In Search of Preferential Macrocyclic Hosts for Sulfur Mustard Sensing and Recognition: A Computational Investigation through the New Composite Method r2SCAN-3c of the Key Factors Influencing the Host-Guest Interactions
Abstract
:1. Introduction
2. Computational Methods
3. Results and Discussion
3.1. Interaction of SM and Its Simulants with EtP[5]
3.1.1. Structural and Energetic Properties
3.1.2. Electronic Properties of SM, S1, S2, S3, S4, and S5@EtP[5] Complexes
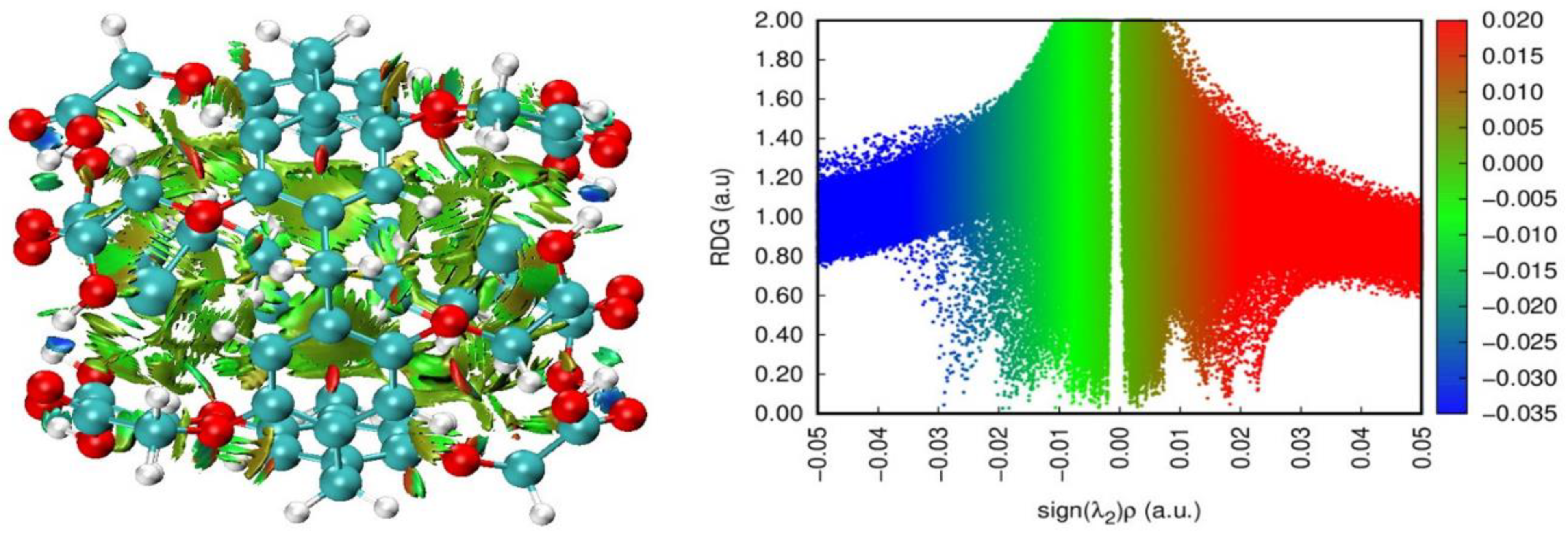
3.1.3. NCI-RDG and IGMH Analysis of the Host-Guest Interactions
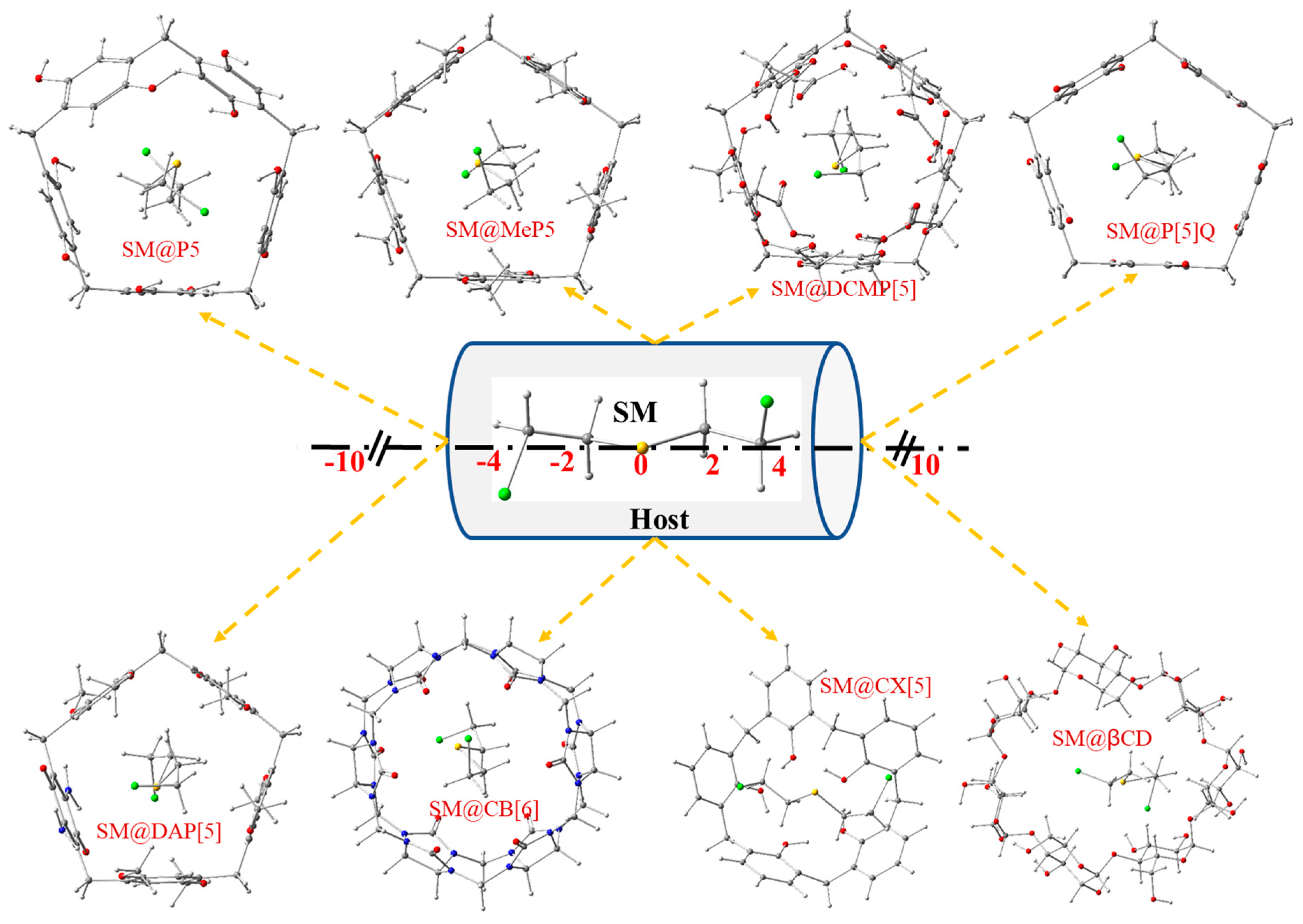
3.2. Interactions between SM and Different Macrocyclic Hosts
3.2.1. Electronic and Chemical Sensing Properties
3.2.2. NCI-RDG Analysis of SM@DCMP[5] Complex
4. Conclusions
- The r2SCAN-3c method can reproduce satisfactorily the crystalline structures of SM@EtP[5], S1@EtP[5], S2@EtP[5], S3@EtP[5], S4@EtP[5], and S5@EtP[5] complexes.
- The complexation energies calculated using r2SCAN-3c correlate with the experimental association constants.
- The major forces that contribute to the stability of the formed complexes involve C-H···H-C, C-H···O intramolecular interactions and C-H···H-C, S···π, C-H···Cl, C-H···S, C-H···O and C-H···π intermolecular interactions as revealed by NCI-RDG and IGM analysis.
- The macrocycles CB[6], β-CD, CX[5] and particularly P[5]Q show great potential as sensors for sulfur mustard.
- Among the studied complexes, SM@DCMP[5] was the most stable with the highest complexation energy of −155.26 kJ/mol, its high stability is due to the occurrence of additional intramolecular hydrogen bonds in DCMP[5].
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Facts About Sulfur Mustard. Available online: https://emergency.cdc.gov/agent/sulfurmustard/basics/facts.asp (accessed on 29 December 2021).
- Wormser, U. Toxicology of mustard gas. Trends Pharmacol. Sci. 1991, 12, 164–167. [Google Scholar] [CrossRef]
- Smith, S.L. Toxic Exposures: Sulfur Mustard and the Health Consequences of World War II in the United States; Rutgers University Press: New Brunswick, NJ, USA, 2017. [Google Scholar]
- Duchovic, R.J.; Vilensky, J.A. Mustard sulfur: Its pre-World War I history. J. Chem. Educ. 2007, 84, 944. [Google Scholar] [CrossRef]
- Norman, J.E., Jr. Lung cancer mortality in World War I veterans with mustard-gas injury: 1919–1965. J. Natl. Cancer Inst. 1975, 54, 311–317. [Google Scholar] [CrossRef]
- Razavi, S.M.; Ghanei, M.; Salamati, P.; Safiabadi, M. Long-term effects of mustard gas on respiratory system of Iranian veterans after Iraq-Iran war: A review. Chin. J. Traumatol. 2013, 16, 163–168. [Google Scholar] [PubMed]
- Liu, Y.; Howarth, A.J.; Vermeulen, N.A.; Moon, S.-Y.; Hupp, J.T.; Farha, O.K. Catalytic degradation of chemical warfare agents and their simulants by metal-organic frameworks. Coord. Chem. Rev. 2017, 346, 101–111. [Google Scholar] [CrossRef]
- Wang, Q.-Q.; Begum, R.A.; Day, V.W.; Bowman-James, K. Chemical mustard containment using simple palladium pincer complexes: The influence of molecular walls. J. Am. Chem. Soc. 2013, 135, 17193–17199. [Google Scholar] [CrossRef]
