One-Pot Synthesis of HRP&SA/ZIF-8 Nanocomposite and Its Application in the Detection of Insecticidal Crystalline Protein Cry1Ab
Abstract
:1. Introduction
2. Material and Methods
2.1. Materials and Reagents
2.2. Instruments
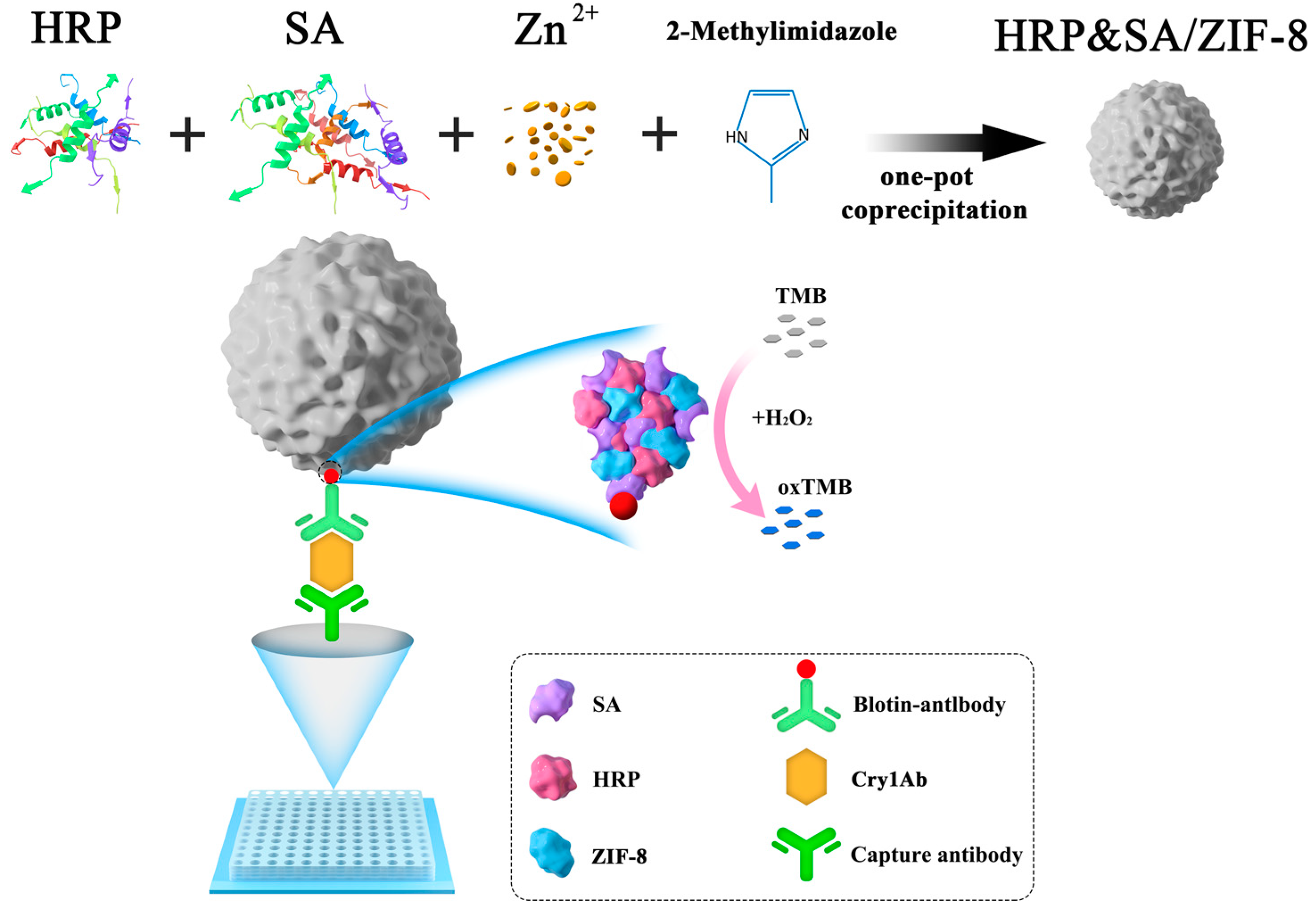
2.3. Synthesis and Characterization of HRP&SA/ZIF-8 Nanocomposite
2.4. Optimization of the Experimental Conditions
2.5. Procedure for Cry1Ab Detection
2.6. Selectivity and Recovery Experiment
3. Results and Discussion
3.1. Principle of Nanocomposites’ Preparation and Detection
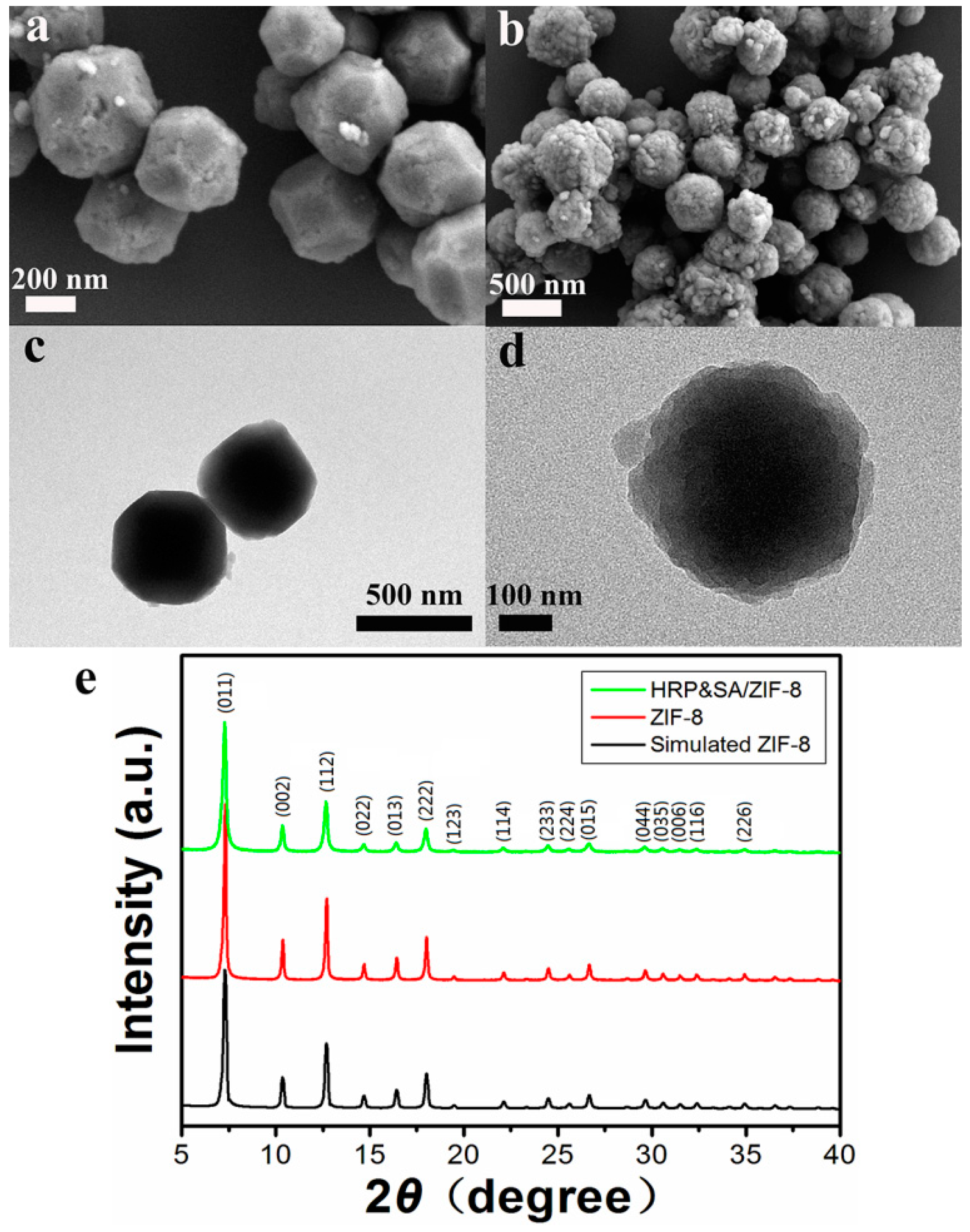
3.2. Characterization of HRP&SA/ZIF-8 Nanocomposites
3.3. Optimization for the Detection
3.4. Detection of Cry1Ab Using HRP&SA/ZIF-8 Nanocomposite
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biggel, M.; Jessberger, N.; Kovac, J.; Johler, S. Recent paradigm shifts in the perception of the role of Bacillus thuringiensis in foodborne disease. Food Microbiol. 2022, 105, 104025. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.B.; Shu, C.L.; Geng, L.L.; Song, F.F.; Zhang, J. Cry78Ba1, One novel crystal protein from Bacillus thuringiensis with high insecticidal activity against rice planthopper. J. Agric. Food Chem. 2020, 68, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Liu, W.; Lu, X.Z.; Chen, W.; Yang, J.B.; Zheng, L. Nondestructive determination of transgenic Bacillus thuringiensis rice seeds (Oryza sativa L.) using multispectral imaging and chemometric methods. Food Chem. 2014, 153, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, M.; Rocca, E.; Bøhn, T.; Wikmark, O.G.; Berg, J.V.D.; Løvik, M.; Traavik, T.; Nygaard, U.C. Humoral and cellular immune responses in mice after airway administration of Bacillus thuringiensis Cry1Ab and MON810 cry1Ab-transgenic maize. Food Agr. Immunol. 2015, 26, 521–537. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, K.R.; Martin, A.; Gowda, L.R.; Prakash, V. Detection of genetically modified soya and maize: Impact of heat processing. Food Chem. 2009, 117, 514–521. [Google Scholar] [CrossRef]
