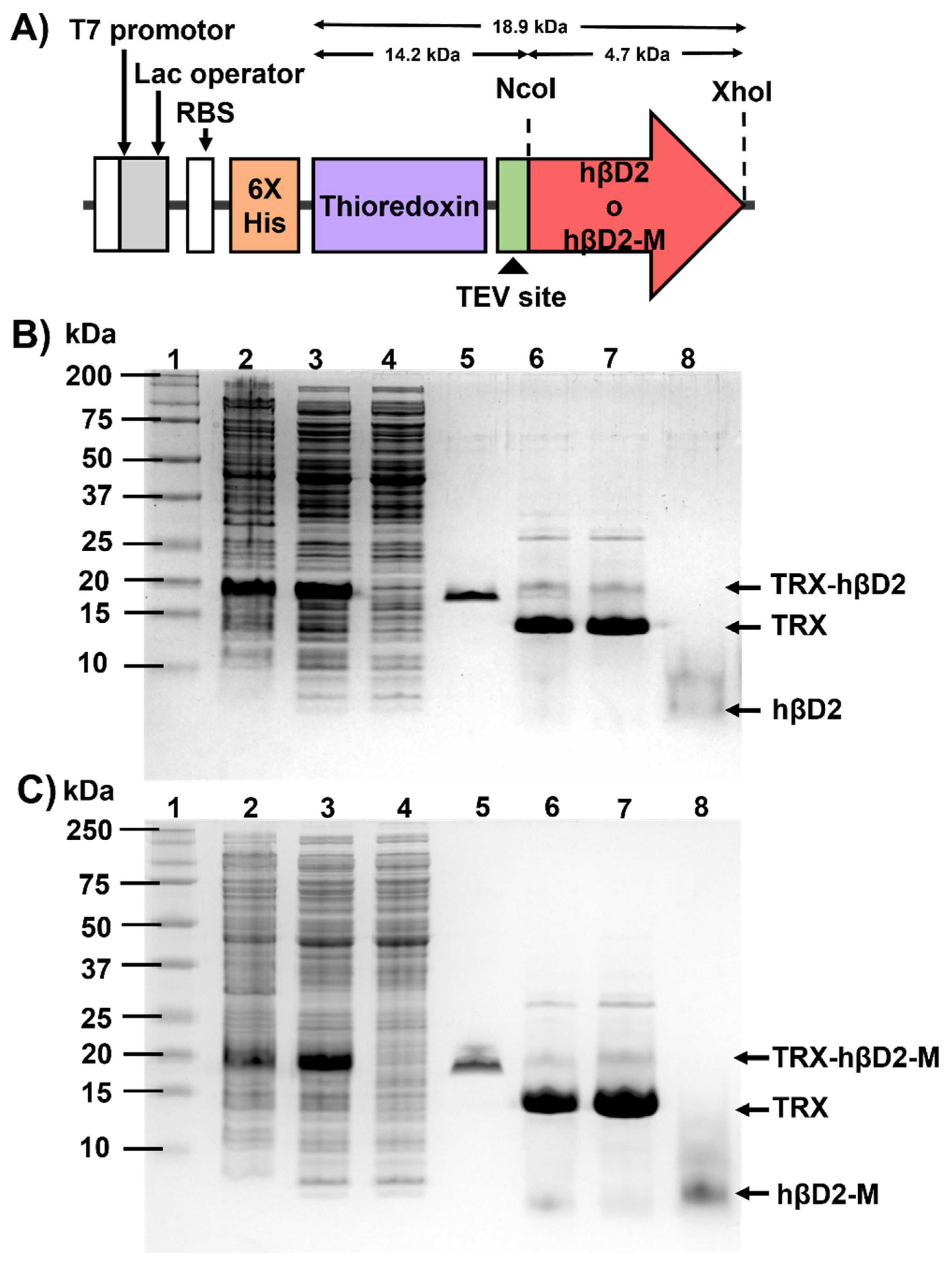
2.2. Plasmids
2.3. Expression of TRX-hβD2 and TrxA-hβD2-M
2.4. Purification of TRX-hβD2 and TrxA-hβD2-M
2.5. Synthesis of Mesoporous Silica Nanoparticles (MSN)
2.6. MSN Characterization
2.7. Antimicrobial Activity Assay and Determination of IC50
2.8. Adsorption/Release of AMPs by MSN5.4
2.9. Antimicrobial Activity of MSN/AMPs Complexes
2.10. MSN against Enzymatic Degradation of AMPs
2.11. Statistical Analysis
3. Results
3.1. Construction, Expression and Purification of Recombinant Proteins
3.2. Antimicrobial Activity Assays against Cmm
3.3. MSN Characterization
3.4. Adsorption and Release of AMPs
3.5. Bioactivity of MSN Loaded with AMPs against Cmm
3.6. Effect of Peptidase on AMPs and the MSN/AMPs Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization. FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 3 March 2021).
