Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Graphene Oxide
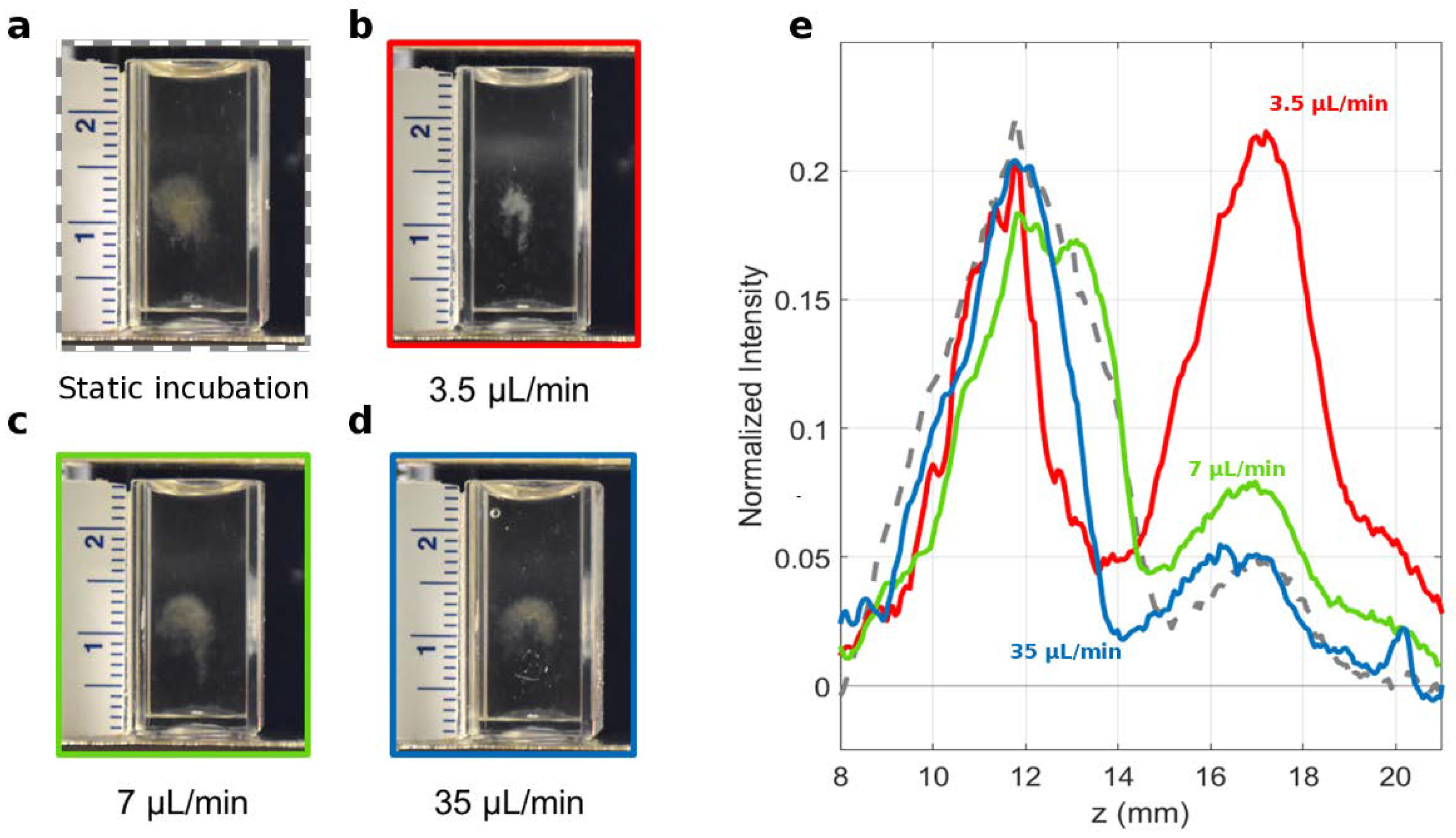
2.2. Preparation of GO-HP Samples
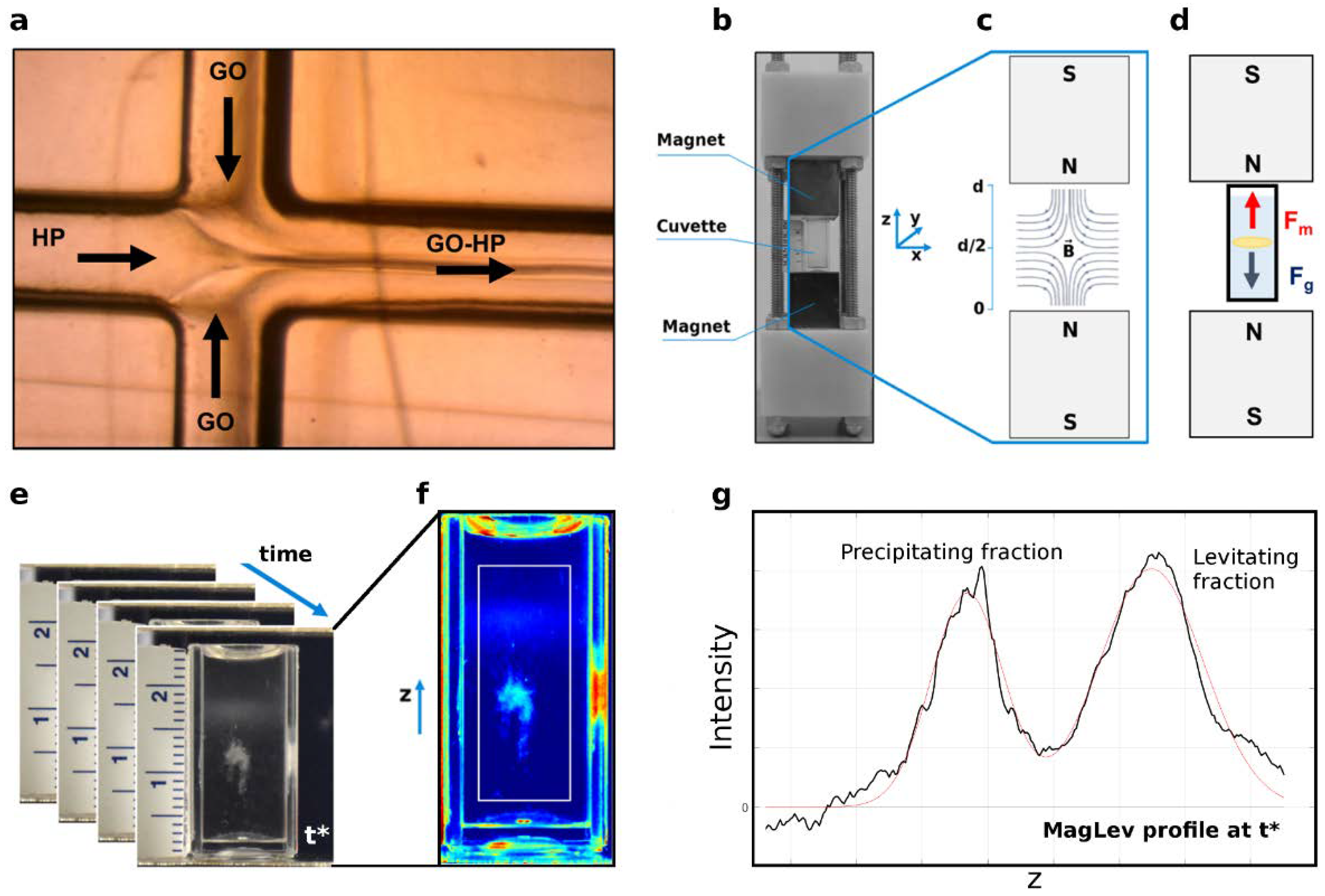
2.3. MagLev Device
2.4. Fundamentals of MagLev
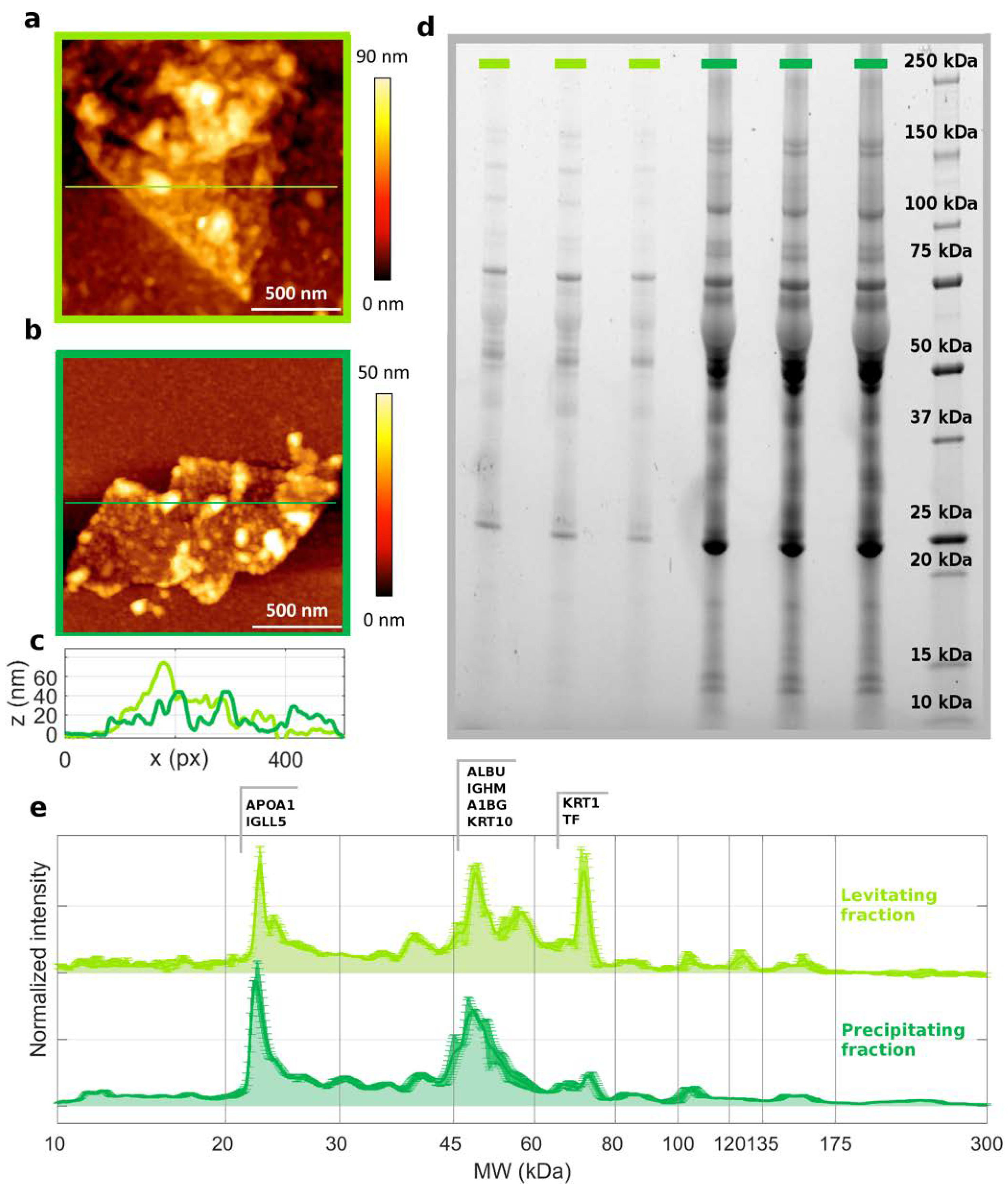
2.5. Atomic Force Microscopy (AFM)
2.6. Bicinchoninic Acid Assay (BCA)
2.7. Sodium Dodecyl Sulphate–Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.8. Nanoliquid Chromatography–Tandem Mass Spectrometry
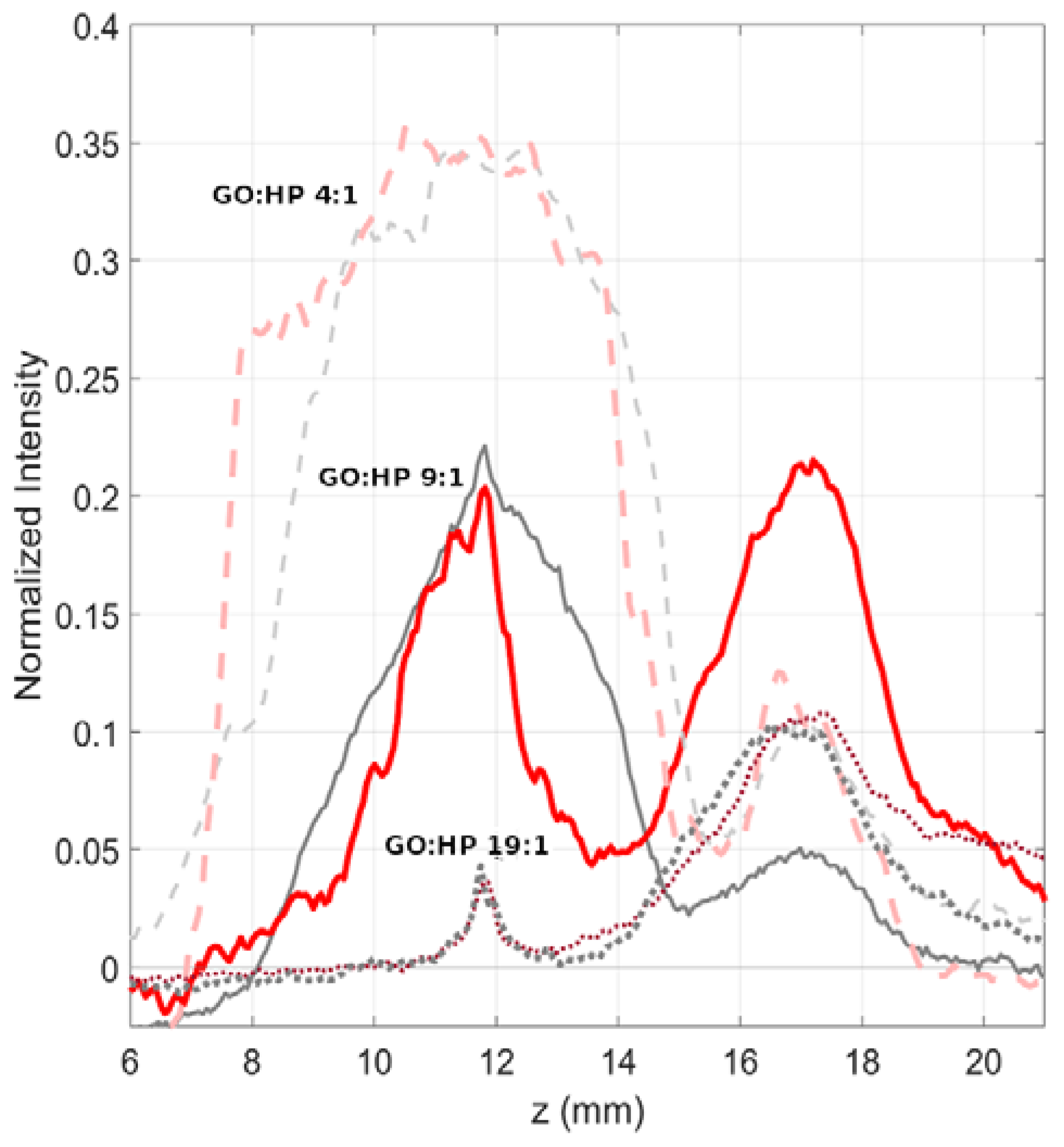
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mirica, K.A.; Shevkoplyas, S.S.; Phillips, S.T.; Gupta, M.; Whitesides, G.M. Measuring densities of solids and liquids using magnetic levitation: Fundamentals. J. Am. Chem. Soc. 2009, 131, 10049–10058. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Raheb, J.; Akhavan, O.; Arjmand, S.; Mashinchian, O.; Rahman, M.; Abdolahad, M.; Serpooshan, V.; Laurent, S.; Mahmoudi, M. Personalized disease-specific protein corona influences the therapeutic impact of graphene oxide. Nanoscale 2015, 7, 8978–8994. [Google Scholar] [CrossRef]
- Ren, J.; Cai, R.; Wang, J.; Daniyal, M.; Baimanov, D.; Liu, Y.; Yin, D.; Liu, Y.; Miao, Q.; Zhao, Y.; et al. Precision nanomedicine development based on specific opsonization of human cancer patient-personalized protein coronas. Nano Lett. 2019, 19, 4692–4701. [Google Scholar] [CrossRef] [PubMed]