- Islamoglu, T.; Chen, Z.; Wasson, M.C.; Buru, C.T.; Kirlikovali, K.O.; Afrin, U.; Mian, M.R.; Farha, O.K. Metal-Organic Frameworks against Toxic Chemicals. Chem. Rev. 2020, 120, 8130–8160. [Google Scholar] [CrossRef]
- Plass, W. Supramolecular interactions of vanadate species: Vanadium (V) complexes with N-salicylidenehydrazides as versatile models. Coord. Chem. Rev. 2003, 237, 205–212. [Google Scholar] [CrossRef]
- Zhao, D.; Yu, S.; Jiang, W.-J.; Cai, Z.-H.; Li, D.-L.; Liu, Y.-L.; Chen, Z.-Z. Recent Progress in Metal-Organic Framework Based Fluorescent Sensors for Hazardous Materials Detection. Molecules 2022, 27, 2226. [Google Scholar] [CrossRef] [PubMed]
- Litim, A.; Belhocine, Y.; Benlecheb, T.; Ghoniem, M.G.; Kabouche, Z.; Ali, F.A.M.; Abdulkhair, B.Y.; Seydou, M.; Rahali, S. DFT-D4 Insight into the Inclusion of Amphetamine and Methamphetamine in Cucurbit [7] uril: Energetic, Structural and Biosensing Properties. Molecules 2021, 26, 7479. [Google Scholar] [CrossRef]
- Novikov, A.S. Non-Covalent Interactions in Organic, Organometallic, and Inorganic Supramolecular Systems Relevant for Medicine, Materials Science, and Catalysis. Crystals 2022, 12, 246. [Google Scholar] [CrossRef]
- Bouhadiba, A.; Rahali, S.; Belhocine, Y.; Allal, H.; Nouar, L.; Rahim, M. Structural and energetic investigation on the host/guest inclusion process of benzyl isothiocyanate into β-cyclodextrin using dispersion-corrected DFT calculations. Carbohydr. Res. 2020, 491, 107980. [Google Scholar] [CrossRef] [PubMed]
- Maharramov, A.M.; Mahmudov, K.T.; Kopylovich, M.N.; Pombeiro, A.J.L. (Eds.) Noncovalent Interactions in the Synthesis and Design of New Compounds; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Várnai, B.; Zsila, F.; Szakács, Z.; Garádi, Z.; Malanga, M.; Béni, S. Sulfobutylation of Beta-Cyclodextrin Enhances the Complex Formation with Mitragynine: An NMR and Chiroptical Study. Int. J. Mol. Sci. 2022, 23, 3844. [Google Scholar] [CrossRef]
- Cid-Samamed, A.; Rakmai, J.; Mejuto, J.C.; Simal-Gandara, J.; Astray, G. Cyclodextrins inclusion complex: Preparation methods, analytical techniques and food industry applications. Food Chem. 2022, 384, 132467. [Google Scholar] [CrossRef]
- Assaba, I.M.; Rahali, S.; Belhocine, Y.; Allal, H. Inclusion complexation of chloroquine with α and β-cyclodextrin: Theoretical insights from the new B97-3c composite method. J. Mol. Struct. 2021, 1227, 129696. [Google Scholar] [CrossRef]
- Wdowiak, K.; Rosiak, N.; Tykarska, E.; Żarowski, M.; Płazińska, A.; Płaziński, W.; Cielecka-Piontek, J. Amorphous Inclusion Complexes: Molecular Interactions of Hesperidin and Hesperetin with HP-Β-CD and Their Biological Effects. Int. J. Mol. Sci. 2022, 23, 4000. [Google Scholar] [CrossRef]
- Baudin, C.; Pean, C.; Perly, B.; Gosselin, P. Inclusion of organic pollutants in cyclodextrin and derivatives. Int. J. Environ. Anal. Chem. 2000, 77, 233–242. [Google Scholar] [CrossRef]
- Del Valle, E.M.M. Cyclodextrins and their uses: A review. Process Biochem. 2004, 39, 1033–1046. [Google Scholar] [CrossRef]
- Crini, G. Review: A history of cyclodextrins. Chem. Rev. 2014, 114, 10940–10975. [Google Scholar] [CrossRef]
- Masson, E.; Ling, X.X.; Joseph, R.; Kyeremeh-Mensah, L.; Lu, X.Y. Cucurbituril chemistry: A tale of supramolecular success. RSC Adv. 2012, 2, 1213–1247. [Google Scholar] [CrossRef]
