One-Chip Isolation of Drug-Resistant Acute Myeloid Leukemia Cells with CXCR4-Targeted Magnetic Fluorescent Nanoprobes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
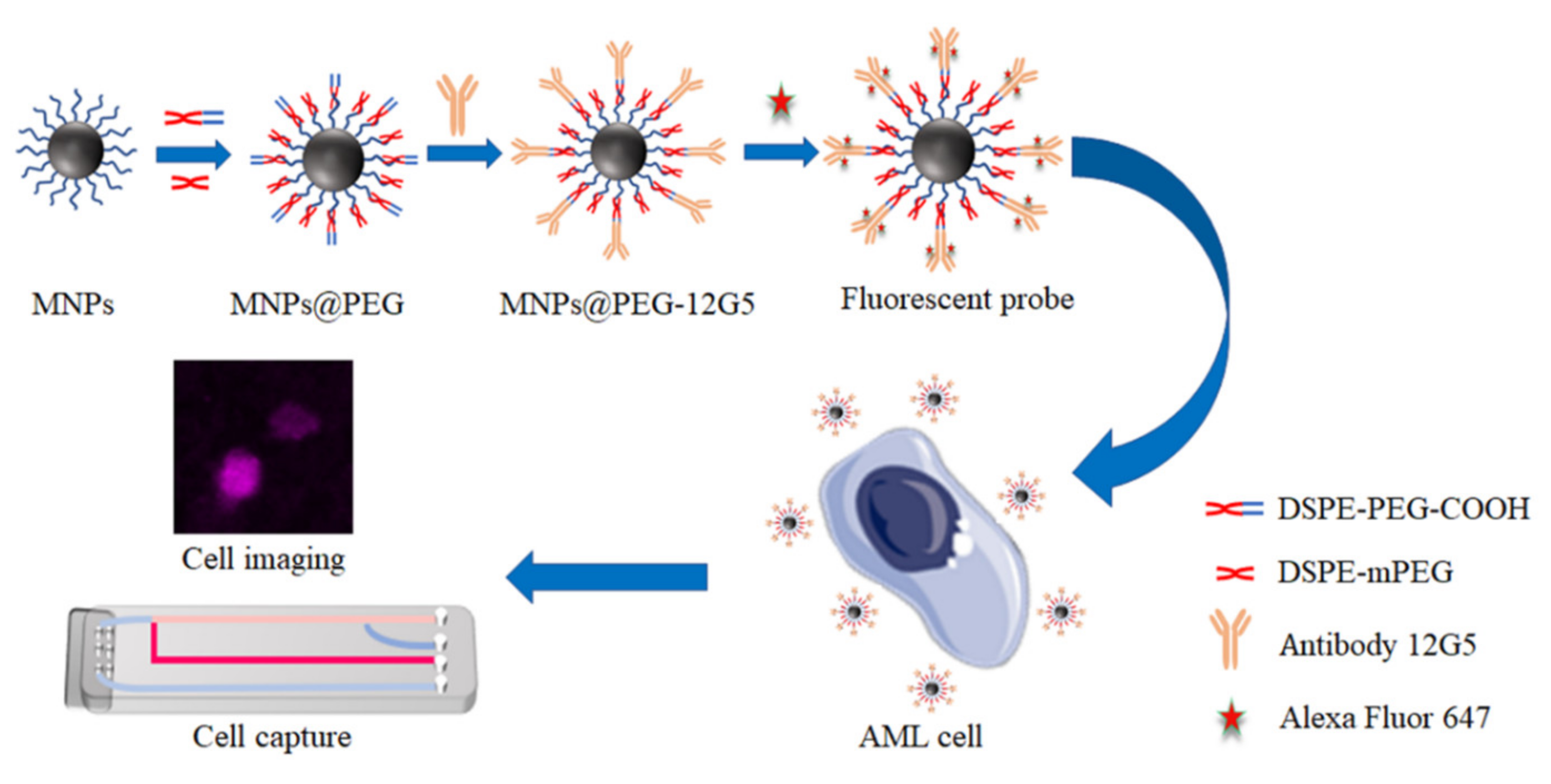
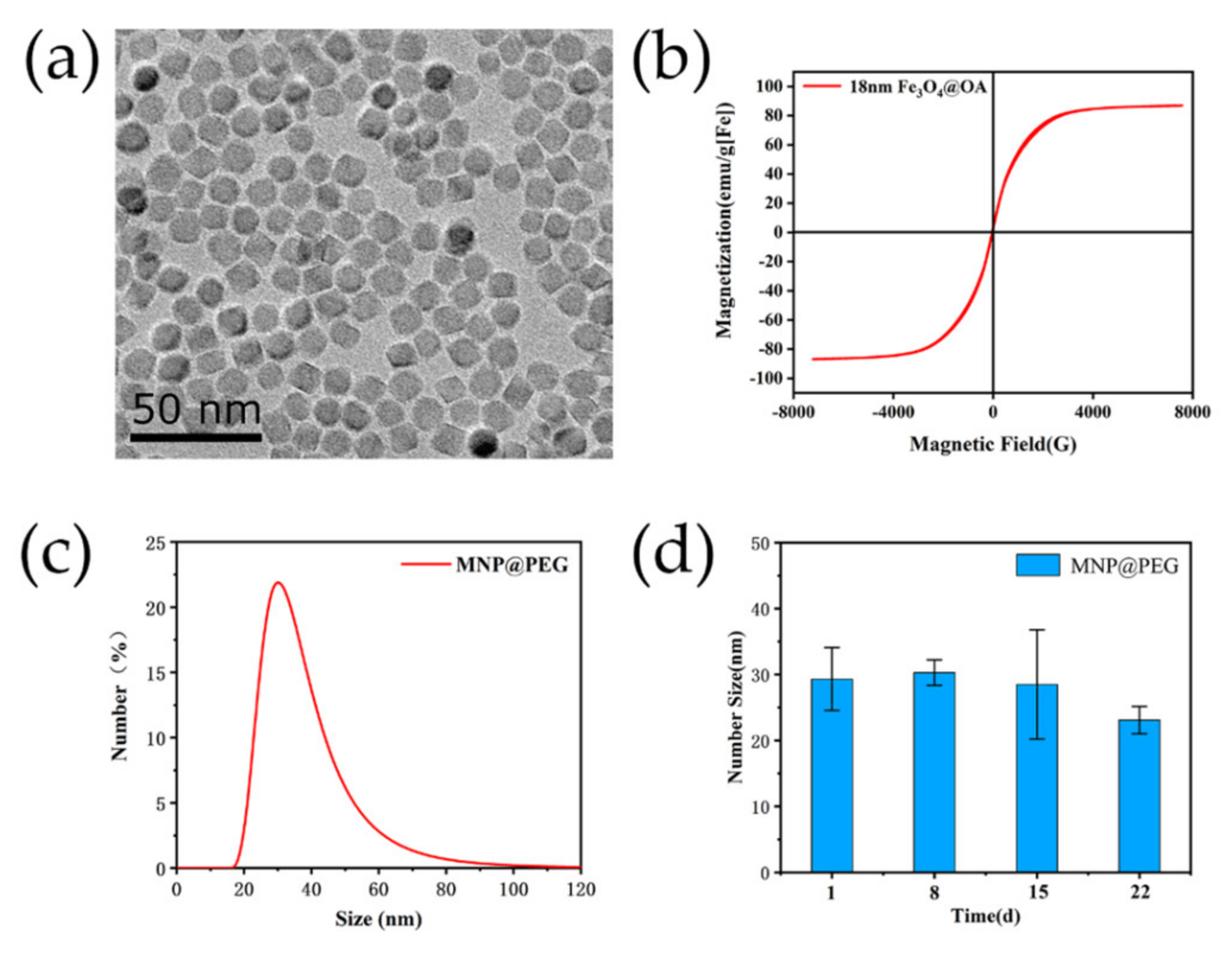
2.2. The Synthesis and Characteration of MNP@PEG
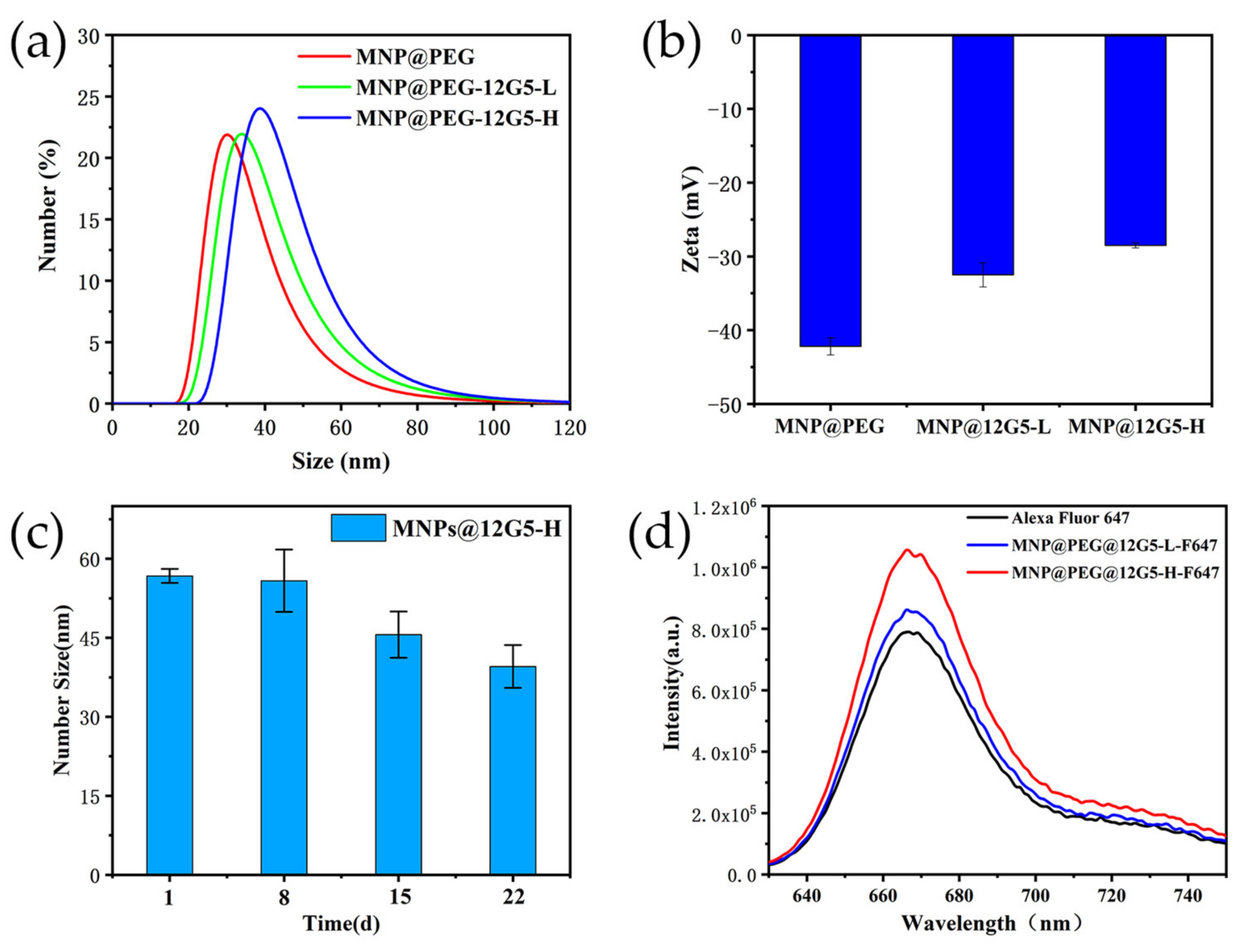
2.3. The Synthesis of MNP@PEG-12G5-F647
2.4. Cell Culture
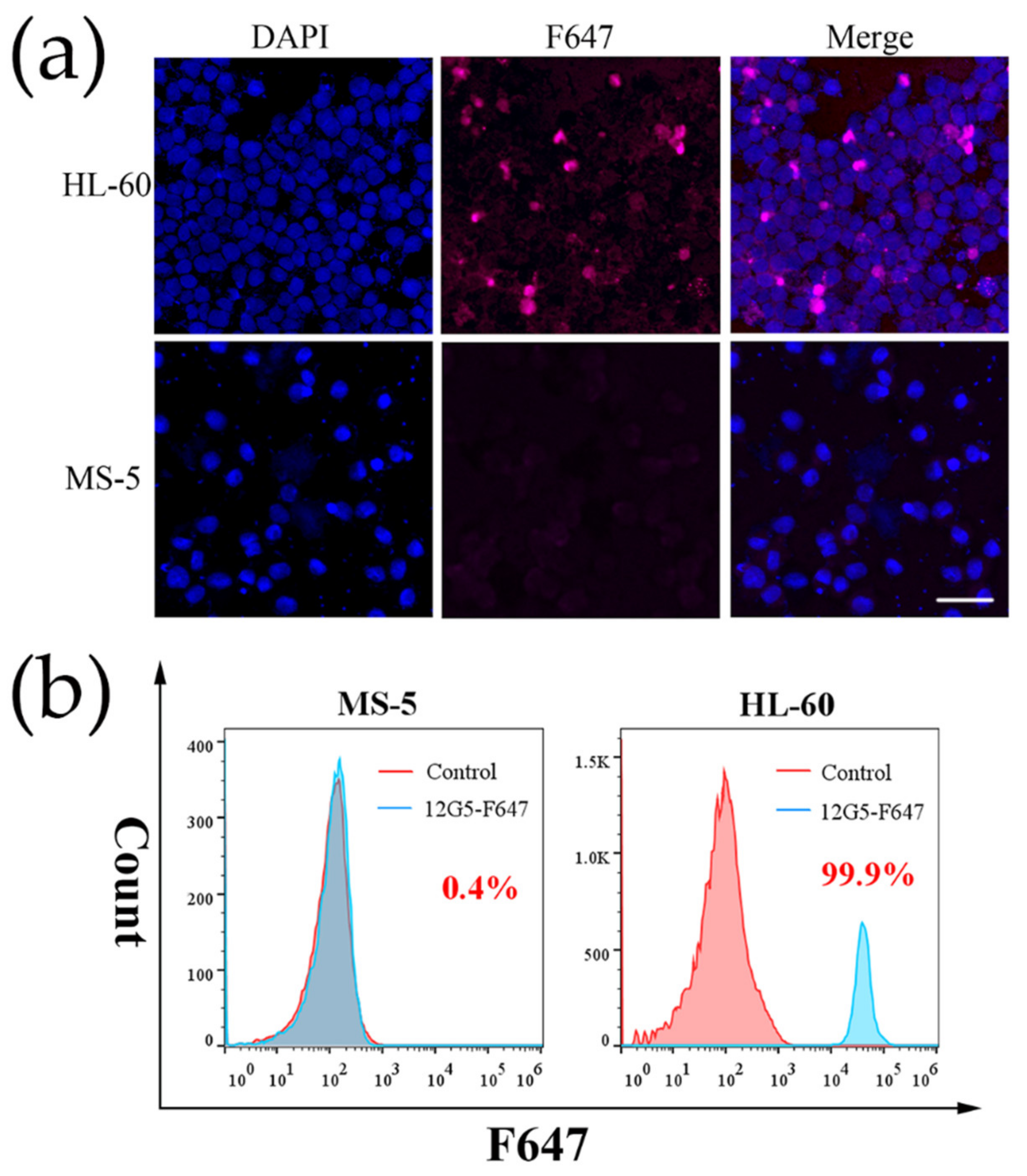
2.5. Cell Labeling with 12G5-F647 and MNP@PEG-12G5-F647
2.6. Microfluidic Array Fabrication
2.7. Cell Capture
3. Results and Discussion
3.1. Characterization of Magnetic Fluorescent Nanoprobes
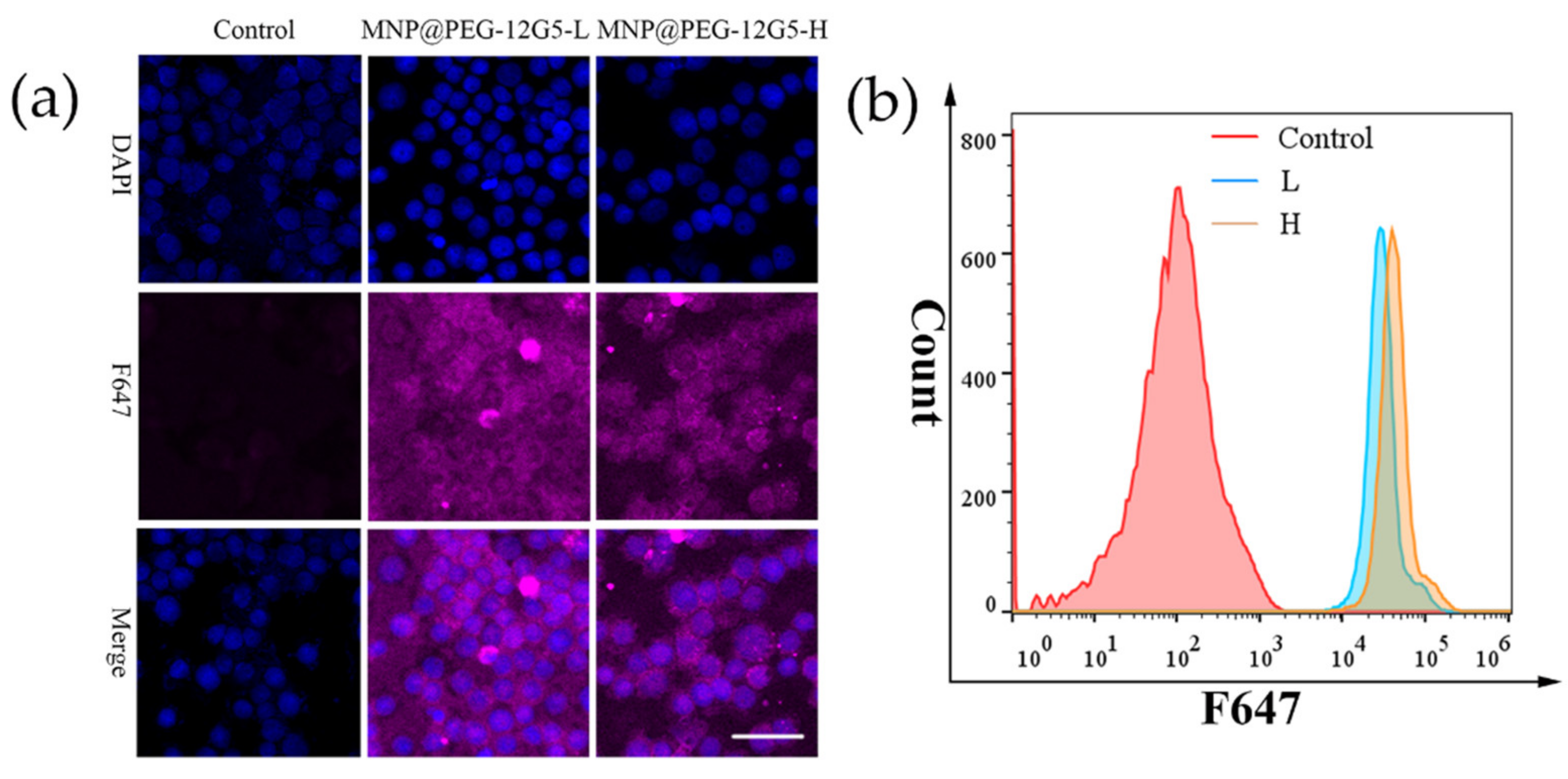
3.2. Cell Labeling with 12G5-F647 and MNP@PEG -12G5-F647
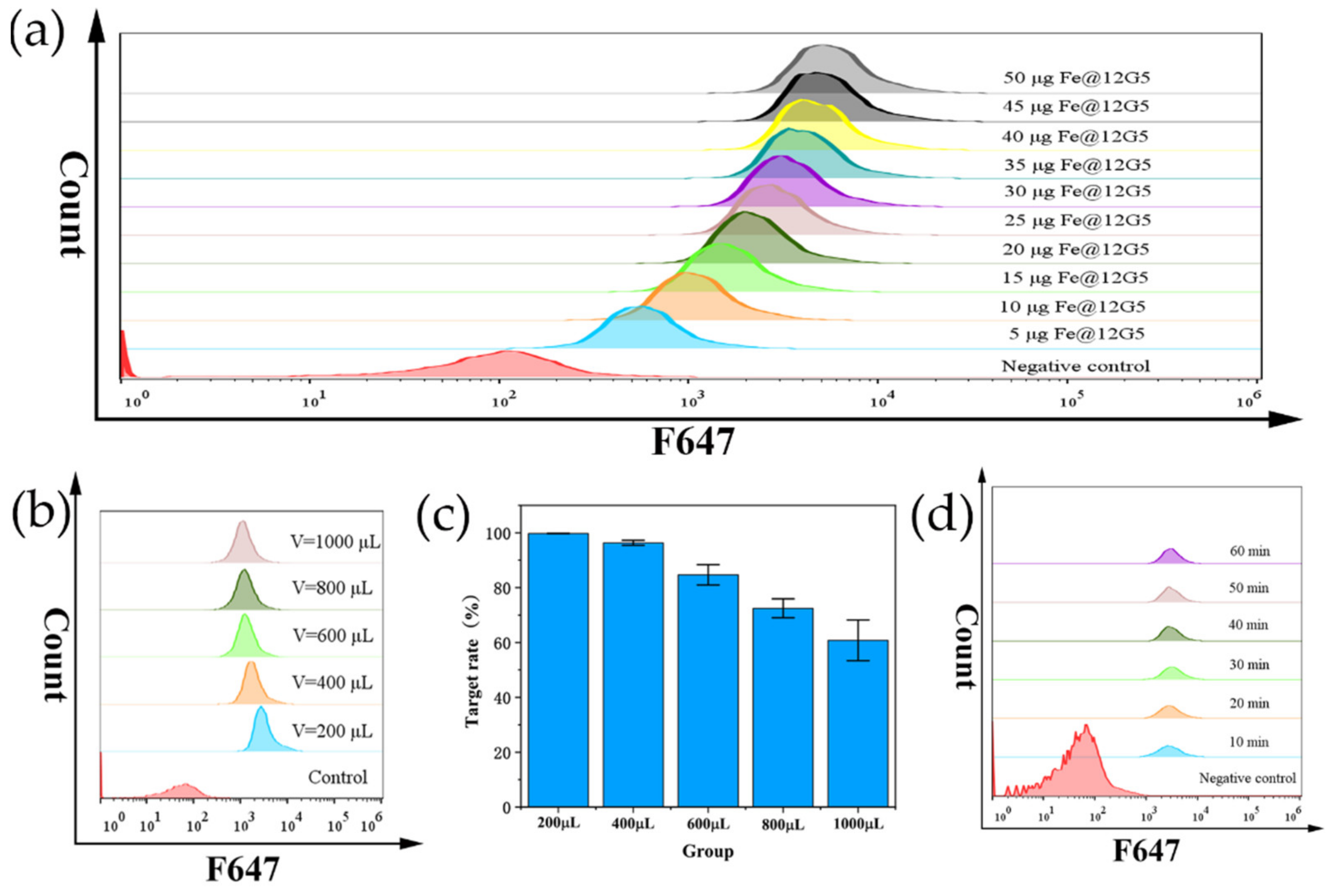
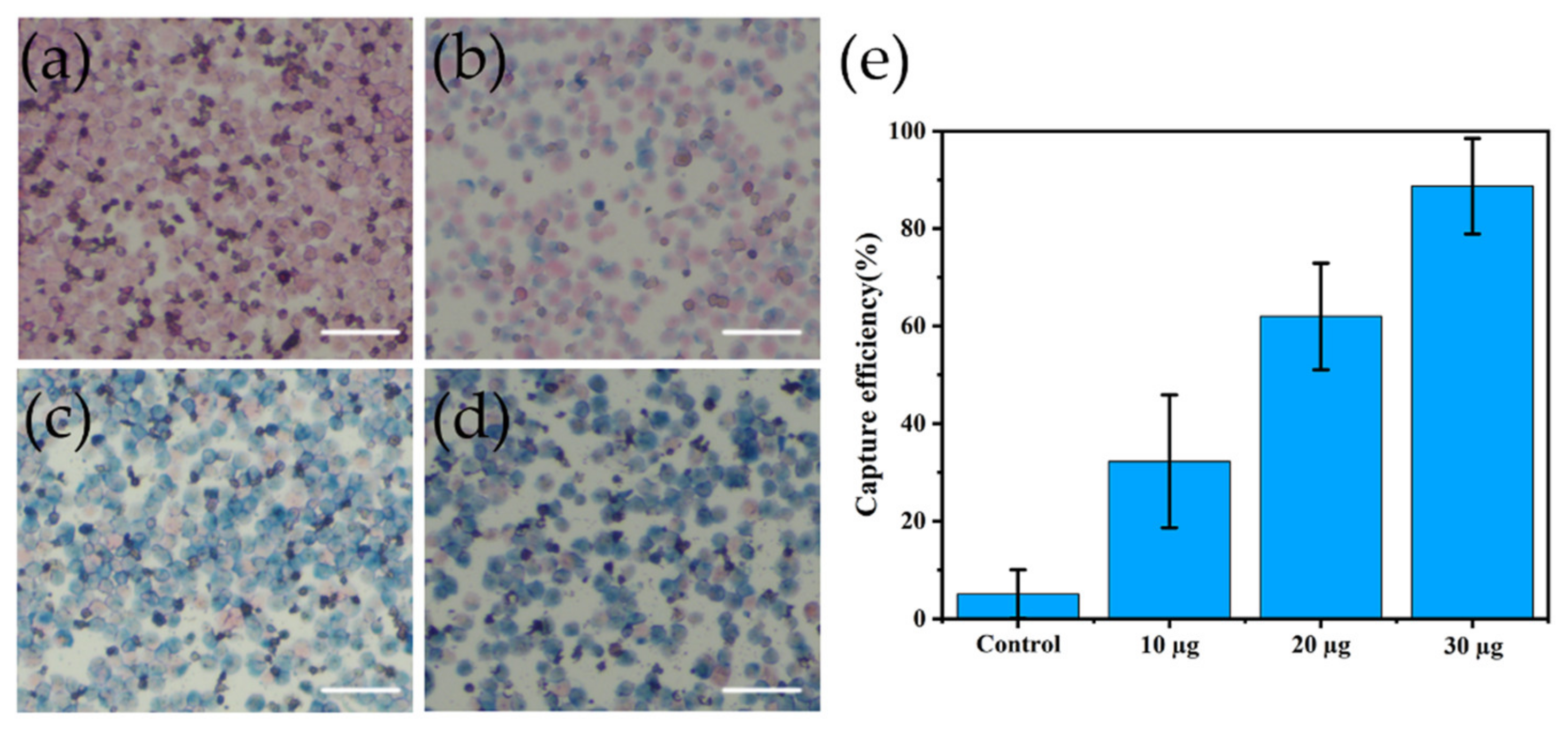
3.3. Cell Capture
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Robak, T.; Wierzbowska, A. Current and emerging therapies for acute myeloid leukemia. Clin. Ther. 2009, 31, 2349–2370. [Google Scholar] [CrossRef] [PubMed]
- Noguera, N.; Catalano, G.; Banella, C.; Divona, M.; Faraoni, I.; Ottone, T.; Arcese, W.; Voso, M. Acute Promyelocytic Leukemia: Update on the Mechanisms of Leukemogenesis, Resistance and on Innovative Treatment Strategies. Cancers 2019, 11, 1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naina, H.V.; Patnaik, M.M.; Harris, S. Anthracycline