- Tsatsakis, A.M.; Nawaz, M.A.; Kouretas, D.; Balias, G.; Savolainen, K.; Tutelyan, V.A.; Golokhvast, K.S.; Lee, J.D.; Yang, S.H.; Chung, G. Environmental impacts of genetically modified plants: A review. Environ. Res. 2017, 156, 818–833. [Google Scholar] [CrossRef]
- Li, Y.L.; Du, J.; Fang, Z.X.; You, J. Dissipation of insecticidal Cry1Ac protein and its toxicity to nontarget aquatic organisms. J. Agric. Food Chem. 2013, 61, 10864–10871. [Google Scholar] [CrossRef] [PubMed]
- Permingeat, H.R.; Reggiardo, M.I.; Vallejos, R.H. Detection and quantification of transgenes in grains by multiplex and real-time PCR. J. Agric. Food Chem. 2002, 50, 4431–4436. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.; Jiang, S.M.; Zhang, M.; Wang, J.; Guo, S.J.; Li, Y.; Jiang, H.W.; Liu, C.X.; Zhang, D.B.; Yang, L.T.; et al. MACRO: A combined microchip-PCR and microarray system for high-throughput monitoring of genetically modified organisms. Anal. Chem. 2014, 86, 1269–1276. [Google Scholar]
- Albright, V.C.; Hellmich, R.L.; Coats, J.R. A review of Cry protein detection with enzyme-linked immunosorbent assays. J. Agric. Food Chem. 2016, 64, 2175–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.J.; Wang, J.B.; Jia, J.W.; Wu, G.G.; Yang, Q.W.; Liu, X.F.; Tang, X.M. Development of a lateral flow test strip for simultaneous detection of BT-Cry1Ab, BT-Cry1Ac and CP4 EPSPS proteins in genetically modified crops. Food Chem. 2021, 335, 127627. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Yan, H.Y.; Xu, W.Q.; Wu, Y.; Gu, W.L.; Li, H.; Du, D.; Lin, Y.H.; Zhu, C.Z. Self-assembly of all-inclusive allochroic nanoparticles for the improved ELISA. Anal. Chem. 2019, 91, 8461–8465. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hao, Y.Q.; Deng, D.H.; Xia, N. Nanomaterials-based colorimetric immunoassays. Nanomaterials 2019, 9, 316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenzl, C.; Hirsch, T.; Baeumner, A.J. Nanomaterials as versatile tools for signal amplification in (bio)analytical applications. Trac. Trends Anal. Chem. 2016, 79, 306–316. [Google Scholar] [CrossRef]
- Lei, J.P.; Ju, H.X. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Pelt, S.V. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [Green Version]
- Ye, R.F.; Xu, H.W.; Gu, J.J.; Chen, H. Bioinspired synthesis of protein-posnjakite organic-inorganic nanobiohybrid for biosensing applications. Anal. Chim. Acta 2021, 1143, 31–36. [Google Scholar] [CrossRef]
- Chen, G.S.; Huang, S.M.; Kou, X.X.; Wei, S.B.; Huang, S.Y.; Jiang, S.Q.; Shen, J.; Zhu, F.; Ouyang, G.F.A. Convenient and versatile amino-acid-boosted biomimetic strategy for the non-destructive encapsulation of biomacromolecules within metal-organic frameworks. Angew. Chem. Int. Ed. 2019, 58, 1463–1467. [Google Scholar] [CrossRef]
- Poddar, A.; Conesa, J.J.; Liang, K.; Dhakal, S.; Reineck, P.; Bryant, G.; Pereiro, E.; Ricco, R.; Amenitsch, H.; Doonan, C.; et al. Encapsulation, visualization and expression of genes with biomimetically mineralized zeolitic imidazolate framework-8 (ZIF-8). Small 2019, 15, 1902268. [Google Scholar] [CrossRef] [Green Version]
- Troyano, J.; Carné-Sánchez, A.; Avci, C.; Imaz, I.; Maspoch, D. Colloidal metal–organic framework particles: The pioneering case of ZIF-8. Chem. Soc. Rev. 2019, 48, 5534–5546. [Google Scholar] [CrossRef]
- Carraro, F.; Williams, J.D.; Linares-Moreau, M.; Parise, C.; Liang, W.; Amenitsch, H.; Doonan, C.; Kappe, C.O.; Falcaro, P. Continuous-Flow Synthesis of ZIF-8 Biocomposites with Tunable Particle Size. Angew. Chem. Int. Ed. 2020, 132, 8200–8204. [Google Scholar] [CrossRef] [Green Version]
