Direct Bactericidal Comparison of Metal Nanoparticles and Their Salts against S. aureus Culture by TEM and FT-IR Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
3. Results
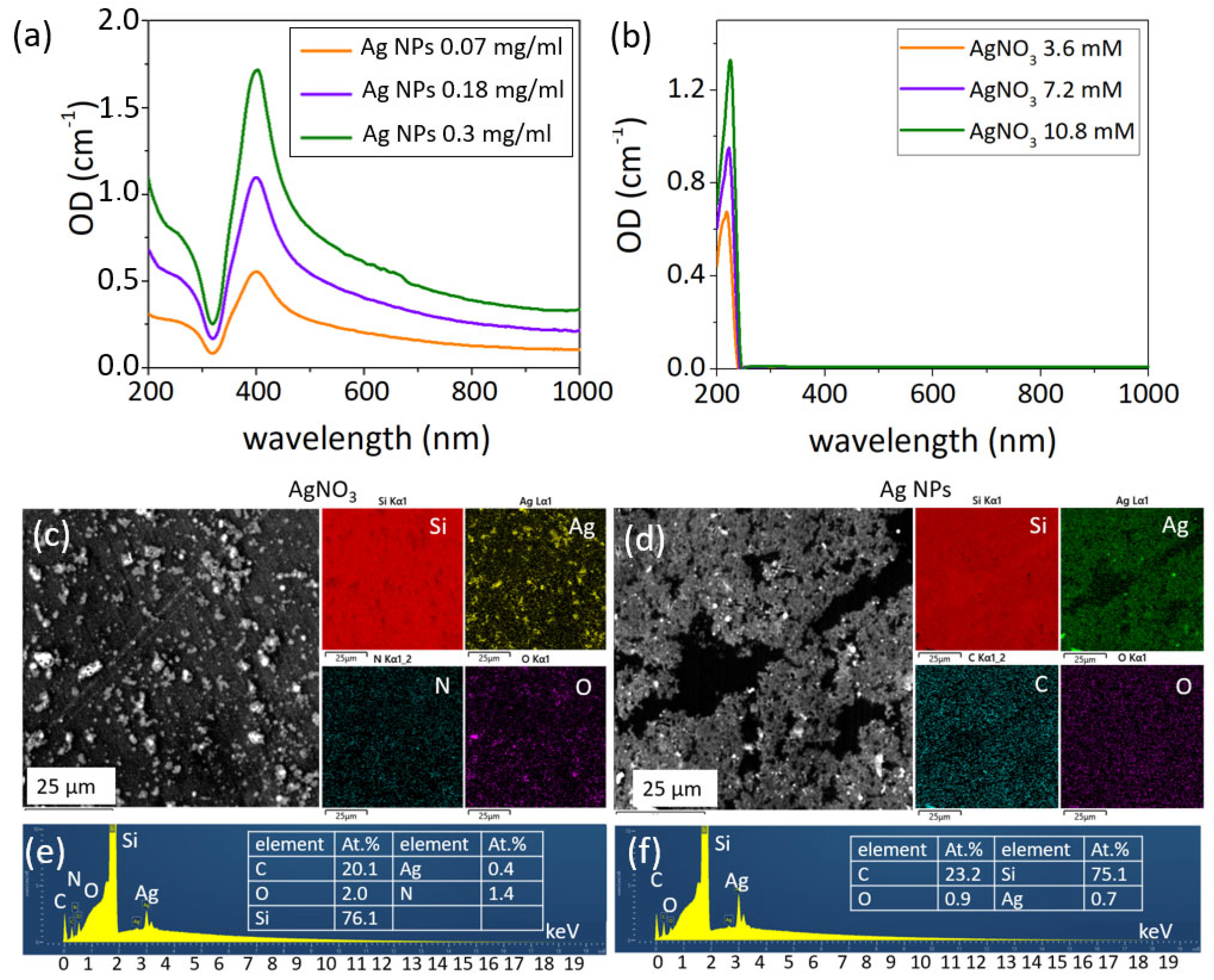
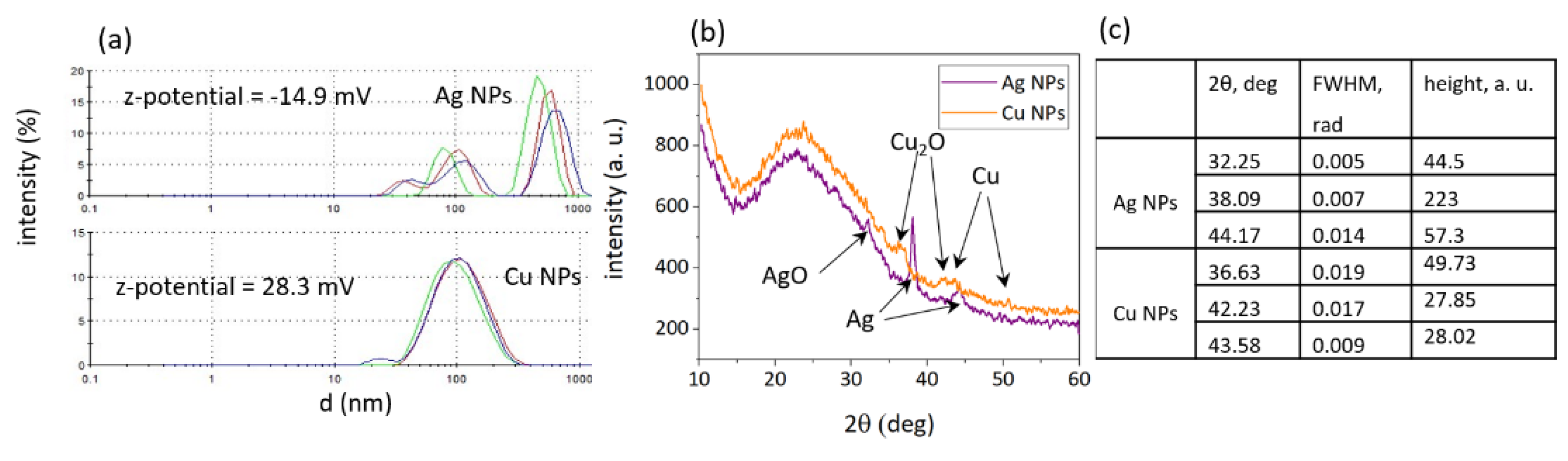
3.1. Characterization of NP Colloids and Solutions
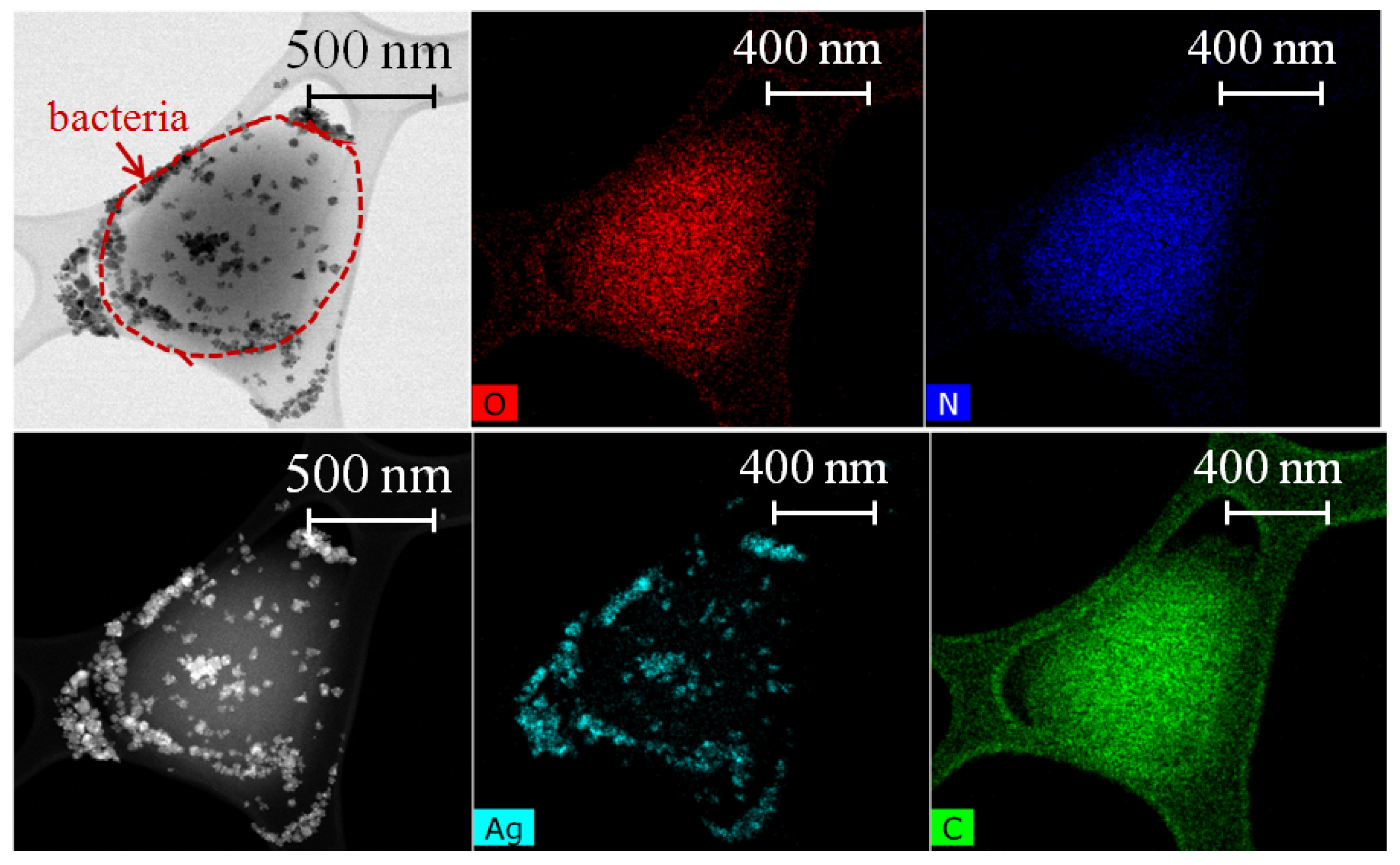
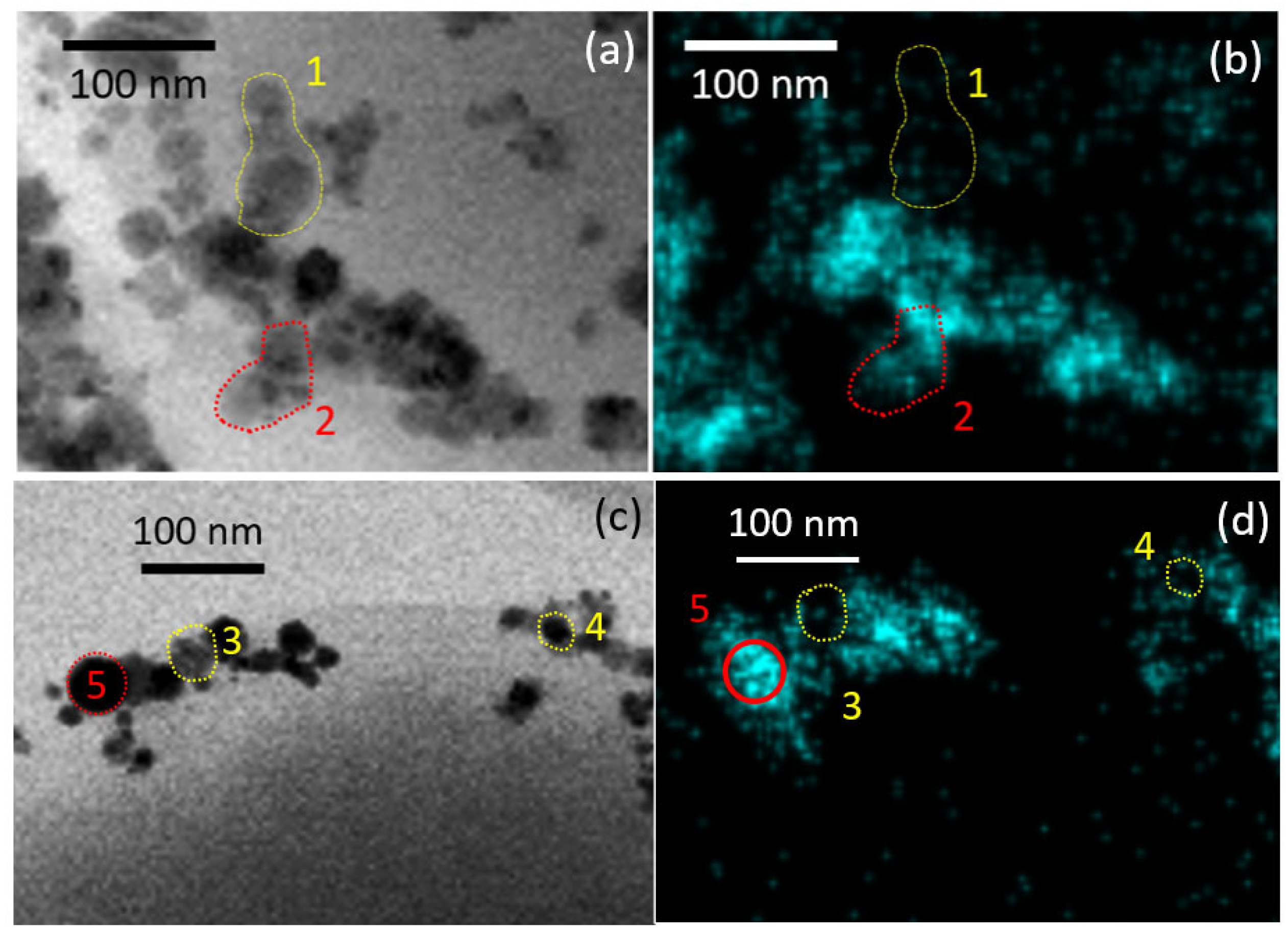
3.2. TEM Characterization of NP-Treated Cells
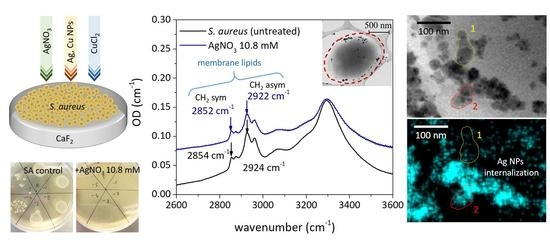
3.3. FT-IR Spectral Measurements
3.4. Microbiological Tests
- (1)
- Ag NPs release ions and ROS, which, due to the close contact of NPs and bacteria in a concentrated (0.3 mg/mL) colloidal solution, and despite negative z-potential, result in lipid peroxidation and cell destruction;
- (2)
- Cu NPs with high positive z-potential are adjacent to bacterial cells via electrostatic attraction and also cause ROS-related lipid peroxidation;
- (3)