dose intensification in acute myeloid leukemia. N. Engl. J. Med. 2009, 361, 2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehninger, A.; Kramer, M.; Röllig, C.; Thiede, C.; Bornhäuser, M.; Von Bonin, M.; Feldmann, A.; Bachmann, M.F.; Ehninger, G.; Oelschlägel, U.; et al. Distribution and levels of cell surface expression of CD33 and CD123 in acute myeloid leukemia. Blood Cancer J. 2014, 4, e218. [Google Scholar] [CrossRef] [Green Version]
- Passaro, D.; Di Tullio, A.; Abarrategi, A.; Rouault-Pierre, K.; Foster, K.; Ariza-McNaughton, L.; Montaner, B.; Chakravarty, P.; Bhaw, L.; Diana, G.; et al. Increased Vascular Permeability in the Bone Marrow Microenvironment Contributes to Disease Progression and Drug Response in Acute Myeloid Leukemia. Cancer Cell 2017, 32, 324–341. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Li, F.; Huang, J.; Jin, J.; Wang, H. Leukemia stem cell-bone marrow microenvironment interplay in acute myeloid leukemia development. Exp. Hematol. Oncol. 2021, 10, 39. [Google Scholar] [CrossRef]
- Yazdani, Z.; Mousavi, Z.; Moradabadi, A.; Hassanshahi, G. Significance of CXCL12/CXCR4 Ligand/Receptor Axis in Various Aspects of Acute Myeloid Leukemia. Cancer Manag. Res. 2020, 12, 2155–2165. [Google Scholar] [CrossRef] [Green Version]