- Dey, R.; Raghuwanshi, R. Comprehensive assessment of growth parameters for screening endophytic bacterial strains in Solanum lycopersicum (tomato). Heliyon 2020, 6, e05325. [Google Scholar] [CrossRef]
- Parrotta, L.; Aloisi, I.; Faleri, C.; Romi, M.; Del Duca, S.; Cai, G. Chronic heat stress affects the photosynthetic apparatus of Solanum lycopersicum L. cv Micro-Tom. Plant Physiol. Biochem. 2020, 154, 463–475. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Barash, I.; Reuven, M.; Dror, O.; Sharabani, G.; Gartemann, K.H.; Eichenlaub, R.; Sessa, G.; Manulis-Sasson, S. Differential contribution of Clavibacter michiganensis ssp. michiganensis virulence factors to systemic and local infection in tomato. Mol. Plant Pathol. 2017, 18, 336–346. [Google Scholar] [CrossRef]
- Ramachandran, S.; Dobhal, S.; Alvarez, A.M.; Arif, M. Improved multiplex TaqMan qPCR assay with universal internal control offers reliable and accurate detection of Clavibacter michiganensis. J. Appl. Microbiol. 2021, 1–12. [Google Scholar] [CrossRef]
- Sai-Anand, G.; Sivanesan, A.; Benzigar, M.R.; Singh, G.; Gopalan, A.I.; Baskar, A.V.; Ilbeygi, H.; Ramadass, K.; Kambala, V.; Vinu, A. Recent progress on the sensing of pathogenic bacteria using advanced nanostructures. Bull. Chem. Soc. Jpn. 2019, 92, 216–244. [Google Scholar] [CrossRef] [Green Version]
- Nandi, M.; Macdonald, J.; Liu, P.; Weselowski, B.; Yuan, Z.C. Clavibacter michiganensis ssp. michiganensis: Bacterial canker of tomato, molecular interactions and disease management. Mol. Plant Pathol. 2018, 19, 2036–2050. [Google Scholar] [CrossRef] [Green Version]
- Gartemann, K.H.; Kirchner, O.; Engemann, J.; Gräfen, I.; Eichenlaub, R.; Burger, A. Clavibacter michiganensis subsp. michiganensis: First steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J. Biotechnol. 2003, 106, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.M.; Vega, D.; Pizzorno, R.; Cordon, G.; Correa, O.S. Hydraulic and leaf reflectance alterations induced by Clavibacter michiganensis subsp. michiganensis on tomato plants. Eur. J. Plant Pathol. 2018, 152, 567–572. [Google Scholar] [CrossRef]
- Vega, D.; Romero, A.M. Survival of Clavibacter michiganensis subsp. michiganensis in tomato debris under greenhouse conditions. Plant Pathol. 2016, 65, 545–550. [Google Scholar] [CrossRef]
- Ombiro, G.S.; Sawai, T.; Noutoshi, Y.; Nishina, Y.; Matsui, H.; Yamamoto, M.; Toyoda, K.; Ichinose, Y. Specific growth inhibitors of Ralstonia solanacearum, Xanthomonas oryzae pv. oryzae, X. campestris pv. campestris, and Clavibacter michiganensis subsp. michiganensis. Microbiol. Res. 2018, 215, 29–35. [Google Scholar] [CrossRef]
- Lahiri, S.; Orr, D. Biological Control in Tomato Production Systems: Theory and Practice; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128135082. [Google Scholar]
- Wittmann, J.; Brancato, C.; Berendzen, K.W.; Dreiseikelmann, B. Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant Pathol. 2016, 65, 496–502. [Google Scholar] [CrossRef]
- Choi, J.; Baek, K.H.; Moon, E. Antimicrobial effects of a hexapetide KCM21 against Pseudomonas syringae pv. tomato DC3000 and Clavibacter michiganensis subsp. michiganensis. Plant Pathol. J. 2014, 30, 245–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-based nanoparticles as antimicrobial agents: An overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Singh, P.; Kashyap, P.; Singh, M.; Azmal, S. Microbial pathogenesis promising role of defensins peptides as therapeutics to combat against viral infection. Microb. Pathog. 2021, 155, 104930. [Google Scholar] [CrossRef]