- Assaf, K.I.; Nau, W.M. Cucurbiturils: From synthesis to high-affinity binding and catalysis. Chem. Soc. Rev. 2015, 44, 394–418. [Google Scholar] [CrossRef] [Green Version]
- Barrow, S.J.; Kasera, S.; Rowland, M.J.; del Barrio, J.; Scherman, O.A. Cucurbituril-based molecular recognition. Chem. Rev. 2015, 115, 12320–12406. [Google Scholar] [CrossRef] [Green Version]
- Español, E.S.; Villamil, M.M. Calixarenes: Generalities and Their Role in Improving the Solubility, Biocompatibility, Stability, Bioavailability, Detection, and Transport of Biomolecules. Biomolecules 2019, 9, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Sharma, A.; Singh, H.; Suating, P.; Kim, H.S.; Sunwoo, K.; Shim, I.; Gibb, B.C.; Kim, J.S. Revisiting Fluorescent Calixarenes: From Molecular Sensors to Smart Materials. Chem. Rev. 2019, 119, 9657–9721. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chawla, S.; Zou, M.C. Calixarenes based materials for gas sensing applications: A review. J. Incl. Phenom. Macrocycl. Chem. 2017, 88, 129–158. [Google Scholar] [CrossRef]
- Koifman, O.I.; Ageeva, T.A.; Beletskaya, I.P.; Averin, A.D.; Yakushev, A.A.; Tomilova, L.G.; Dubinina, T.V.; Tsivadze, A.Y.; Gorbunova, Y.G.; Stuzhin, P.A.; et al. Macroheterocyclic Compounds—A Key Building Block in New Functional Materials and Molecular Devices. Macroheterocycles 2020, 13, 311–467. [Google Scholar] [CrossRef]
- Xue, M.; Yang, Y.; Chi, X.; Zhang, Z.; Huang, F. Pillararenes, A New Class of Macrocycles for Supramolecular Chemistry. Acc. Chem. Res. 2012, 45, 1294–1308. [Google Scholar] [CrossRef]
- Guo, F.; Sun, Y.; Xi, B.; Diao, G. Recent progress in the research on the host-guest chemistry of pillar[n]arenes. Supramol. Chem. 2018, 30, 81–92. [Google Scholar] [CrossRef]
- Ogoshi, T.; Kanai, S.; Fujinami, S.; Yamagishi, T.A.; Nakamoto, Y. para-Bridged symmetrical Pillar[5]arenes: Their Lewis acid catalyzed synthesis and host-guest property. J. Am. Chem. Soc. 2008, 130, 5022–5023. [Google Scholar] [CrossRef]
- Gu, R.; Lehn, J.-M. Constitutional Dynamic Selection at Low Reynolds Number in a Triple Dynamic System: Covalent Dynamic Adaptation Driven by Double Supramolecular Self-Assembly. J. Am. Chem. Soc. 2021, 143, 14136–14146. [Google Scholar] [CrossRef]
- Wang, S.; Li, X.; Zhang, X.; Huang, P.; Fang, P.; Wang, J.; Yang, S.; Wu, K.; Du, P. A supramolecular polymeric heterojunction composed of an all-carbon conjugated polymer and fullerenes. Chem. Sci. 2021, 12, 10506–10513. [Google Scholar] [CrossRef]
- Lou, X.Y.; Song, N.; Yang, Y.W. A stimuli-responsive pillararene-based hybrid material with enhanced tunable multicolor luminescence and ion-sensing ability. Natl. Sci. Rev. 2021, 8, nwaa281. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Li, Q.-H.; Shu, L.; Wan, J.-H. A Shape-Persistent Arylene Ethynylene Macrocycle with a Multiple Acetamide Modified Cavity: Synthesis and Gelation. Soft Matter 2021, 17, 3242–3249. [Google Scholar] [CrossRef] [PubMed]
- Xing, R.; Liu, K.; Jiao, T.; Zhang, N.; Ma, K.; Zhang, R.; Zou, Q.; Ma, G.; Yan, X. An injectable self-assembling collagen-gold hybrid hydrogel for combinatorial antitumor photothermal/photodynamic therapy. Adv. Mater. 2016, 28, 3669–3676. [Google Scholar] [CrossRef]
