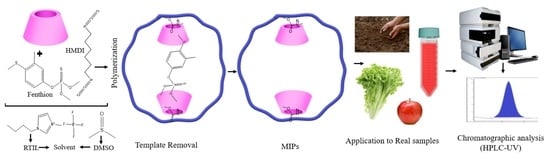
Development of Molecularly Imprinted Polymers for Fenthion Detection in Food and Soil Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instruments Used
2.3. Chromatographic Analysis
2.4. Preparation of Molecularly Imprinted Polymers
2.5. MIP Characterization Analysis
2.6. Adsorption Isotherm
2.7. Adsorption Kinetics
2.8. Selective Recognition Experiment
2.9. Extraction of Template
2.10. Reusability
2.11. FNT Detection in Real Samples
3. Results and Discussion
3.1. Polymer Synthesis
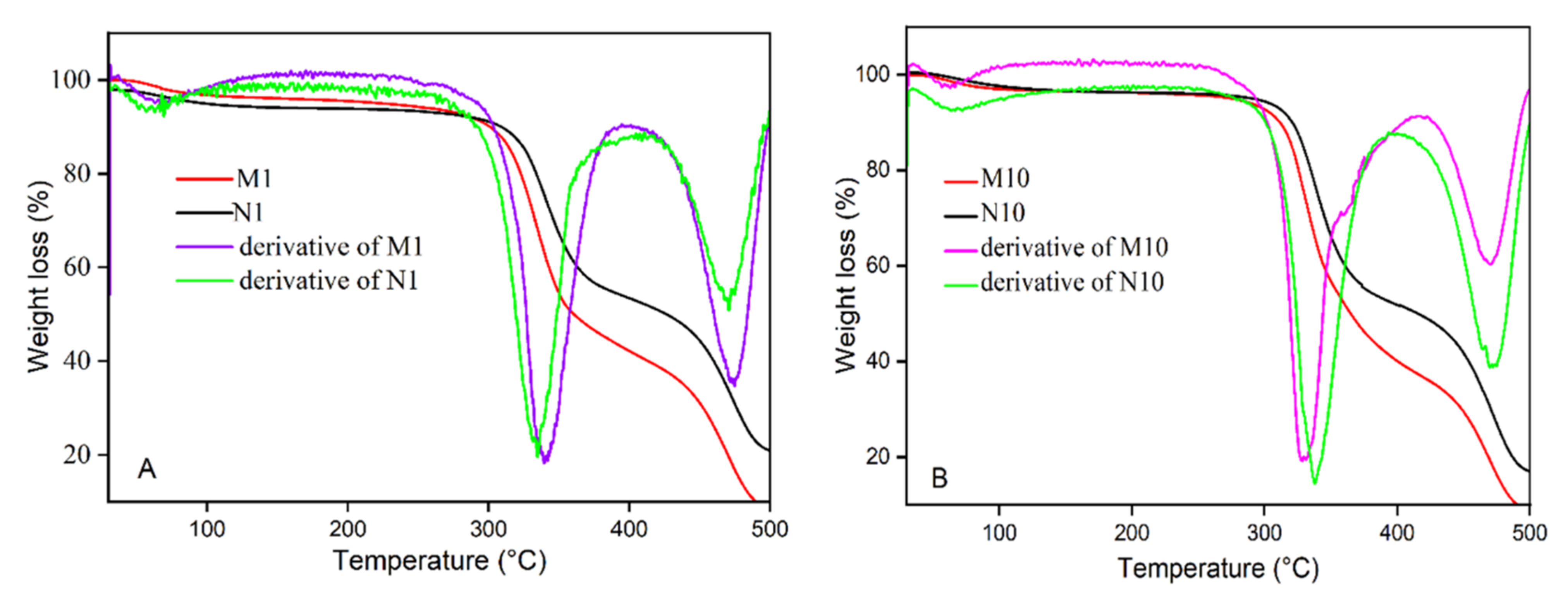
3.2. Characterization Study
3.3. Adsorption Isotherm
3.4. Adsorption Kinetics
3.5. Selectivity
3.6. Template Extraction
3.7. pH Optimization
3.8. Reusability
3.9. FNT Detection in Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Weng, S.; Qiu, M.; Dong, R.; Wang, F.; Huang, L.; Zhang, D.; Zhao, J. Fast detection of fenthion on fruit and vegetable peel using dynamic surface-enhanced Raman spectroscopy and random forests with variable selection. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2018, 200, 20–25. [Google Scholar] [CrossRef]
- Gao, Q.; Liu, W.; Zhu, X. Choline proline ionic liquid-modified magnetic graphene oxide combined with HPLC for analysis of fenthion. J. Iran. Chem. Soc. 2021, 18, 129–137. [Google Scholar] [CrossRef]
- Bakas, I.; Ben Oujji, N.; Istamboulié, G.; Piletsky, S.; Piletska, E.; Ait-Addi, E.; Ait-Ichou, I.; Noguer, T.; Rouillon, R. Molecularly imprinted polymer cartridges coupled to high performance liquid chromatography (HPLC-UV) for simple and rapid analysis of fenthion in olive oil. Talanta 2014, 125, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cao, Y.; Sun, B.; Chen, H.; Dang, X.; Liu, X.; Ai, Y. Magnetic Fe3O4@SiO2@β-cyclodextrin for solid phase extraction of methyl parathion and fenthion in lettuce samples. Anal. Methods 2021, 13, 2974–2980. [Google Scholar] [CrossRef] [PubMed]
- Picó, Y.; Farré, M.; Soler, C.; Barceló, D. Confirmation of fenthion metabolites in oranges by IT-MS and QqTOF-MS. Anal. Chem. 2007, 79, 9350–9363. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, J.H. Simultaneous analysis of fenthion and its five metabolites in produce using ultra-high performance liquid chromatography-tandem mass spectrometry. Molecules 2020, 25, 1938. [Google Scholar] [CrossRef] [Green Version]
- Farooq, S.; Nie, J.; Cheng, Y.; Yan, Z.; Li, J.; Bacha, S.A.S.; Mushtaq, A.; Zhang, H. Molecularly imprinted polymers’ application in pesticide residue detection. Analyst 2018, 143, 3971–3989. [Google Scholar] [CrossRef]
- Farooq, S.; Wu, H.; Nie, J.; Ahmad, S.; Muhammad, I.; Zeeshan, M.; Khan, R.; Asim, M. Application, advancement and green aspects of magnetic molecularly imprinted polymers in pesticide residue detection. Sci. Total Environ. 2022, 804, 150293. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.A.; Yuesuo, Y.; Ao, Q.; Adeel, M.; Hui, Z.Y.; Javed, R. Appraisal of comparative therapeutic potential of undoped and nitrogen-doped titanium dioxide nanoparticles. Molecules 2019, 24, 3916. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.X.; Gao, H.J.; Zhang, L.M.; Chen, X.Q.; Qiao, X.G. The biomimetic immunoassay based on molecularly imprinted polymer: A comprehensive review of recent progress and future prospects. J. Food Sci. 2011, 76, R69–R75. [Google Scholar] [CrossRef]