- Cho, B.S.; Kim, H.J.; Konopleva, M. Targeting the CXCL12/CXCR4 axis in acute myeloid leukemia: From bench to bedside. Korean J. Intern. Med. 2017, 32, 248–257. [Google Scholar] [CrossRef]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin. Cancer Res. 2008, 14, 2519–2526. [Google Scholar] [CrossRef] [Green Version]
- Rombouts, E.J.C.; Pavic, B.; Löwenberg, B.; Ploemacher, R.E. Relation between CXCR-4 expression, Flt3 mutations, and unfavorable prognosis of adult acute myeloid leukemia. Blood 2004, 104, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Tavernier-Tardy, E.; Cornillon, J.; Campos, L.; Flandrin, P.; Duval, A.; Nadal, N.; Guyotat, D. Prognostic value of CXCR4 and FAK expression in acute myelogenous leukemia. Leuk. Res. 2009, 33, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Mannelli, F.; Cutini, I.; Gianfaldoni, G.; Bencini, S.; Scappini, B.; Pancani, F.; Ponziani, V.; Bonetti, M.I.; Biagiotti, C.; Longo, G.; et al. CXCR4 expression accounts for clinical phenotype and outcome in acute myeloid leukemia. Cytom. B Clin. Cytom. 2014, 86, 340–349. [Google Scholar] [CrossRef]
- Matsuo, H.; Ms, N.N.; Tomizawa, D.; Saito, A.M.; Kiyokawa, N.; Horibe, K.; Ms, Y.N.; Tokumasu, M.; Itoh, H.; Kamikubo, Y.; et al. CXCR4 Overexpression is a Poor Prognostic Factor in Pediatric Acute Myeloid Leukemia with Low Risk: A Report from the Japanese Pediatric Leukemia/Lymphoma Study Group. Pediatr. Blood Cancer 2016, 63, 1394–1399. [Google Scholar] [CrossRef]