- Song, J.; Yuan, C.; Jiao, T.; Xing, R.; Yang, M.; Adams, D.J.; Yan, X. Multifunctional antimicrobial biometallohydrogels based on amino Acid coordinated self-assembly. Small 2020, 16, 1907309. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. Capture of sulfur mustard by pillar[5]arene: From host-guest complexation to efficient adsorption using nonporous adaptive crystals. iScience 2020, 23, 101443. [Google Scholar] [CrossRef]
- Zhou, S.; Li, W.; Zhao, Q.; Dong, H.; Wang, Y.; Lu, F.; Zhao, J.; Liu, S.; Chen, H.; Wang, L.; et al. Detoxification of the Toxic Sulfur Mustard Simulant by a Supramolecular Antidote in Vitro and in Vivo. ACS Appl. Mater. Interfaces 2021, 13, 58291–58300. [Google Scholar] [CrossRef] [PubMed]
- De Souza, L.A.; Nogueira, C.A.S.; Lopes, J.F.; Dos Santos, H.F.; De Almeida, W.B. DFT study of cisplatin@carbon nanohorns complexes. J. Inorg. Biochem. 2013, 129, 71–83. [Google Scholar] [CrossRef]
- Belhocine, Y.; Bouhadiba, A.; Rahim, M.; Nouar, L.; Djilani, I.; Khatmi, D.I. Inclusion Complex Formation of β-Cyclodextrin with the Nonsteroidal Anti-inflammatory Drug Flufenamic Acid: Computational Study. Macroheterocycles 2018, 11, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Aree, T.; Jongrungruangchok, S. Structure–antioxidant activity relationship of β-cyclodextrin inclusion complexes with olive tyrosol, hydroxytyrosol and oleuropein: Deep insights from X-ray analysis, DFT calculation and DPPH assay. Carbohydr. Polym. 2018, 199, 661–669. [Google Scholar] [CrossRef]
- Buta, M.C.; Toader, A.M.; Frecus, B.; Oprea, C.I.; Cimpoesu, F.; Ionita, G. Molecular and Supramolecular Interactions in Systems with Nitroxide-Based Radicals. Int. J. Mol. Sci. 2019, 20, 4733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Shen, C.; Shi, H.; Luo, S.; He, M.; Chen, M. Theoretical prediction of structures and inclusion properties of heteroatom-bridged pillar[n]arenes. Struct. Chem. 2020, 31, 329–337. [Google Scholar] [CrossRef]
- Nikolova, V.; Velinova, A.; Dobrev, S.; Kircheva, N.; Angelova, S.; Dudev, T. Host–Guest Complexation of Cucurbit[7]Uril and Cucurbit[8]Uril with the Antineoplastic and Multiple Sclerosis Agent Mitoxantrone (Novantrone). J. Phys. Chem. A 2021, 125, 536–542. [Google Scholar] [CrossRef]
- Venkataramanan, N.S.; Suvitha, A.; Mizuseki, H.; Kawazoe, Y. Computational study on the interactions of mustard gas with cucurbituril macrocycles. Int. J. Quantum Chem. 2015, 115, 1515–1525. [Google Scholar] [CrossRef]
- Venkataramanan, N.S.; Ambigapathy, S. Encapsulation of sulfur, oxygen, and nitrogen mustards by cucurbiturils: A DFT study. J. Incl. Phenom. Macrocycl. Chem. 2015, 83, 387–400. [Google Scholar] [CrossRef]
- Grimme, S.; Hansen, A.; Ehlert, S.; Mewes, J.M. r2SCAN-3c: A “Swiss army knife” composite electronic-structure method. J. Chem. Phys. 2021, 154, 064103. [Google Scholar] [CrossRef]
- Furness, J.W.; Kaplan, A.D.; Ning, J.; Perdew, J.P.; Sun, J. Accurate and Numerically Efficient r2SCAN Meta-Generalized Gradient Approximation. J. Phys. Chem. Lett. 2020, 11, 8208–8215, Erratum in J. Phys. Chem. Lett. 2020, 11, 9248. [Google Scholar] [CrossRef]