- Farooq, S.; Nie, J.; Cheng, Y.; Bacha, S.A.S.; Chang, W. Selective extraction of fungicide carbendazim in fruits using β-cyclodextrin based molecularly imprinted polymers. J. Sep. Sci. 2020, 43, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jin, P.; Zhu, F.; Nie, L.; Qiu, H. A review on the use of ionic liquids in preparation of molecularly imprinted polymers for applications in solid-phase extraction. TrAC Trends Anal. Chem. 2021, 134, 116132. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Sun, C.; Sun, J. Comprehensive evaluation of ionic liquid [Bmim][PF6] for absorbing toluene and acetone. Environ. Pollut. 2021, 285, 117675. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, S.; Hu, X.; Chen, C. Phytotoxicity and oxidative stress effect of 1-octyl-3-methylimidazolium chloride ionic liquid on rice seedlings. Environ. Pollut. 2013, 181, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Farooq, S.; Chen, B.; Ahmad, S.; Muhammad, I.; Hussain, Q.; Wu, H. Room-temperature, ionic-liquid-enhanced, beta-cyclodextrin-based, molecularly imprinted polymers for the selective extraction of abamectin. Nanomaterials 2022, 12, 1017. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, X.L.; Ma, L.; Shang, P.P.; Huang, Y.P.; Liu, Z.S. Improving affinity of β-cyclodextrin-based molecularly imprinted polymer using room temperature ionic liquid. Eur. Polym. J. 2019, 116, 275–282. [Google Scholar] [CrossRef]
- Tsarpali, V.; Dailianis, S. Toxicity of two imidazolium ionic liquids, [bmim][BF4] and [omim][BF4], to standard aquatic test organisms: Role of acetone in the induced toxicity. Ecotoxicol. Environ. Saf. 2015, 117, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Qi, L.; Li, X.; Zhang, W.; Cheng, G. Stimuli-responsive recognition of BSA-imprinted poly vinyl acetate grafted calcium alginate core-shell hydrogel microspheres. J. Appl. Polym. Sci. 2013, 127, 3898–3909. [Google Scholar] [CrossRef]
- Ozmen, E.Y.; Sezgin, M.; Yilmaz, A.; Yilmaz, M. Synthesis of β-cyclodextrin and starch based polymers for sorption of azo dyes from aqueous solutions. Bioresour. Technol. 2008, 99, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Booker, K.; Bowyer, M.C.; Holdsworth, C.I.; McCluskey, A. Efficient preparation and improved sensitivity of molecularly imprinted polymers using room temperature ionic liquids. Chem. Commun. 2006, 16, 1730–1732. [Google Scholar] [CrossRef]
- Chen, L.; Xu, S.; Li, J.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.A.; Li, Z.H.; Du, J.M.; Han, B.X.; Li, G.Z.; Hou, W.G.; Shen, D.; Zheng, L.Q.; Zhang, G.Y. Preparation and characterization of inclusion complexes of β-cyclodextrin with ionic liquid. Chem.—A Eur. J. 2005, 11, 5875–5880. [Google Scholar] [CrossRef]
- Jing, T.; Wang, Y.; Dai, Q.; Xia, H.; Niu, J.; Hao, Q.; Mei, S.; Zhou, Y. Preparation of mixed-templates molecularly imprinted polymers and investigation of the recognition ability for tetracycline antibiotics. Biosens. Bioelectron. 2010, 25, 2218–2224. [Google Scholar] [CrossRef]
