pH-Driven Intracellular Nano-to-Molecular Disassembly of Heterometallic [Au2L2]{Re6Q8} Colloids (L = PNNP Ligand; Q = S2− or Se2−)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Methods
2.2.1. Dynamic Light Scattering
2.2.2. UV-Vis Absorption Spectra
2.2.3. Fluorescence Spectroscopy
2.2.4. IR Spectroscopy
2.2.5. ICP-OES
2.2.6. TEM Measurements
2.2.7. Powder X-ray Diffraction (PXRD)
2.2.8. Confocal Laser Microscopy
2.2.9. Statistical Analyses
2.2.10. Cytotoxicity Assay
2.2.11. Cellular Uptake Study
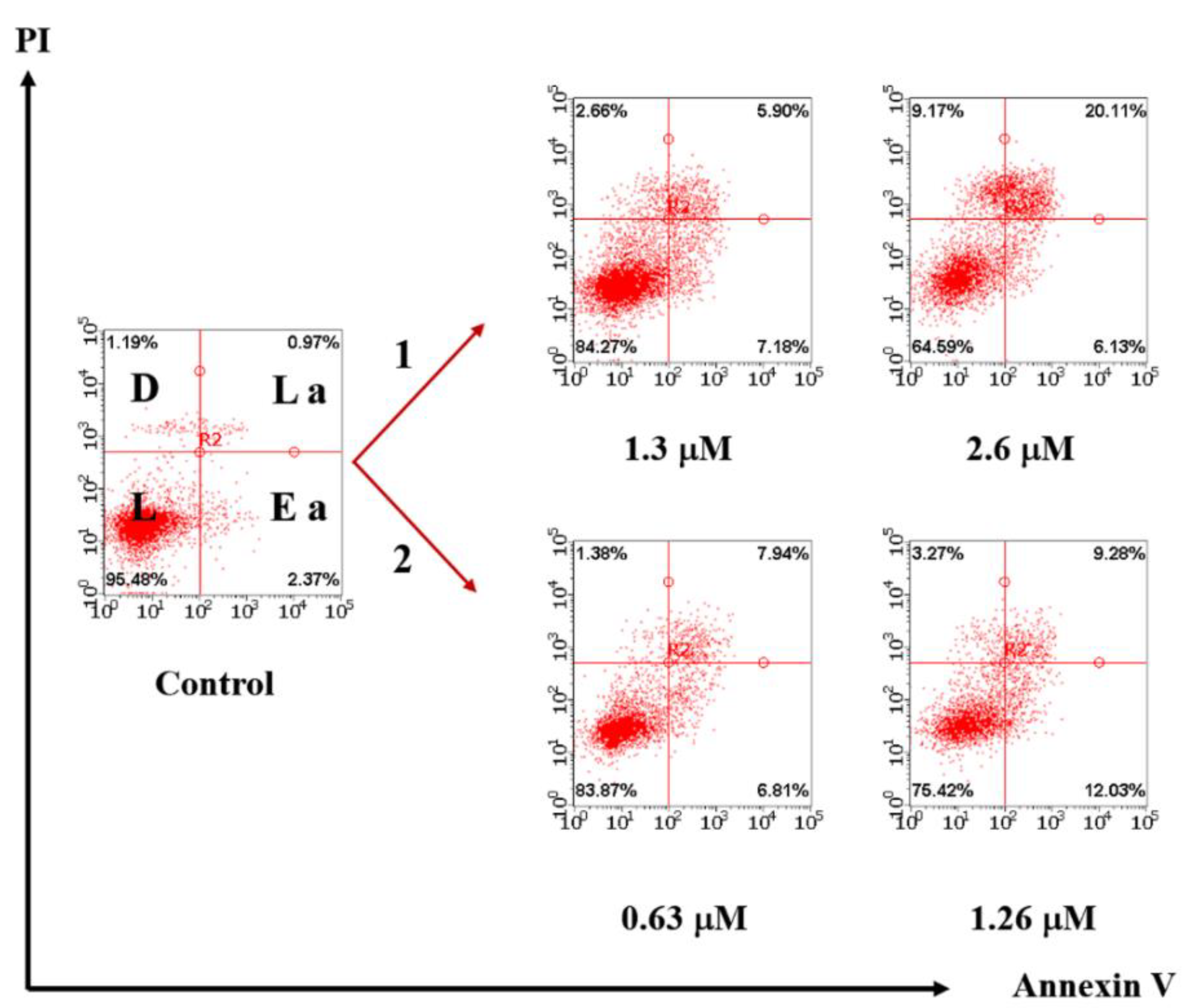
2.2.12. Cell Apoptosis Analysis
3. Results and Discussion
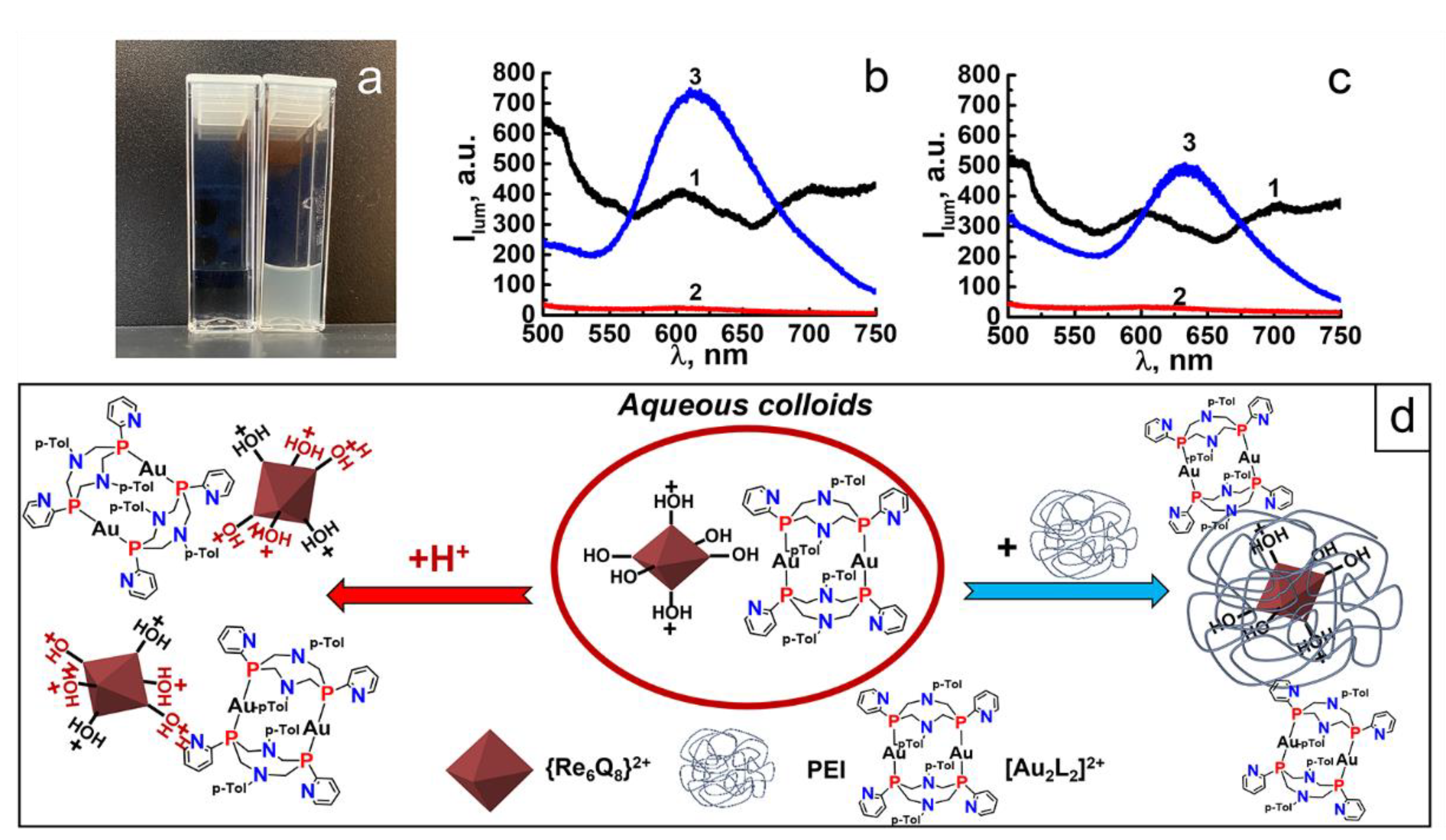
3.1. Synthesis and Characterization of Au2Re6-Q Colloids
3.2. pH-Dependent Leaching of Au2 and Re6-Q Units
3.3. PEI-Induced Disassembly of Au2Re6-Q Colloids and Their Surface Decoration by LSZ
3.4. Cytotoxicity and Cell Internalization of Au2Re6-Q and LSZ-Au2Re6-Q Colloids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gwili, N.; Jones, S.J.; Amri, W.A.; Carr, I.M.; Harris, S.; Hogan, B.V.; Hughes, W.E.; Kim, B.; Langlands, F.E.; Millican-Slater, R.A.; et al. Transcriptome profiles of stem-like cells from primary breast cancers allow identification of ITGA7 as a predictive marker of chemotherapy response. Br. J. Cancer 2021, 125, 983–993. [Google Scholar] [CrossRef]
- Hutchinson, S.A.; Websdale, A.; Cioccoloni, G.; Røberg-Larsen, H.; Lianto, P.; Kim, B.; Rose, A.; Soteriou, C.; Pramanik, A.; Wastall, L.M.; et al. Liver x receptor alpha drives chemoresistance in response to side-chain hydroxycholesterols in triple negative breast cancer. Oncogene 2021, 40, 2872–2883. [Google Scholar] [CrossRef]
- Artzi, N. Trojan siRNA Opens the Door to Chemotherapy. Sci. Transl. Med. 2013, 5, ec186–ec211. [Google Scholar] [CrossRef]
- Teng, L.; Song, G.; Liu, Y.; Han, X.; Li, Z.; Wang, Y.; Huan, S.; Zhang, X.-B.; Tan, W. Nitric oxide-activated “Dual-Key–One-Lock” nanoprobe for in vivo molecular imaging and high-specificity cancer therapy. J. Am. Chem. Soc. 2019, 141, 13572–13581. [Google Scholar] [CrossRef]