- Santra, G.; Martin, J.M.L. Pure and Hybrid SCAN, rSCAN, and r2SCAN: Which One Is Preferred in KS- and HF-DFT Calculations, and How Does D4 Dispersion Correction Affect This Ranking? Molecules 2022, 27, 141. [Google Scholar] [CrossRef]
- Kingsbury, R.; Gupta, A.; Bartel, C.; Munro, J.; Dwaraknath, S.; Horton, M.; Persson, K. Performance comparison of r2SCAN and SCAN metaGGA density functionals for solid materials via an automated, high-throughput computational workflow. Phys. Rev. Mater. 2022, 6, 013801. [Google Scholar] [CrossRef]
- Ehlert, S.; Grimme, S.; Hanson, A. Conformational Energy Benchmark for Longer n-Alkane Chains. J. Phys. Chem. A 2022, 126, 3521–3535. [Google Scholar] [CrossRef]
- Furness, J.W.; Kaplan, A.D.; Ning, J.; Perdew, J.P.; Sun, J. Construction of meta-GGA functionals through restoration of exact constraint adherence to regularized SCAN functionals. J. Chem. Phys. 2021, 156, 034109. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, S.; Huniar, U.; Ning, J.; Furness, J.W.; Sun, J.; Kaplan, A.D.; Perdew, J.P.; Brandenburg, J.G. r2SCAN-D4: Dispersion corrected meta-generalized gradient approximation for general chemical applications. J. Chem. Phys. 2021, 154, 061101. [Google Scholar] [CrossRef] [PubMed]
- Grimm, L.M.; Spicher, S.; Tkachenko, B.; Schreiner, P.R.; Grimme, S.; Biedermann, F. The Role of Packing, Dispersion, Electrostatics, and Solvation in High-Affinity Complexes of Cucurbit[n ]urils with Uncharged Polar Guests. Chem. A Eur. J. 2022, 28, e202200529. [Google Scholar] [CrossRef] [PubMed]
- Bursch, M.; Mewes, J.-M.; Hansen, A.; Grimme, S. Best Practice DFT Protocols for Basic Molecular Computational Chemistry; Cambridge Open Engage: Cambridge, UK, 2022. [Google Scholar]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Phys. Chem. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Kruse, H.; Grimme, S. A geometrical correction for the inter- and intra-molecular basis set superposition error in Hartree-Fock and density functional theory calculations for large systems. J. Chem. Phys. 2012, 136, 154101. [Google Scholar] [CrossRef] [Green Version]
- Caldeweyher, E.; Bannwarth, C.; Grimme, S. Extension of the D3 dispersion coefficient model. J. Chem. Phys. 2017, 147, 034112. [Google Scholar] [CrossRef]
- Caldeweyher, E.; Ehlert, S.; Hansen, A.; Neugebauer, H.; Spicher, S.; Bannwarth, C.; Grimme, S. A generally applicable atomic-charge dependent London dispersion correction. J. Chem. Phys. 2019, 150, 154122. [Google Scholar] [CrossRef]
- Caldeweyher, E.; Mewes, J.-M.; Ehlert, S.; Grimme, S. Extension and evaluation of the D4 London-dispersion model for periodic systems. Phys. Chem. Chem. Phys. 2020, 22, 8499–8512. [Google Scholar] [CrossRef]
- Neese, F. The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The ORCA program system, version 4.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 8, e1327. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef] [PubMed]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Cossi, M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A 1998, 102, 1995–2001. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1831237: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1884850: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1831239: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1884851: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1831240: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- Li, B.; Li, S.; Wang, B.; Meng, Z.; Wang, Y.; Meng, Q.; Li, C. CCDC 1884852: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2020. [Google Scholar] [CrossRef]