- Vence, M.G.; del Pilar Chantada-Vazquez, M.; Vázquez-Estévez, S.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Nunez, C. Potential clinical applications of the personalized, disease-specific protein corona on nanoparticles. Clin. Chim. Acta 2020, 501, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Winzen, S.; Schoettler, S.; Baier, G.; Rosenauer, C.; Mailaender, V.; Landfester, K.; Mohr, K. Complementary analysis of the hard and soft protein corona: Sample preparation critically effects corona composition. Nanoscale 2015, 7, 2992–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faserl, K.; Chetwynd, A.J.; Lynch, I.; Thorn, J.A.; Lindner, H.H. Corona isolation method matters: Capillary electrophoresis mass spectrometry based comparison of protein corona compositions following on-particle versus in-solution or in-gel digestion. Nanomaterials 2019, 9, 898. [Google Scholar] [CrossRef] [Green Version]
- Papi, M.; Palmieri, V.; Digiacomo, L.; Giulimondi, F.; Palchetti, S.; Ciasca, G.; Perini, G.; Caputo, D.; Cartillone, M.C.; Cascone, C.; et al. Converting the personalized biomolecular corona of graphene oxide nanoflakes into a high-throughput diagnostic test for early cancer detection. Nanoscale 2019, 11, 15339–15346. [Google Scholar] [CrossRef]