- Li, K.; Li, W.; Chen, X.; Luo, T.; Mu, Y.; Chen, X. Molecular and functional identification of a β-defensin homolog in large yellow croaker (Larimichthys crocea). J. Fish Dis. 2021, 44, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Pazgier, M.; Hoover, D.M.; Yang, D.; Lu, W.; Lubkowski, J. Human β-defensins. Cell. Mol. Life Sci. CMLS 2006, 63, 1294–1313. [Google Scholar] [CrossRef]
- Semple, F.; Dorin, J.R. β-Defensins: Multifunctional modulators of infection, inflammation and more? J. Innate Immun. 2012, 4, 337–348. [Google Scholar] [CrossRef]
- Izadpanah, A.; Gallo, R.L. Antimicrobial peptides. J. Am. Acad. Dermatol. 2005, 52, 381–390. [Google Scholar] [CrossRef]
- Järvå, M.; Phan, T.K.; Lay, F.T.; Caria, S.; Kvansakul, M.; Hulett, M.D. Human-defensin 2 kills Candida albicans through phosphatidylinositol 4,5-bisphosphate–mediated membrane permeabilization. Sci. Adv. 2018, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Pept. Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Zhuang, C.; Huo, H.; Yang, N.; Fu, Q.; Xue, T.; Zhu, Q.; Wang, B.; Liu, X.; Li, C. Characterization of antibacterial activities and the related mechanisms of a β-defensin in turbot (Scophthalmus maximus). Aquaculture 2021, 541, 736839. [Google Scholar] [CrossRef]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. Mini review: A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walia, N.; Dasgupta, N.; Ranjan, S.; Ramalingam, C.; Gandhi, M. Methods for nanoemulsion and nanoencapsulation of food bioactives. Environ. Chem. Lett. 2019, 17, 1471–1483. [Google Scholar] [CrossRef]
- Luo, Z.; Deng, Y.; Zhang, R.; Wang, M.; Bai, Y.; Zhao, Q.; Lyu, Y.; Wei, J.; Wei, S. Peptide-laden mesoporous silica nanoparticles with promoted bioactivity and osteo-differentiation ability for bone tissue engineering. Colloids Surf. B Biointerfaces 2015, 131, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.; Mallen, S.; Durack, E.; O’Connor, P.M.; Hudson, S.P. Mesoporous matrices for the delivery of the broad spectrum bacteriocin, nisin A. J. Colloid Interface Sci. 2019, 537, 396–406. [Google Scholar] [CrossRef]
- Selvarajan, V.; Obuobi, S.; Lai, P.; Ee, R. Silica nanoparticles—A versatile tool for the treatment of bacterial infections. Front. Chem. 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Tenland, E.; Pochert, A.; Krishnan, N.; Rao, K.U.; Kalsum, S.; Braun, K.; Glegola-madejska, I.; Id, M.L. Effective delivery of the anti-mycobacterial peptide NZX in mesoporous silica nanoparticles. PLoS ONE 2019, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Diosa, J.; Guzman, F.; Bernal, C.; Mesa, M. Formation mechanisms of chitosan-silica hybrid materials and its performance as solid support for KR-12 peptide adsorption: Impact on KR-12 antimicrobial activity and proteolytic stability. Integr. Med. Res. 2019, 9, 890–901. [Google Scholar] [CrossRef]