- MacRae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piya, A.A.; Shamim, S.U.D.; Uddin, M.N.; Munny, K.N.; Alam, A.; Hossain, M.K.; Ahmed, F. Adsorption behavior of cisplatin anticancer drug on the pristine, Al-and Ga-doped BN nanosheets: A comparative DFT study. Comput. Theor. Chem. 2021, 1200, 113241. [Google Scholar] [CrossRef]
- Rezaei, A.; Ghiasi, R.; Marjani, A. Strong chemisorption of E2H2 and E2H4 (E = C, Si) on B12N12 nano-cage. J. Nanostruct. Chem. 2020, 10, 179–191. [Google Scholar] [CrossRef]
- Ding, S.; Gu, W. Evaluate the potential utilization of B24N24 fullerene in the recognition of COS, H2S, SO2, and CS2 gases (environmental pollution). J. Mol. Liq. 2022, 345, 117041. [Google Scholar] [CrossRef]
- Fifere, A.; Marangoci, N.; Maier, S.S.; Coroaba, A.; Maftei, D.; Pinteala, M. Theoretical study on β-cyclodextrin inclusion complexes with propiconazole and protonated propiconazole. Beilstein J. Org. Chem. 2012, 8, 2191–2201. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, F.; Schneider, H.-J. Experimental binding energies in supramolecular complexes. Chem. Rev. 2016, 116, 5216–5300. [Google Scholar] [CrossRef]
- Saleh, G.; Gatti, C.; Lo Presti, L. Non-covalent interaction via the reduced density gradient: Independent atom model vs experimental multipolar electron densities. Comput. Theor. Chem. 2012, 998, 148–163. [Google Scholar] [CrossRef]
- Lu, T.; Chen, Q. Independent gradient model based on Hirshfeld partition: A new method for visual study of interactions in chemical systems. J. Comput. Chem. 2022, 43, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, S. CH/π Interactions. Annual Reports Section “C”. Phys. Chem. 2012, 108, 69–95. [Google Scholar]
- Ogoshi, T.; Aoki, T.; Kitajima, K.; Fujinami, S.; Yamagishi, T.-A.; Nakamoto, Y. Facile, rapid, and high-yield synthesis of pillar[5]arene from commercially available reagents and its X-ray crystal structure. J. Org. Chem. 2011, 76, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shu, X.; Li, J.; Chen, S.; Han, K.; Xu, M.; Jia, X. Complexation of 1,4-Bis(pyridinium)butanes by negatively charged carboxylatopillar[5]arene. J. Org. Chem. 2011, 76, 8458–8465. [Google Scholar] [CrossRef]
- Shurpik, D.N.; Yakimova, L.S.; Makhmutova, L.I.; Makhmutova, A.R.; Rizvanov, I.K.; Plemenkov, V.V.; Stoikov, I.I. Pillar[5]arenes with morpholide and pyrrolidide substituents: Synthesis and complex formation with alkali metal ions. Macroheterocycles 2014, 7, 351–357. [Google Scholar] [CrossRef]
- Yakimova, L.S.; Shurpik, D.N.; Makhmutova, A.R.; Stoikov, I.I. pillar[5]arenes bearing amide and carboxylic groups as synthetic receptors for alkali metal ions. Macroheterocycles 2017, 10, 226–232. [Google Scholar] [CrossRef]
- Shivakumar, K.I.; Yan, Y.; Hughes, C.E.; Apperley, D.C.; Harris, K.D.M.; Sanjayan, G.J. Exploiting powder X-ray diffraction to establish the solvent-assisted solid-state supramolecular assembly of pillar[5]quinone. Cryst. Growth Des. 2015, 15, 1583–1587. [Google Scholar] [CrossRef]