- Lyu, F.J.; Zhang, Y.F.; Zare, R.N.; Ge, J.; Liu, Z. One-pot synthesis of protein-embedded metal-organic frameworks with enhanced biological activities. Nano Lett. 2014, 14, 5761–5765. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Q.; Zhang, X.P.; Huang, L.; Zhang, Z.Q.; Dong, S.J. GOx@ZIF-8(NiPd) Nanoflower: An artificial enzyme system for tandem catalysis. Angew. Chem. Int. Ed. 2017, 129, 16298–16301. [Google Scholar] [CrossRef]
- Ricco, R.; Wied, P.; Nidetzky, B.; Amenitsch, H.; Falcaro, P. Carbonic magnetically responsive horseradish peroxidase@ZIF-8 for biocatalysis. Chem. Commun. 2020, 56, 5775–5778. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhi, W.J.; Lian, D.S.; Wang, Y.; Han, J.; Wang, Y. HRP@ZIF-8/DNA hybrids: Functionality integration of ZIF-8 via biomineralization and surface absorption. ACS Sustain. Chem. Eng. 2019, 7, 14611–14620. [Google Scholar] [CrossRef]
- Xiong, J.; Li, Z.; Tan, J.; Ji, S.; Sun, J.; Li, X.; Huo, Y. Two new quinoline-based regenerable fluorescent probes with AIE characteristics for selective recognition of Cu2+ in aqueous solution and test strips. Analyst 2018, 143, 4870–4886. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Li, M.; Li, G.H.; Zhou, Z.K.; Liu, H.; Lei, H.T.; Shen, Y.F.; Wan, Y.K. Nanobody-based electrochemical immunoassay for Bacillus thuringiensis Cry1Ab toxin by detecting the enzymatic formation of polyaniline. Microchim. Acta 2015, 182, 2451–2459. [Google Scholar] [CrossRef]
- Dong, S.; Liu, Y.; Zhang, X.; Xu, C.X.; Liu, X.J.; Zhang, C.Z. Development of an immunochromatographic assay for the specific detection of Bacillus thuringiensis (Bt) Cry1Ab toxin. Anal. Biochem. 2019, 567, 1–7. [Google Scholar] [CrossRef]
- Qiu, Y.L.; Li, P.; Dong, S.; Zhang, X.S.; Yang, Q.R.; Wang, Y.L.; Ge, J.; Hammock, B.D.; Zhang, C.Z.; Liu, X.J. Phage-mediated competitive chemiluminescent immunoassay for detecting Cry1Ab Toxin by Using an anti-Idiotypic camel nanobody. J. Agric. Food Chem. 2018, 66, 950–956. [Google Scholar] [CrossRef]