- Di Domenico, M.; Pozzi, D.; Palchetti, S.; Digiacomo, L.; Iorio, R.; Astarita, C.; Fiorelli, A.; Pierdiluca, M.; Santini, M.; Barbarino, M.; et al. Nanoparticle-biomolecular corona: A new approach for the early detection of non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 9378–9386. [Google Scholar] [CrossRef]
- Papi, M.; Palmieri, V.; Palchetti, S.; Pozzi, D.; Digiacomo, L.; Guadagno, E.; del Basso De Caro, M.; Di Domenico, M.; Ricci, S.; Pani, R.; et al. Exploitation of nanoparticle-protein interactions for early disease detection. Appl. Phys. Lett. 2019, 114, 163702. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Whitesides, G.M. “Axial” magnetic levitation using ring magnets enables simple density-based analysis, separation, and manipulation. Anal. Chem. 2018, 90, 12239–12245. [Google Scholar] [CrossRef]
- Digiacomo, L.; Quagliarini, E.; La Vaccara, V.; Coppola, A.; Coppola, R.; Caputo, D.; Pozzi, D. Detection of Pancreatic Ductal Adenocarcinoma by Ex Vivo Magnetic Levitation of Plasma Protein-Coated Nanoparticles. Cancers 2021, 13, 5155. [Google Scholar] [CrossRef]
- Faria, M.; Björnmalm, M.; Thurecht, K.J.; Kent, S.J.; Parton, R.G.; Kavallaris, M.; Johnston, A.P.; Gooding, J.J.; Corrie, S.R.; Boyd, B.J. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol. 2018, 13, 777–785. [Google Scholar] [CrossRef]