- Strutt, N.L.; Zhang, H.; Schneebeli, S.T.; Stoddart, J.F. Amino-functionalized pillar[5]arene. Chemistry 2014, 20, 10996–11004. [Google Scholar] [CrossRef]
- Chen, K.; Kang, Y.S.; Zhao, Y.; Yang, J.M.; Lu, Y.; Sun, W.Y. Cucurbit[6]uril-based supramolecular assemblies: Possible application in radioactive cesium cation capture. J. Am. Chem. Soc. 2014, 136, 16744–16747. [Google Scholar] [CrossRef] [PubMed]
- Dalgarno, S.J.; Tian, J.; Warren, J.E.; Clark, T.E.; Makha, M.; Raston, C.L.; Atwood, J.L. Calix[5]arene: A versatile sublimate that displays gas sorption properties. Chem. Commun. 2007, 46, 4848–4850. [Google Scholar] [CrossRef] [PubMed]
- Dalgarno, S.J.; Tian, J.; Warren, J.E.; Clark, T.E.; Makha, M.; Raston, C.L.; Atwood, J.L. CCDC 637165: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2008. [Google Scholar] [CrossRef]
- Aree, T.; Chaichit, N. Crystal structure of β-cyclodextrin–benzoic acid inclusion complex. Carbohydr. Res. 2003, 338, 439–446. [Google Scholar] [CrossRef]
- Aree, T.; Chaichit, N. CCDC 191347: Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2003. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Q.-X. Use of quantum chemical methods to study cyclodextrin chemistry. J. Incl. Phenom. Macrocycl. Chem. 2004, 50, 95–103. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.; Millam, J. GaussView; Version 5; Semichem Inc.: Shawnee Mission, KS, USA, 2009. [Google Scholar]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avogadro: An Open-Source Molecular Builder and Visualization Tool, Version 1.2.0. Available online: http://avogadro.cc/ (accessed on 24 May 2022).





| Interaction | Interaction Distance | r2SCAN-3c | Exp. [39] | Gas-Phase Optimized Geometries |
|---|---|---|---|---|
| C–H···π | Ba | 2.82 (0.01) | 2.81 | 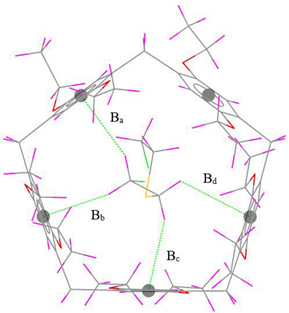 |
| Bb | 2.55 (0.13) | 2.68 | ||
| Bc | 2.56 (0.18) | 2.74 | ||
| Bd | 2.70 (0.26) | 2.96 | ||
| ∆B | 0.58 | - | ||
| C–H···Cl | Be | 3.02 (0.20) | 3.22 | 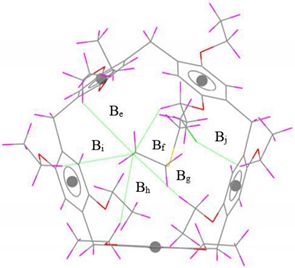 |
| Bf | 3.05 (0.03) | 3.08 | ||
| Bg | 3.35 (0.01) | 3.34 | ||
| Bh | 3.14 (0.06) | 3.20 | ||
| Bi | 3.19 (0.04) | 3.15 | ||
| Bj | 3.34 (0.02) | (3.32) | ||
| ∆B | 0.36 | - | ||
| C–H···S | Bk | 3.24 (0.08) | 3.32 | 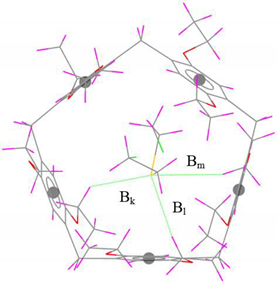 |
| Bl | 3.01 (0.11) | 3.12 | ||
| Bm | 3.18 (0.02) | 3.16 | ||
| ∆B | 0.21 | - |
| Complex | r2SCAN-3c (Gas Phase) | r2SCAN-3c (O-Xylene) | r2SCAN-3c Gas Phase Dispersion Energy | Association Constants (M−1) [39] |
|---|---|---|---|---|
| SM@EtP[5] | −141 | −114.44 | −55.48 | 6.2 × 103 |
| S1@EtP[5] | −122.74 | −105.69 | −51.03 | 2.9 × 102 |
| S2@EtP[5] | −133.76 | −113.01 | −50.31 | 1.3 × 103 |
| S3@EtP[5] | −110.36 | −97.43 | −46.66 | 67 |
| S4@EtP[5] | −144.92 | −118.35 | −53.72 | 1.8 × 104 |
| S5@EtP[5] | −119.69 | −103.09 | −50.09 | 7.9 × 102 |
| Complex | HOMO (eV) | LUMO (eV) | |H-L| gap (eV) | |ΔEg| % | µ (Debye) |
|---|---|---|---|---|---|
| EtP[5] | −4.38 | −0.87 | 3.51 | - | 0.06 |
| SM@EtP[5] | −4.44 | −1.06 | 3.38 | 3.85 | 3.44 |
| S1@EtP[5] | −4.43 | −0.95 | 3.48 | 0.84 | 1.87 |
| S2@EtP[5] | −4.47 | −1.01 | 3.46 | 1.42 | 0.88 |