- Degors, I.M.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers break barriers in drug delivery: Endocytosis and endosomal escape of gene delivery vectors. Acc. Chem. Res. 2019, 52, 1750–1760. [Google Scholar] [CrossRef]
- Saftig, P.; Sandhoff, K. Killing from the inside. Nature 2013, 502, 312–313. [Google Scholar] [CrossRef]
- Hu, Y.; Litwin, T.; Nagaraja, A.R.; Kwong, B.; Katz, J.; Watson, N.; Irvine, D.J. Cytosolic delivery of membrane-impermeable molecules in dendritic cells using pH-responsive core− shell nanoparticles. Nano Lett. 2007, 7, 3056–3064. [Google Scholar] [CrossRef]
- Siek, M.; Kandere-Grzybowska, K.; Grzybowski, B.A. Mixed-Charge, pH-Responsive Nanoparticles for Selective Interactions with Cells, Organelles, and Bacteria. Acc. Mater. Res. 2020, 1, 188–200. [Google Scholar] [CrossRef]
- Manshian, B.B.; Soenen, S.J.; Brown, A.; Hondow, N.; Wills, J.; Jenkins, G.J.; Doak, S.H. Genotoxic capacity of Cd/Se semiconductor quantum dots with differing surface chemistries. Mutagenesis 2016, 31, 97–106. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, H.R.; Dong, L.; Xu, M.R.; Zhang, L.; Ding, W.P.; Zhang, J.-Q.; Lin, J.; Zhang, Y.-J.; Qiu, B.-S.; et al. Enhancing tumor chemotherapy and overcoming drug resistance through autophagy-mediated intracellular dissolution of zinc oxide nanoparticles. Nanoscale 2019, 11, 11789–11807. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.X. Uptake, intracellular dissolution, and cytotoxicity of silver nanowires in cell models. Chemosphere 2021, 281, 130762. [Google Scholar] [CrossRef]
- Abdelkhaliq, A.; van der Zande, M.; Undas, A.K.; Peters, R.J.; Bouwmeester, H. Impact of in vitro digestion on gastrointestinal fate and uptake of silver nanoparticles with different surface modifications. Nanotoxicology 2020, 14, 111–126. [Google Scholar] [CrossRef]
- De Jong, W.H.; De Rijk, E.; Bonetto, A.; Wohlleben, W.; Stone, V.; Brunelli, A.; Badetti, E.; Marcomini, A.; Gosens, I.; Cassee, F.R. Toxicity of copper oxide and basic copper carbonate nanoparticles after short-term oral exposure in rats. Nanotoxicology 2019, 13, 50–72. [Google Scholar] [CrossRef]