- Di Santo, R.; Digiacomo, L.; Quagliarini, E.; Capriotti, A.L.; Laganà, A.; Zenezini Chiozzi, R.; Caputo, D.; Cascone, C.; Coppola, R.; Pozzi, D. Personalized graphene oxide-protein corona in the human plasma of pancreatic cancer patients. Front. Bioeng. Biotechnol. 2020, 8, 491. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.B.; Vishwanath, A.; Brody, J.P.; Austin, R.H. Hydrodynamic focusing on a silicon chip: Mixing nanoliters in microseconds. Phys. Rev. Lett. 1998, 80, 3863. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 2013 8, 772–781. [Google Scholar] [CrossRef]
- Palmieri, V.; Bugli, F.; Cacaci, M.; Perini, G.; Maio, F.D.; Delogu, G.; Papi, M. Graphene oxide coatings prevent Candida albicans biofilm formation with a controlled release of curcumin-loaded nanocomposites. Nanomedicine 2018, 13, 2867–2879. [Google Scholar] [CrossRef]
- Digiacomo, L.; Giulimondi, F.; Capriotti, A.L.; Piovesana, S.; Montone, C.M.; Chiozzi, R.Z.; Caracciolo, G. Optimal centrifugal isolating of liposome–protein complexes from human plasma. Nanoscale Adv. 2021, 3, 3824–3834. [Google Scholar] [CrossRef]