- Farooq, S.; Nie, J.; Cheng, Y.; Yan, Z.; Bacha, S.A.S.; Zhang, J.; Nahiyoon, R.A.; Hussain, Q. Synthesis of core-shell magnetic molecularly imprinted polymer for the selective determination of imidacloprid in apple samples. J. Sep. Sci. 2019, 42, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, Y.; Liu, Y.; Zhang, C.; Zhou, Y. Magnetic-graphene based molecularly imprinted polymer nanocomposite for the recognition of bovine hemoglobin. Talanta 2015, 144, 411–419. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, J.; Liu, H. Rapid determination of lambda-cyhalothrin using a fluorescent probe based on ionic-liquid-sensitized carbon dots coated with molecularly imprinted polymers. Anal. Bioanal. Chem. 2019, 411, 5309–5316. [Google Scholar] [CrossRef]
- Cheng, Y.; Nie, J.; Li, Z.; Yan, Z.; Xu, G.; Li, H.; Guan, D. A molecularly imprinted polymer synthesized using β-cyclodextrin as the monomer for the efficient recognition of forchlorfenuron in fruits. Anal. Bioanal. Chem. 2017, 409, 5065–5072. [Google Scholar] [CrossRef]
- Tan, I.A.W.; Ahmad, A.L.; Hameed, B.H. Adsorption isotherms, kinetics, thermodynamics and desorption studies of 2,4,6-trichlorophenol on oil palm empty fruit bunch-based activated carbon. J. Hazard. Mater. 2009, 164, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Narayanan, N.; Gupta, S. Application of magnetic molecularly imprinted polymers for extraction of imidacloprid from eggplant and honey. Food Chem. 2018, 255, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tang, Q.; Wang, Q.; Qiao, X.; Xu, Z. Study of a molecularly imprinted solid-phase extraction coupled with high-performance liquid chromatography for simultaneous determination of trace trichlorfon and monocrotophos residues in vegetables. J. Sci. Food Agric. 2014, 94, 1409–1415. [Google Scholar] [CrossRef]
- Li, X.; Zhang, B.; Li, W.; Lei, X.; Fan, X.; Tian, L.; Zhang, H.; Zhang, Q. Preparation and characterization of bovine serum albumin surface-imprinted thermosensitive magnetic polymer microsphere and its application for protein recognition. Biosens. Bioelectron. 2014, 51, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dang, X.; Ai, Y.; Chen, H. Preparation of an acryloyl β-cyclodextrin-silica hybrid monolithic column and its application in pipette tip solid-phase extraction and HPLC analysis of methyl parathion and fenthion. J. Sep. Sci. 2018, 41, 3508–3514. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; He, M.; Chen, B.; Hu, B. Metal organic frameworks-derived magnetic nanoporous carbon for preconcentration of organophosphorus pesticides from fruit samples followed by gas chromatography-flame photometric detection. J. Chromatogr. A 2019, 1583, 19–27. [Google Scholar] [CrossRef]
- Heravizadeh, O.R.; Khadem, M.; Dehghani, F.; Shahtaheri, S.J. Determination of fenthion in urine samples using molecularly imprinted nanoparticles: Modelling and optimisation by response surface methodology. Int. J. Environ. Anal. Chem. 2020, 1–15. [Google Scholar] [CrossRef]



| Polymers | Fenthion (mmol) | β-CD (mmol) | HMDI (mmol) | RTIL (mL) | DMSO (mL) |