- Latvala, S.; Hedberg, J.; Di Bucchianico, S.; Möller, L.; Odnevall Wallinder, I.; Elihn, K.; Karlsson, H.L. Nickel release, ROS generation and toxicity of Ni and NiO micro-and nanoparticles. PLoS ONE 2016, 11, e0159684. [Google Scholar] [CrossRef]
- Zhu, J.; He, K.; Dai, Z.; Gong, L.; Zhou, T.; Liang, H.; Liu, J. Self-assembly of luminescent gold nanoparticles with sensitive pH-stimulated structure transformation and emission response toward lysosome escape and intracellular imaging. Anal. Chem. 2019, 91, 8237–8243. [Google Scholar] [CrossRef]
- Naatz, H.; Manshian, B.B.; Rios Luci, C.; Tsikourkitoudi, V.; Deligiannakis, Y.; Birkenstock, J.; Pokhrel, S.; Madler, L.; Soenen, S.S. Model-based nanoengineered pharmacokinetics of iron-doped copper oxide for nanomedical applications. Angew. Chem. Int. Ed. 2020, 59, 1828–1836. [Google Scholar] [CrossRef]
- Sousa, C.A.; Soares, H.M.; Soares, E.V. Nickel oxide (NiO) nanoparticles disturb physiology and induce cell death in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2018, 102, 2827–2838. [Google Scholar] [CrossRef]
- Cai, X.; Liu, X.; Jiang, J.; Gao, M.; Wang, W.; Zheng, H.; Xu, S.; Li, R. Molecular mechanisms, characterization methods, and utilities of nanoparticle biotransformation in nanosafety assessments. Small 2020, 16, 1907663. [Google Scholar] [CrossRef]
- Xiong, Y.; Xiao, C.; Li, Z.; Yang, X. Engineering nanomedicine for glutathione depletion-augmented cancer therapy. Chem. Soc. Rev. 2021, 50, 6013–6041. [Google Scholar] [CrossRef]
- Zhang, K.; Meng, X.; Yang, Z.; Dong, H.; Zhang, X. Enhanced cancer therapy by hypoxia-responsive copper metal-organic frameworks nanosystem. Biomaterials 2020, 258, 120278. [Google Scholar] [CrossRef]
- Zhao, R.M.; Guo, Y.; Yang, H.Z.; Zhang, J.; Yu, X.Q. Zn-Promoted gene transfection efficiency for non-viral vectors: A mechanism study. New J. Chem. 2021, 45, 13549–13557. [Google Scholar] [CrossRef]