- Ag NO3 solution forms precipitates on the bacterial cell surface, which lead to the mechanical rupture of the membrane and subsequent penetration of the precipitates in the emerged damaged spots, and to the complete destruction of the membrane and bacterial death;
- (4)
- Cu ions from the CuCl2 solution cause damage to phosphorus and sulfur-containing biomolecules, which leads to disruption of intracellular biochemical processes.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

References
- Angenent, L.T.; Mau, M.; George, U.; Zahn, J.A.; Raskin, L. Effect of the presence of the antimicrobial tylosin in swine waste on anaerobic treatment. Wat. Res. 2008, 42, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Kemper, N.; Färber, H.; Skutlarek, D.; Krieter, J. Analysis of antibiotic residues in liquid manure and leachate of dairy farms in northern Germany. Agr. Wat. Manag. 2008, 95, 1288–1292. [Google Scholar] [CrossRef]
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, J.L. Antibiotics and antibiotic resistance genes in natural environments. Science 2008, 321, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Mackie, R.I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Vassallo, E.; Pedroni, M.; Silvetti, T.; Morandi, S.; Toffolatti, S.; Angella, G.; Brasca, M. Bactericidal performance of nanostructured surfaces by fluorocarbon plasma. Mat. Sci. Eng. C 2017, 80, 117–121. [Google Scholar] [CrossRef]
- Jiang, R.; Hao, L.; Song, L.; Tian, L.; Fan, Y.; Zhao, J.; Ren, L. Lotus-leaf-inspired hierarchical structured surface with non-fouling and mechanical bactericidal performances. Chem. Eng. J. 2020, 398, 125609. [Google Scholar] [CrossRef]
- Das, B.; Dash, S.K.; Mandal, D.; Ghosh, T.; Chattopadhyay, S.; Tripathy, S.; Roy, S. Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage. Arab. J. Chem. 2017, 10, 862–876. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhao, L.; Ma, H.; Zhang, H.; Guo, L.H. Quantitative analysis of reactive oxygen species photogenerated on metal oxide nanoparticles and their bacteria toxicity: The role of superoxide radicals. Env. Sci. Tech. 2017, 51, 10137–10145. [Google Scholar] [CrossRef]