- Sison, E.A.R.; McIntyre, E.; Magoon, D.; Brown, P. Dynamic chemotherapy-induced upregulation of CXCR4 expression: A mechanism of therapeutic resistance in pediatric AML. Mol. Cancer Res. 2013, 11, 1004–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Meng, F.; Lu, Q. Expression Profile Screening and Bioinformatics Analysis of circRNA, LncRNA, and mRNA in Acute Myeloid Leukemia Drug-Resistant Cells. Turk. J. Hematol. 2020, 37, 104–110. [Google Scholar] [CrossRef]
- Lai, Q.Y.; He, Y.Z.; Peng, X.W.; Zhou, X.; Liang, D.; Wang, L. Histone deacetylase 1 induced by neddylation inhibition contributes to drug resistance in acute myelogenous leukemia. Cell Commun. Signal. 2019, 17, 86. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Ju, Y.; Xu, J.; Yang, Z.; Gao, S.; Hou, Y. Magnetic Nanomaterials: Chemical Design, Synthesis, and Potential Applications. Acc. Chem. Res. 2018, 51, 404–413. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Chen, F.; Zhong, Z.; Qi, L. Detection of K-ras gene mutations in feces by magnetic nanoprobe in patients with pancreatic cancer: A preliminary study. Exp. Ther. Med. 2018, 15, 527–531. [Google Scholar] [CrossRef]
- Karimi, N.; Oloomi, M.; Orafa, Z. Circulating Tumor Cells Detection in Patients with Early Breast Cancer Using MACS Immunomagnetic Flow Cytometry. Avicenna J. Med. Biotechnol. 2020, 12, 148–156. [Google Scholar]