- Lin, G.; Zhang, Y.; Zhang, L.; Wang, J.; Tian, Y.; Cai, W.; Tang, S.; Chu, C.; Zhou, J.; Mi, P.; et al. Metal-organic frameworks nanoswitch: Toward photo-controllable endo/lysosomal rupture and release for enhanced cancer RNA interference. Nano Res. 2020, 13, 238–245. [Google Scholar] [CrossRef]
- Tsai, J.L.L.; Zou, T.; Liu, J.; Chen, T.; Chan, A.O.Y.; Yang, C.; Lok, C.N.; Che, C.M. Luminescent platinum (II) complexes with self-assembly and anti-cancer properties: Hydrogel, pH dependent emission color and sustained-release properties under physiological conditions. Chem. Sci. 2015, 6, 3823–3830. [Google Scholar] [CrossRef]
- Pramanik, A.; Xu, Z.; Shamsuddin, S.H.; Khaled, Y.S.; Ingram, N.; Maisey, T.; Tomlinson, D.; Coletta, P.L.; Jayne, D.; Hughes, T.A.; et al. Affimer Tagged Cubosomes: Targeting of Carcinoembryonic Antigen Expressing Colorectal Cancer Cells Using In Vitro and In Vivo Models. ACS Appl. Mater. Interfaces 2022, 14, 11078–11091. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, R.; Liu, J.; Kong, H.; Zhang, K.; Zhang, Y.N.; Kong, X.; Zhang, Q.; Zhao, Y. Hierarchical nano-to-molecular disassembly of boron dipyrromethene nanoparticles for enhanced tumor penetration and activatable photodynamic therapy. Biomaterials 2021, 275, 120945. [Google Scholar] [CrossRef]
- Kongkatigumjorn, N.; Smith, S.A.; Chen, M.; Fang, K.; Yang, S.; Gillies, E.R.; Johnston, A.P.R.; Such, G.K. Controlling endosomal escape using pH-responsive nanoparticles with tunable disassembly. ACS Appl. Nano Mater. 2018, 1, 3164–3173. [Google Scholar] [CrossRef]
- Zhang, M.; Saint-Germain, C.; He, G.; Sun, R.W.Y. Drug delivery systems for anti-cancer active complexes of some coinage metals. Curr. Med. Chem. 2018, 25, 493–505. [Google Scholar] [CrossRef]
- Gou, Y.; Huang, G.; Li, J.; Yang, F.; Liang, H. Versatile delivery systems for non-platinum metal-based anticancer therapeutic agents. Coord. Chem. Rev. 2021, 441, 213975. [Google Scholar] [CrossRef]