|---|---|---|---|---|---|
| M1 * | 1 | 4 | 16 | 10 | 20 |
| M2 | 1 | 4 | 16 | 15 | 15 |
| M3 | 1 | 4 | 16 | 20 | 10 |
| M4 | 1 | 8 | 20 | 10 | 20 |
| M5 | 1 | 8 | 20 | 15 | 15 |
| M6 | 1 | 8 | 20 | 20 | 10 |
| M7 | 1 | 12 | 24 | 10 | 20 |
| M6 | 1 | 12 | 24 | 15 | 15 |
| M9 | 1 | 12 | 24 | 20 | 10 |
| M10 * | 1 | 4 | 16 | 0 | 20 |
| Langmuir Isotherm Model | |||
|---|---|---|---|
| Polymers | Qmax (mg g−1) | KL (L mL−1) | R2 |
| M1 | 29.70 | 0.010 | 0.997 |
| N1 | 7.92 | 0.287 | 0.992 |
| M10 | 19.35 | 0.041 | 0.994 |
| N10 | 6.96 | 0.485 | 0.984 |
| Freundlich isotherm model | |||
| Polymers | N | Kf (L mg−1) | R2 |
| M1 | 3.905 | 9.089 | 0.946 |
| N1 | 2.461 | 1.164 | 0.955 |
| M10 | 2.650 | 3.322 | 0.949 |
| N10 | 1.201 | 0.660 | 0.944 |
| Pseudo-second-order model | |||
| Polymers | Qe (mg g−1) | K (mg g−1 s−1) | R2 |
| M1 | 28.01 | 0.010 | 0.998 |
| N1 | 6.83 | 0.034 | 0.999 |
| M10 | 17.01 | 0.018 | 0.999 |
| N10 | 5.56 | 0.075 | 0.999 |
| Sample | Spiked Levels (mg L−1) | Found Concentration (mg L−1) | RSD (%) | Recovery (%) |
|---|---|---|---|---|
| Soil | 0.20 | 0.186 | 1.61 | 93.00 |
| 0.10 | 0.091 | 2.08 | 91.20 | |
| 0.05 | 0.044 | 2.80 | 87.44 | |
| Lettuce | 0.20 | 0.198 | 1.31 | 99.11 |
| 0.10 | 0.097 | 1.95 | 97.41 | |
| 0.05 | 0.047 | 2.11 | 94.64 | |
| Grapes | 0.20 | 0.203 | 1.28 | 101.25 |
| 0.10 | 0.099 | 2.23 | 98.77 | |
| 0.05 | 0.048 | 2.30 | 95.60 |
| Sample | Method | Recovery (%) | RSD (%) | LOD (mg L−1) | LOQ (mg L−1) | Linear Range (mg L−1) | Reference |
|---|---|---|---|---|---|---|---|
| Olive oil | MIP-HPLC-UV | 96.1 | – | 0.005 | 0.023 | – | [3] |
| Water and apple | MIP-HPLC-UV | 94.0–100.4 | 1.7–4.2 | 0.0018 | – | 0.02–10 | [2] |
| Lettuce | MSPE-HPLC-UV | 89.2–101.2 | 9.1 | 0.0005 | 0.0015 | 0.0015–2.0 | [4] |
| Lettuce | SPE HPLC-PDA | 96.0–104.2 | 6.2 | 0.006 | 0.02 | 0.02–0.40 | [32] |
| Fruits | GC-FPD | 91–112 | 3.7 | 0.000033 | 0.00198 | 0.0001–0.1 | [33] |
| Urine | SPE HPLC-UV | 92.69–95.64 | 3.75 | 0.00458 | – | 0.02–0.12 | [34] |
| Soil, lettuce and grapes | MIP-HPLC-UV | 87.44–101.25 | 1.28–2.80 | 0.04 | 0.11 | 0.02–3.0 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooq, S.; Chen, B.; Gao, F.; Muhammad, I.; Ahmad, S.; Wu, H. Development of Molecularly Imprinted Polymers for Fenthion Detection in Food and Soil Samples. Nanomaterials 2022, 12, 2129. https://doi.org/10.3390/nano12132129
Farooq S, Chen B, Gao F, Muhammad I, Ahmad S, Wu H. Development of Molecularly Imprinted Polymers for Fenthion Detection in Food and Soil Samples. Nanomaterials. 2022; 12(13):2129. https://doi.org/10.3390/nano12132129
Chicago/Turabian StyleFarooq, Saqib, Bochang Chen, Fukun Gao, Ihsan Muhammad, Shakeel Ahmad, and Haiyan Wu. 2022. "Development of Molecularly Imprinted Polymers for Fenthion Detection in Food and Soil Samples" Nanomaterials 12, no. 13: 2129. https://doi.org/10.3390/nano12132129
APA StyleFarooq, S., Chen, B., Gao, F., Muhammad, I., Ahmad, S., & Wu, H. (2022). Development of Molecularly Imprinted Polymers for Fenthion Detection in Food and Soil Samples. Nanomaterials, 12(13), 2129. https://doi.org/10.3390/nano12132129