- Gao, H.F.; Wen, L.K.; Wu, Y.H.; Fu, Z.F.; Wu, G. An ultrasensitive label-free electrochemiluminescent immunosensor for measuring CrylAb level and genetically modified crops content. Biosens. Bioelectron. 2017, 97, 122–127. [Google Scholar] [CrossRef]
- Freitas, M.; Correr, W.; Cancino-Bernardi, J.; Barroso, M.F.; Delerue-Matos, C.; Zucolotto, V. Impedimetric immunosensors for the detection of Cry1Ab protein from genetically modified maize seeds. Sens. Actuat. B-Chem. 2016, 237, 702–709. [Google Scholar] [CrossRef]


| Methods | Linear Range ng/mL | LOD ng/mL | Reference |
|---|---|---|---|
| Electrochemical immunoassay | 0–1000 | 0.07 | [27] |
| Immunochromatographic assay | - | 100 | [28] |
| Chemiluminescent immunoassay | 10.49–307.1 | 6.45 | [29] |
| Electrochemiluminescent immunoassay | 0.010–1.0 | 0.03 | [30] |
| Impedimetric immunoassay | 1–10 | 0.37 | [31] |
| This work | 0.05–16 | 0.0048 | This work |
| Cry1Ab Added (ng/mL) | Cry1Ab Found (ng/mL) | Recovery (%) |
|---|---|---|
| 0 | not been found | - |
| 1 | 0.92 ± 0.05 | 92.0 ± 5.0 |
| 4 | 4.17 ± 0.38 | 104.3 ± 9.5 |
| 8 | 7.46 ± 0.61 | 93.4 ± 7.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, R.; Chen, H.; Li, H. One-Pot Synthesis of HRP&SA/ZIF-8 Nanocomposite and Its Application in the Detection of Insecticidal Crystalline Protein Cry1Ab. Nanomaterials 2022, 12, 2679. https://doi.org/10.3390/nano12152679
Ye R, Chen H, Li H. One-Pot Synthesis of HRP&SA/ZIF-8 Nanocomposite and Its Application in the Detection of Insecticidal Crystalline Protein Cry1Ab. Nanomaterials. 2022; 12(15):2679. https://doi.org/10.3390/nano12152679
Chicago/Turabian StyleYe, Ranfeng, Hao Chen, and He Li. 2022. "One-Pot Synthesis of HRP&SA/ZIF-8 Nanocomposite and Its Application in the Detection of Insecticidal Crystalline Protein Cry1Ab" Nanomaterials 12, no. 15: 2679. https://doi.org/10.3390/nano12152679