- Rousselle, B.; Bouyer, F.; Bayardon, J.; Laly, M.; Ghiringhelli, F.; Rousselin, Y.; Bodio, E.; Malacea-Kabbara, R. Development of a novel highly anti-proliferative family of gold complexes: Au (i)-phosphonium-phosphines. Dalton Trans. 2021, 50, 4880–4889. [Google Scholar] [CrossRef]
- Rouco, L.; Sánchez-González, Á.; Alvariño, R.; Alfonso, A.; Vázquez-López, E.M.; García-Martínez, E.; Maneiro, M. Combined effect of caspase-dependent and caspase-independent apoptosis in the anticancer activity of gold complexes with phosphine and benzimidazole derivatives. Pharmaceuticals 2020, 14, 10. [Google Scholar] [CrossRef]
- Kim, J.H.; Reeder, E.; Parkin, S.; Awuah, S.G. Gold (I/III)-phosphine complexes as potent antiproliferative agents. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Quero, J.; Ruighi, F.; Osada, J.; Gimeno, M.C.; Cerrada, E.; Rodriguez-Yoldi, M.J. Gold (I) Complexes Bearing Alkylated 1, 3, 5-Triaza-7-phosphaadamantane Ligands as Thermoresponsive Anticancer Agents in Human Colon Cells. Biomedicines 2021, 9, 1848. [Google Scholar] [CrossRef]
- Mirzadeh, N.; Reddy, T.S.; Bhargava, S.K. Advances in diphosphine ligand-containing gold complexes as anticancer agents. Coord. Chem. Rev. 2019, 388, 343–359. [Google Scholar] [CrossRef]
- Barnard, P.J.; Berners-Price, S.J. Targeting the mitochondrial cell death pathway with gold compounds. Coord. Chem. Rev. 2007, 251, 1889–1902. [Google Scholar] [CrossRef]
- Faizullin, B.A.; Strelnik, I.D.; Dayanova, I.R.; Gerasimova, T.P.; Kholin, K.V.; Nizameev, I.R.; Voloshina, A.D.; Gubaidullin, A.T.; Fedosimova, S.V.; Mikhailov, M.A.; et al. Structure impact on photodynamic therapy and cellular contrasting functions of colloids constructed from dimeric Au (I) complex and hexamolybdenum clusters. Mat. Sci. Eng. C 2021, 128, 112355. [Google Scholar] [CrossRef]
- Brylev, K.A.; Mironov, Y.V.; Yarovoi, S.S.; Naumov, N.G.; Fedorov, V.E.; Kim, S.-J.; Kitamura, N.; Kuwahara, Y.; Yamada, K.; Ishizaka, S.; et al. A Family of Octahedral Rhenium Cluster Complexes [Re6Q8(H2O)n(OH)6−n]n−4 (Q = S, Se; n = 0–6): Structural and pH−Dependent Spectroscopic Studies. Inorg. Chem. 2007, 46, 7414–7422. [Google Scholar] [CrossRef]