- Ulbrich, K.; Holá, K.; Šubr, V.; Bakandritsos, A.; Tuček, J.; Zbořil, R. Targeted Drug Delivery with Polymers and Magnetic Nanoparticles: Covalent and Noncovalent Approaches, Release Control, and Clinical Studies. Chem. Rev. 2016, 116, 5338–5431. [Google Scholar] [CrossRef] [PubMed]
- Manohar, S.M.; Shah, P.; Nair, A. Flow cytometry: Principles, applications and recent advances. Bioanalysis 2021, 13, 181–198. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.O.; Huh, Y.J.; Jang, J. Selective Depletion of SSEA-3-and TRA-1-60-Positive Undifferentiated Human Embryonic Stem Cells by Magnetic Activated Cell Sorter (MACS). Tissue Eng. Regen. Med. 2011, 8, 253–261. [Google Scholar]
- Shields, C.W., 4th; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myklatun, A.; Cappetta, M.; Winklhofer, M.; Ntziachristos, V.; Westmeyer, G.G. Microfluidic sorting of intrinsically magnetic cells under visual control. Sci. Rep. 2017, 7, 6942. [Google Scholar] [CrossRef] [Green Version]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.-I.; Morgan, B.; Trautwein, J.; et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef] [Green Version]
- Herhaus, P.; Lipkova, J.; Lammer, F.; Slotta-Huspenina, J.; Wiestler, B.; Vag, T.; Habringer, S.; Yakushev, I.; Lapa, C.; Pukrop, T.; et al. CXCR4-targeted positron emission tomography imaging of central nervous system B-cell lymphoma. Soc. Nucl. Med. 2020, 61, 1765–1771. [Google Scholar] [CrossRef]
- Poschenrieder, A.; Osl, T.; Schottelius, M.; Hoffmann, F.; Wirtz, M.; Schwaiger, M.; Wester, H.-J. First 18F-Labeled Pentixafor-Based Imaging Agent for PET Imaging of CXCR4 Expression In Vivo. Tomography 2016, 2, 85–93. [Google Scholar] [CrossRef]
- Béné, M.C. Microfluidics in flow cytometry and related techniques. Int. J. Lab. Hematol. 2017, 39 (Suppl. 1), 93. [Google Scholar] [CrossRef] [Green Version]
- Plouffe, B.D.; Murthy, S.K.; Lewis, L.H. Fundamentals and application of magnetic particles in cell isolation and enrichment: A review. Rep. Prog. Phys. 2015, 78, 016601. [Google Scholar] [CrossRef]
- Thi, T.T.H.; Pilkington, E.H.; Nguyen, D.H.; Lee, J.S.; Park, K.D.; Truong, N.P. The Importance of Poly (ethylene glycol) Alternatives for Overcoming PEG Immunogenicity in Drug Delivery and Bioconjugation. Polymers 2020, 12, 298. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, X.; Lin, Y.; Wang, Y.; Wu, S.; Xia, K.; Xu, C.; Ma, H.; Zheng, J.; Luo, L.; et al. Design, synthesis, and evaluation of pyrrolidine based CXCR4 antagonists with invivo anti-tumor metastatic activity. Eur. J. Med. Chem. 2020, 205, 112537. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, B.; Wei, P.; Wang, X.; Lou, W. Controlled synthesis and size-dependent thermal conductivity of Fe3O4 magnetic nanofluids. Dalton Trans. 2012, 41, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Kong, P.Y.; Shi, Z.Z.; Zeng, D.F.; Chen, X.H.; Chang, C.; Peng, X.G.; Zhang, Y.; Liu, H. Effects of anti-CXCR 4 monoclonal antibody 12G5 on proliferation and apoptosis of human acute myelocytic leukemia cell line HL-60. J. Med. Coll. PLA 2007, 22, 17–22. [Google Scholar] [CrossRef]
- Cho, C.H.; Cho, M.; Park, J.K. Biomarker barcodes: Multiplexed microfluidic immunohistochemistry enables high-throughput analysis of tissue microarray. Lab Chip 2021, 21, 3471–3482. [Google Scholar] [CrossRef]
- Lee, N.; Kim, H.; Choi, S.H.; Park, M.; Kim, D.; Kim, H.C.; Choi, Y.; Lin, S.; Kim, B.H.; Jung, H.S.; et al. Magnetosome-like ferrimagnetic iron oxide nanocubes for highly sensitive MRI of single cells and transplanted pancreatic islets. Proc. Natl. Acad. Sci. USA 2011, 108, 2662–2667. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Wei, X.; Gan, J.; Huang, L.; Shen, T.; Lou, J.; Liu, B.; Zhang, J.X.J.; Qian, K. Multifunctional Magnetic Particles for Combined Circulating Tumor Cells Isolation and Cellular Metabolism Detection. Adv. Funct. Mater. 2016, 26, 4016–4025. [Google Scholar] [CrossRef]