- La Barbera, G.; Capriotti, A.L.; Cavaliere, C.; Ferraris, F.; Montone, C.M.; Piovesana, S.; Chiozzi, R.Z.; Laganà, A. Saliva as a source of new phosphopeptide biomarkers: Development of a comprehensive analytical method based on shotgun peptidomics. Talanta 2018, 183, 245–249. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Quagliarini, E.; Digiacomo, L.; Caputo, D.; Coppola, A.; Amenitsch, H.; Caracciolo, G.; Pozzi, D. Magnetic Levitation of Personalized Nanoparticle-Protein Corona as an Effective Tool for Cancer Detection. Nanomaterials 2022, 12, 1397. [Google Scholar] [CrossRef]
- Chetwynd, A.J.; Lynch, I. The rise of the nanomaterial metabolite corona, and emergence of the complete corona. Env.-Ment. Sci. Nano 2020, 7, 1041–1060. [Google Scholar] [CrossRef]
- Chetwynd, A.J.; Zhang, W.; Thorn, J.A.; Lynch, I.; Ramautar, R. The nanomaterial metabolite corona determined using a quantitative metabolomics approach: A pilot study. Small 2020, 16, 2000295. [Google Scholar] [CrossRef]
- Ashkarran, A.A.; Suslick, K.S.; Mahmoudi, M. Magnetically levitated plasma proteins. Anal. Chem. 2020, 92, 1663–1668. [Google Scholar] [CrossRef] [Green Version]
| Dynamic Incubation | Static Incubation | |||
|---|---|---|---|---|
| GO:HP FRR | GO Flow Rate (μL/min) | HP Flow Rate (μL/min) | GO Amount (μL) | HP Amount (μL) |
| 4:1 | 2.80 | 0.70 | 80 | 20 |