- Elistratova, J.; Strelnik, I.; Brylev, K.; Shestopalov, M.A.; Gerasimova, T.; Babaev, V.; Kholin, K.; Dobrynin, A.; Musina, E.; Katsyuba, S.; et al. Novel Water Soluble Cationic Au (I) Complexes with Cyclic PNNP Ligand as Building Blocks for Heterometallic Supramolecular Assemblies with Anionic Hexarhenium Cluster Units. J. Lumin. 2018, 196, 485–491. [Google Scholar] [CrossRef]
- Yarovoi, S.S.; Mironov, Y.V.; Naumov, D.Y.; Gatilov, Y.V.; Kozlova, S.G.; Kim, S.-J.; Fedorov, V.E. Octahedral Hexahydroxo Rhenium Cluster Complexes [Re6Q8(OH)6]4–·(Q = S, Se): Synthesis, Structure, and Properties. Eur. J. Inorg. Chem. 2005, 2005, 3945–3949. [Google Scholar] [CrossRef]
- Musina, E.I.; Khrizanforova, V.V.; Strelnik, I.D.; Valitov, M.I.; Spiridonova, Y.S.; Krivolapov, D.B.; Litvinov, I.A.; Kadirov, M.K.; Lönnecke, P.; Hey-Hawkins, E.; et al. New functional cyclic aminomethylphosphine ligands for the construction of catalysts for electrochemical hydrogen transformations. Chem. Eur. J. 2014, 20, 3169–3182. [Google Scholar] [CrossRef]
- Delgado, A.V.; Gonzalez-Caballero, F.; Hunter, R.J.; Koopal, L.K.; Lyklema, J. Measurement and interpretation of electrokinetic phenomena. J. Colloid Interface Sci. 2007, 309, 194–224. [Google Scholar] [CrossRef]
- DIFFRAC Plus Evaluation Package EVA, Version 11; User’s Manual; Bruker AXS: Karlsruhe, Germany, 2005; 258p.
- TOPAS V3: General Profile and Structure Analysis Software for Powder Diffraction Data; Technical Reference; Bruker AXS: Karlsruhe, Germany, 2005; 117p.
- Voloshina, A.D.; Sapunova, A.S.; Kulik, N.V.; Belenok, M.G.; Strobykina, I.Y.; Lyubina, A.; Gumerova, S.K.; Kataev, V.E. Antimicrobial and cytotoxic effects of ammonium derivatives of diterpenoids steviol and isosteviol. Bioorg. Med. Chem. 2021, 32, 115974. [Google Scholar] [CrossRef]
- Quest Graph™ IC50 Calculator, AAT Bioquest, Inc. Available online: https://www.aatbio.com/tools/ic50-calculator (accessed on 17 May 2022).