- Içöz, K.; Eken, A.; Çınar, S.; Murat, A.; Özcan, S.; Ünal, E.; Deniz, G. Immunomagnetic separation of B type acute lymphoblastic leukemia cells from bone marrow with flow cytometry validation and microfluidic chip measurements. Sep. Sci. Technol. 2020, 56, 2659–2666. [Google Scholar] [CrossRef]


| Sample | Quantities of Coupled Antibody (µg) | Ratio of Coupled Antibody | Quantities of Coupled Fluorescent Molecular (µg) | Ratio of Coupled Fluorescent Molecular |
|---|---|---|---|---|
| MNP@PEG-12G5-L-F647 | 46 | 92% | 3.24 | 27.0% |
| MNP@PEG-12G5-H-F647 | 92 | 92% | 7.33 | 30.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Jiang, Y.; Wang, L.; Chen, Y.; Zhang, Y.; Ma, M. One-Chip Isolation of Drug-Resistant Acute Myeloid Leukemia Cells with CXCR4-Targeted Magnetic Fluorescent Nanoprobes. Nanomaterials 2022, 12, 1711. https://doi.org/10.3390/nano12101711
Wang F, Jiang Y, Wang L, Chen Y, Zhang Y, Ma M. One-Chip Isolation of Drug-Resistant Acute Myeloid Leukemia Cells with CXCR4-Targeted Magnetic Fluorescent Nanoprobes. Nanomaterials. 2022; 12(10):1711. https://doi.org/10.3390/nano12101711
Chicago/Turabian StyleWang, Fan, Yuqi Jiang, Luhai Wang, Yi Chen, Yu Zhang, and Ming Ma. 2022. "One-Chip Isolation of Drug-Resistant Acute Myeloid Leukemia Cells with CXCR4-Targeted Magnetic Fluorescent Nanoprobes" Nanomaterials 12, no. 10: 1711. https://doi.org/10.3390/nano12101711