- Durack, E.; Mallen, S.; O’Connor, P.M.; Rea, M.C.; Ross, R.P.; Hill, C.; Hudson, S. Protecting bactofencin A to enable its antimicrobial activity using mesoporous matrices. Int. J. Pharm. 2019, 558, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Routsias, J.G.; Karagounis, P.; Parvulesku, G.; Legakis, N.J.; Tsakris, A. In Vitro bactericidal activity of human β-defensin 2 against nosocomial strains. Peptides 2010, 31, 1654–1660. [Google Scholar] [CrossRef]
- Gu, J.; Huang, K.; Zhu, X.; Li, Y.; Wei, J.; Zhao, W.; Liu, C.; Shi, J. Sub-150nm mesoporous silica nanoparticles with tunable pore sizes and well-ordered mesostructure for protein encapsulation. J. Colloid Interface Sci. 2013, 407, 236–242. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, P.; Holtsmark, I.; Skaugen, M.; Eijsink, V.G.H.; Brurberg, M.B. New type of antimicrobial protein produced by the plant pathogen Clavibacter michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 2013, 79, 5721–5727. [Google Scholar] [CrossRef] [Green Version]
- Tay, D.K.S.; Rajagopalan, G.; Li, X.; Chen, Y.; Lua, L.H.L.; Leong, S.S.J. A new bioproduction route for a novel antimicrobial peptide. Biotechnol. Bioeng. 2011, 108, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.F.A.D.M.; Silva, L.V.D.A.T.D.; Nascimento, T.G.D.; Almeida, L.; Calumby, R.J.N.; Nunes, Á.M.; Oliveira, L.M.T.D.M.; Fonseca, E.J.D.S. Antioxidant and antimicrobial activity of red propolis embedded mesoporous silica nanoparticles. Drug Dev. Ind. Pharm. 2020, 46, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Sahare, P.; Ayala, M.; Vazquez-Duhalt, R.; Pal, U.; Loni, A.; Canham, L.T.; Osorio, I.; Agarwal, V. Enhancement of peroxidase stability against oxidative self-inactivation by co-immobilization with a redox-active protein in mesoporous silicon and silica microparticles. Nanoscale Res. Lett. 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Alessi, A.; Agnello, S.; Buscarino, G.; Gelardi, F.M. Structural properties of core and surface of silica nanoparticles investigated by Raman spectroscopy. J. Raman Spectrosc. 2013, 44, 810–816. [Google Scholar] [CrossRef]
- Hoover, D.M.; Rajashankar, K.R.; Blumenthal, R.; Puri, A.; Oppenheim, J.J.; Chertov, O.; Lubkowski, J. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 2000, 275, 32911–32918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, M.; Piccoli, D.; Weselowski, B.; McDowell, T.; Renaud, J.; MacDonald, J.; Yuan, Z.C. Surfactin-producing bacillus velezensis 1B-23 and bacillus sp. 1D-12 protect tomato against bacterial canker caused by Clavibacter michiganensis subsp. michiganensis. J. Plant Pathol. 2020, 102, 451–458. [Google Scholar] [CrossRef]
- Khazigaleeva, R.A.; Vinogradova, S.V.; Petrova, V.L.; Fesenko, I.A.; Arapidi, G.P.; Kamionskaya, A.M.; Govorun, V.M.; Ivanov, V.T. Antimicrobial activity of endogenous peptides of the moss Physcomitrella patens. Russ. J. Bioorganic Chem. 2017, 43, 248–254. [Google Scholar] [CrossRef]
- Yokoyama, S.; Kato, K.; Koba, A.; Minami, Y.; Watanabe, K.; Yagi, F. Purification, characterization, and sequencing of antimicrobial peptides, Cy-AMP1, Cy-AMP2, and Cy-AMP3, from the cycad (Cycas revoluta) seeds. Peptides 2008, 29, 2110–2117. [Google Scholar] [CrossRef]