- Kashapov, R.; Razuvayeva, Y.; Ziganshina, A.; Sapunova, A.; Lyubina, A.; Amerhanova, S.; Kulik, N.; Voloshina, A.; Nizameev, I.; Salnikov, V.; et al. Effect of preorganization and amphiphilicity of calix[4]arene platform on functional properties of viologen derivatives. J. Mol. Liq. 2022, 345, 117801. [Google Scholar] [CrossRef]
- Strobykina, I.Y.; Voloshina, A.D.; Andreeva, O.V.; Sapunova, A.S.; Lyubina, A.P.; Amerhanova, S.K.; Belenok, M.G.; Saifina, L.F.; Semenov, V.E.; Kataev, V.E. Synthesis, antimicrobial activity and cytotoxicity of triphenylphosphonium (TPP) conjugates of 1,2,3-triazolyl nucleoside analogues. Bioorg. Chem. 2021, 116, 105328. [Google Scholar] [CrossRef]
- Elistratova, J.G.; Brylev, K.A.; Solovieva, A.O.; Pozmogova, T.N.; Mustafina, A.R.; Shestopalova, L.V.; Shestopalov, M.A.; Syakayev, V.V.; Karasik, A.A.; Sinyashin, G. Supporting effect of polyethylenimine on hexarhenium hydroxo cluster complex for cellular imaging applications. J. Photochem. Photobiol. A 2017, 340, 46–52. [Google Scholar] [CrossRef]
- Khazieva, A.; Kholin, K.; Nizameev, I.; Brylev, K.; Kashnik, I.; Voloshina, A.; Lyubina, A.; Gubaidullin, A.; Daminova, A.; Petrov, K.; et al. Surface modification of silica nanoparticles by hexarhenium anionic cluster complexes for pH-sensing and staining of cell nuclei. J. Colloid Interface Sci. 2021, 594, 759–769. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Yang, Y.; Sun, B.; Wang, K.; Zhu, L. Impacts of proteins on dissolution and sulfidation of silver nanowires in an aquatic environment: Importance of surface charges. Environ. Sci. Technol. 2020, 54, 5560–5568. [Google Scholar] [CrossRef]
- Elistratova, J.; Faizullin, B.; Strelnik, I.; Gerasimova, T.; Khairullin, R.; Sapunova, A.; Voloshina, A.; Mukhametzyanov, T.; Musina, E.; Karasik, A.; et al. Impact of oppositely charged shell and cores on interaction of core-shell colloids with differently charged proteins as a route for tuning of the colloids cytotoxicity. Colloids Surf. B 2020, 196, 111306. [Google Scholar] [CrossRef]
- Ding, L.; Yao, C.; Yin, X.; Li, C.; Huang, Y.; Wu, M.; Wang, B.; Guo, X.; Wang, Y.; Wu, M. Size, shape, and protein corona determine cellular uptake and removal mechanisms of gold nanoparticles. Small 2018, 14, 1801451. [Google Scholar] [CrossRef]
- Foroozandeh, P.; Aziz, A.A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 2018, 13, 1–12. [Google Scholar] [CrossRef]
- Donahue, N.D.; Acar, H.; Wilhelm, S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Delivery Rev. 2019, 143, 68–96. [Google Scholar] [CrossRef]
- Manzanares, D.; Ceña, V. Endocytosis: The nanoparticle and submicron nanocompounds gateway into the cell. Pharmaceutics 2020, 12, 371. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Bilardo, R.; Traldi, F.; Vdovchenko, A.; Resmini, M. Influence of surface chemistry and morphology of nanoparticles on protein corona formation. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, 14, e1788. [Google Scholar] [CrossRef]