| S3@EtP[5] | −4.41 | −0.97 | 3.44 | 1.99 | 1.87 |
| S4@EtP[5] | −4.44 | −1.07 | 3.37 | 3.99 | 2.11 |
| S5@EtP[5] | −4.43 | −0.98 | 3.45 | 1.71 | 1.82 |
| Position (Å) | SM@P[5] | SM@MeP[5] | SM@DCMP[5] | SM@P[5]Q | SM@DAP[5] | SM@CB[6] | SM@CX[5] | SM@β-CD |
|---|---|---|---|---|---|---|---|---|
| −10 | −54.25 | −29.41 | −83.32 | −90.51 | −44.74 | −72.12 | −32.35 | −56.82 |
| −8 | −77.19 | −109.80 | −121.57 | −74.96 | −71.12 | −68.88 | −32.77 | −52.10 |
| −6 | −77.28 | −110.00 | −121.66 | −90.53 | −114.52 | −75.41 | −51.72 | −51.61 |
| −4 | −123.65 | −109.99 | −118.70 | −87.29 | −112.35 | −96.42 | - | −54.82 |
| −2 | −126.79 | −110.00 | −120.90 | −85.67 | −116.51 | −96.44 | - | −55.36 |
| 0 | −123.62 | −113.16 | −155.26 | −86.63 | −112.37 | −96.80 | - | −79.49 |
| +2 | −101.89 | −109.90 | −128.54 | −90.23 | −114.22 | −95.76 | - | −89.07 |
| +4 | −101.97 | −109.92 | −118.63 | −90.38 | −114.18 | −96.94 | - | −87.11 |
| +6 | −101.67 | −109.93 | −122.04 | −90.33 | −114.32 | −75.28 | −71.41 | −85.58 |
| +8 | −99.42 | −109.66 | −80.91 | −90.77 | −37.20 | −69.12 | −71.42 | −81.01 |
| +10 | −99.40 | −29.50 | −85.44 | −90.69 | −96.49 | −73.44 | −53.95 | −78.78 |
| Host/Complex | Electronic Chemical Parameters | |||
|---|---|---|---|---|
| EHOMO (eV) | ELUMO (eV) | |ΔE|gap (eV) | ∆Eg % | |
| β-CD | −6.14 | −0.03 | 6.11 | 23.73 |
| SM@β-CD | −5.98 | −1.32 | 4.66 | |
| CB[6] | −5.82 | −0.07 | 5.75 | 30.78 |
| SM@CB[6] | −4.21 | −0.23 | 3.98 | |
| P[5] | −4.54 | −1.49 | 3.05 | 0.00 |
| SM@P[5] | −4.70 | −1.65 | 3.05 | |
| MeP[5] | −4.44 | −0.92 | 3.52 | 6.82 |
| SM@MeP[5] | −4.40 | −1.12 | 3.28 | |
| DCMP[5] | −5.10 | −1.79 | 3.31 | 4.83 |
| SM@DCMP[5] | −5.23 | −1.76 | 3.47 | |
| DAP[5] | −4.56 | −3.01 | 1.55 | 9.03 |
| SM@DAP[5] | −4.56 | −2.87 | 1.69 | |
| CX[5] | −5.44 | −1.26 | 4.18 | 14.35 |
| SM@CX[5] | −5.14 | −1.56 | 3.58 | |
| P[5]Q | −6.68 | −4.73 | 1.95 | 51.28 |
| SM@P[5]Q | −5.88 | −4.93 | 0.95 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messiad, F.A.; Ammouchi, N.; Belhocine, Y.; Alhussain, H.; Ghoniem, M.G.; Said, R.B.; Ali, F.A.M.; Rahali, S. In Search of Preferential Macrocyclic Hosts for Sulfur Mustard Sensing and Recognition: A Computational Investigation through the New Composite Method r2SCAN-3c of the Key Factors Influencing the Host-Guest Interactions. Nanomaterials 2022, 12, 2517. https://doi.org/10.3390/nano12152517
Messiad FA, Ammouchi N, Belhocine Y, Alhussain H, Ghoniem MG, Said RB, Ali FAM, Rahali S. In Search of Preferential Macrocyclic Hosts for Sulfur Mustard Sensing and Recognition: A Computational Investigation through the New Composite Method r2SCAN-3c of the Key Factors Influencing the Host-Guest Interactions. Nanomaterials. 2022; 12(15):2517. https://doi.org/10.3390/nano12152517
Chicago/Turabian StyleMessiad, Fatine Ali, Nesrine Ammouchi, Youghourta Belhocine, Hanan Alhussain, Monira Galal Ghoniem, Ridha Ben Said, Fatima Adam Mohamed Ali, and Seyfeddine Rahali. 2022. "In Search of Preferential Macrocyclic Hosts for Sulfur Mustard Sensing and Recognition: A Computational Investigation through the New Composite Method r2SCAN-3c of the Key Factors Influencing the Host-Guest Interactions" Nanomaterials 12, no. 15: 2517. https://doi.org/10.3390/nano12152517