- Cutrona, K.J.; Kaufman, B.A.; Figueroa, D.M.; Elmore, D.E. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett. 2015, 589, 3915–3920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreev, K.; Bianchi, C.; Laursen, J.S.; Citterio, L.; Hein-Kristensen, L.; Gram, L.; Kuzmenko, I.; Olsen, C.A.; Gidalevitz, D. Guanidino groups greatly enhance the action of antimicrobial peptidomimetics against bacterial cytoplasmic membranes. Biochim. Biophys. Acta-Biomembr. 2014, 1838, 2492–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.H.; Pan, Y.C.; Liu, F.W.; Chen, C.Y. Prokaryotic expression and action mechanism of antimicrobial LsGRP1C recombinant protein containing a fusion partner of small ubiquitin-like modifier. Appl. Microbiol. Biotechnol. 2017, 101, 8129–8138. [Google Scholar] [CrossRef] [PubMed]
- Oh, R.; Lee, M.J.; Kim, Y.O.; Nam, B.H.; Kong, H.J.; Kim, J.W.; Park, J.Y.; Seo, J.K.; Kim, D.G. Myticusin-beta, antimicrobial peptide from the marine bivalve, Mytilus coruscus. Fish Shellfish. Immunol. 2020, 99, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Kachbouri, S.; Mnasri, N.; Elaloui, E.; Moussaoui, Y. Tuning particle morphology of mesoporous silica nanoparticles for adsorption of dyes from aqueous solution. J. Saudi Chem. Soc. 2018, 22, 405–415. [Google Scholar] [CrossRef]
- Luo, L.; Liang, Y.; Erichsen, E.S.; Anwander, R. Monodisperse mesoporous silica nanoparticles of distinct topology. J. Colloid Interface Sci. 2017, 495, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Balas, F.; Manzano, M.; Horcajada, P.; Vallet-Regi, M. Confinement and controlled release of bisphosphonates on ordered mesoporous silica-based materials. J. Am. Chem. Soc. 2006, 128, 8116–8117. [Google Scholar] [CrossRef]
- Ding, Y.; Tang, S.; Yu, B.; Yan, Y.; Li, H.; Wei, J.; Su, J. In Vitro degradability, bioactivity and primary cell responses to bone cements containing mesoporous magnesium-calcium silicate and calcium sulfate for bone regeneration. J. R. Soc. Interface 2015, 12. [Google Scholar] [CrossRef]



| Protein | IC50 (µg/mL) |
|---|---|
| hβD2 in Tris | 3.61 ± 0.47 |
| hβD2 in CH3COOH | 3.64 ± 0.33 |
| hβD2-M in Tris | 1.75 ± 0.13 |
| hβD2-M in CH3COOH | 1.56 ± 0.16 |
| TRX-hβD2-M in Tris | 19.99 ± 3.55 |
| TRX-hβD2-M in CH3COOH | 6.17 ± 0.08 |
| Mesoporous Silicates | Pore Diameter a (nm) | Poro Volume (cm3/g) | BET Surface Area (m2/g) |
|---|---|---|---|
| MSN5.4 | 5.4 | 1.77 | 1021.4 |
| MSN4.8 | 4.8 | 1.02 | 711.4 |
| MSN0.9 | 0.9 | 0.31 | 653.2 |
| MSN0.9’ | 0.9 | 0.30 | 688.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcelino-Pérez, G.; Ruiz-Medrano, R.; Gallardo-Hernández, S.; Xoconostle-Cázares, B. Adsorption of Recombinant Human β-Defensin 2 and Two Mutants on Mesoporous Silica Nanoparticles and Its Effect against Clavibacter michiganensis subsp. michiganensis. Nanomaterials 2021, 11, 2144. https://doi.org/10.3390/nano11082144
Marcelino-Pérez G, Ruiz-Medrano R, Gallardo-Hernández S, Xoconostle-Cázares B. Adsorption of Recombinant Human β-Defensin 2 and Two Mutants on Mesoporous Silica Nanoparticles and Its Effect against Clavibacter michiganensis subsp. michiganensis. Nanomaterials. 2021; 11(8):2144. https://doi.org/10.3390/nano11082144
Chicago/Turabian StyleMarcelino-Pérez, Gabriel, Roberto Ruiz-Medrano, Salvador Gallardo-Hernández, and Beatriz Xoconostle-Cázares. 2021. "Adsorption of Recombinant Human β-Defensin 2 and Two Mutants on Mesoporous Silica Nanoparticles and Its Effect against Clavibacter michiganensis subsp. michiganensis" Nanomaterials 11, no. 8: 2144. https://doi.org/10.3390/nano11082144