- Xiao, Q.; Zoulikha, M.; Qiu, M.; Teng, C.; Lin, C.; Li, X.; Sallam, M.A.; Xu, Q.; He, W. The effects of protein corona on in vivo fate of nanocarriers. Adv. Drug Delivery Rev. 2022, 186, 114356. [Google Scholar] [CrossRef]
- Ferreira, L.F.; Picco, A.S.; Galdino, F.E.; Albuquerque, L.J.C.; Berret, J.F.; Cardoso, M.B. Nanoparticle-Protein Interaction: Demystifying the Correlation Between Protein Corona and Aggregation Phenomena. ACS Appl. Mater. Interfaces 2022, 14, 28559–28569. [Google Scholar] [CrossRef]




| dnum, nm | PDI | ζ, mV | |
| Au2Re6-S | 498–520 | ~1 | –10.8 ± 0.6 |
| Au2Re6-Se | 400–460 | 0.846 | –17.1 ± 0.8 |
| Phosphate buffer solutions (pH = 7): | |||
| Au2Re6-S | 630–1100 | ~1 | –5.0 ± 0.5 |
| LSZ-Au2Re6-S | 620–1040 | ~1 | –6.0 ± 0.6 |
| Au2Re6-Se | 340–530 | 0.9 | –6.9 ± 0.6 |
| LSZ-Au2-Re6-Se | 460–530 | 0.9 | –4.2 ± 0.7 |
| IC50, μM | ||
|---|---|---|
| M-HeLa | Chang Liver | |
| [Au2L2]Cl2 | 2.0 * | 3.0 * |
| Au2Re6-S | 2.24 ± 0.17 ** | 1.85 ± 0.20 ** |
| LSZ-Au2Re6-S | 1.20 ± 0.10 ** | 2.59 ± 0.18 ** |
| Au2Re6-Se | 1.70 ± 0.13 ** | 1.70 ± 0.11 ** |
| LSZ-Au2Re6-Se | 3.40 ± 0.24 ** | 2.90 ± 0.17 ** |
| LSZ | >30 | >30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faizullin, B.; Dayanova, I.; Strelnik, I.; Kholin, K.; Nizameev, I.; Gubaidullin, A.; Voloshina, A.; Gerasimova, T.; Kashnik, I.; Brylev, K.; et al. pH-Driven Intracellular Nano-to-Molecular Disassembly of Heterometallic [Au2L2]{Re6Q8} Colloids (L = PNNP Ligand; Q = S2− or Se2−). Nanomaterials 2022, 12, 3229. https://doi.org/10.3390/nano12183229
Faizullin B, Dayanova I, Strelnik I, Kholin K, Nizameev I, Gubaidullin A, Voloshina A, Gerasimova T, Kashnik I, Brylev K, et al. pH-Driven Intracellular Nano-to-Molecular Disassembly of Heterometallic [Au2L2]{Re6Q8} Colloids (L = PNNP Ligand; Q = S2− or Se2−). Nanomaterials. 2022; 12(18):3229. https://doi.org/10.3390/nano12183229
Chicago/Turabian StyleFaizullin, Bulat, Irina Dayanova, Igor Strelnik, Kirill Kholin, Irek Nizameev, Aidar Gubaidullin, Alexandra Voloshina, Tatiana Gerasimova, Ilya Kashnik, Konstantin Brylev, and et al. 2022. "pH-Driven Intracellular Nano-to-Molecular Disassembly of Heterometallic [Au2L2]{Re6Q8} Colloids (L = PNNP Ligand; Q = S2− or Se2−)" Nanomaterials 12, no. 18: 3229. https://doi.org/10.3390/nano12183229
APA StyleFaizullin, B., Dayanova, I., Strelnik, I., Kholin, K., Nizameev, I., Gubaidullin, A., Voloshina, A., Gerasimova, T., Kashnik, I., Brylev, K., Sibgatullina, G., Samigullin, D., Petrov, K., Musina, E., Karasik, A., & Mustafina, A. (2022). pH-Driven Intracellular Nano-to-Molecular Disassembly of Heterometallic [Au2L2]{Re6Q8} Colloids (L = PNNP Ligand; Q = S2− or Se2−). Nanomaterials, 12(18), 3229. https://doi.org/10.3390/nano12183229