| 9:1 | 3.14 | 0.35 | 90 | 10 |
| 19:1 | 3.30 | 0.17 | 95 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Digiacomo, L.; Quagliarini, E.; Marmiroli, B.; Sartori, B.; Perini, G.; Papi, M.; Capriotti, A.L.; Montone, C.M.; Cerrato, A.; Caracciolo, G.; et al. Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes. Nanomaterials 2022, 12, 2376. https://doi.org/10.3390/nano12142376
Digiacomo L, Quagliarini E, Marmiroli B, Sartori B, Perini G, Papi M, Capriotti AL, Montone CM, Cerrato A, Caracciolo G, et al. Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes. Nanomaterials. 2022; 12(14):2376. https://doi.org/10.3390/nano12142376
Chicago/Turabian StyleDigiacomo, Luca, Erica Quagliarini, Benedetta Marmiroli, Barbara Sartori, Giordano Perini, Massimiliano Papi, Anna Laura Capriotti, Carmela Maria Montone, Andrea Cerrato, Giulio Caracciolo, and et al. 2022. "Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes" Nanomaterials 12, no. 14: 2376. https://doi.org/10.3390/nano12142376
APA StyleDigiacomo, L., Quagliarini, E., Marmiroli, B., Sartori, B., Perini, G., Papi, M., Capriotti, A. L., Montone, C. M., Cerrato, A., Caracciolo, G., & Pozzi, D. (2022). Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle–Protein Complexes. Nanomaterials, 12(14), 2376. https://doi.org/10.3390/nano12142376