- Cho, K.H.; Park, J.E.; Osaka, T.; Park, S.G. The study of antimicrobial activity and preservative effects of nanosilver ingredient. Electrochim. Act. 2005, 51, 956–960. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, V.K.; Yngard, R.A.; Lin, Y. Silver nanoparticles: Green synthesis and their antimicrobial activities. Adv. Coll. Int. Sci. 2009, 145, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Raffi, M.; Mehrwan, S.; Bhatti, T.M.; Akhter, J.I.; Hameed, A.; Yawar, W.; ul Hasan, M.M. Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli. Ann. Microbiol. 2010, 60, 75–80. [Google Scholar] [CrossRef]
- Esteban-Cubillo, A.; Pecharromán, C.; Aguilar, E.; Santarén, J.; Moya, J.S. Antibacterial activity of copper monodispersed nanoparticles into sepiolite. J. Mat. Sci. 2006, 41, 5208–5212. [Google Scholar] [CrossRef]
- Ruparelia, J.P.; Chatterjee, A.K.; Duttagupta, S.P.; Mukherji, S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Act. Biomater. 2008, 4, 707–716. [Google Scholar] [CrossRef]
- Theivasanthi, T.; Alagar, M. Studies of copper nanoparticles effects on micro-organisms. arXiv 2011, arXiv:1110.1372. [Google Scholar]
- Lin YS, E.; Vidic, R.D.; Stout, J.E.; Victor, L.Y. Individual and combined effects of copper and silver ions on inactivation of Legionella pneumophila. Wat. Res. 1996, 30, 1905–1913. [Google Scholar] [CrossRef]
- Yu-sen, E.L.; Vidic, R.D.; Stout, J.E.; McCartney, C.A.; Victor, L.Y. Inactivation of Mycobacterium avium by copper and silver ions. Wat. Res. 1998, 32, 1997–2000. [Google Scholar] [CrossRef]
- Yoon, K.Y.; Byeon, J.H.; Park, J.H.; Hwang, J. Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci. Total Environ. 2007, 373, 572–575. [Google Scholar] [CrossRef]
- Jeon, H.J.; Yi, S.C.; Oh, S.G. Preparation and antibacterial effects of Ag–SiO2 thin films by sol–gel method. Biomaterials 2003, 24, 4921–4928. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial activity of silver nanoparticles: Structural effects. Adv. Healthc. Mat. 2018, 7, 1701503. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mat. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Nanda, A.; Majeed, S. Comparative Analysis of Antibiotic Effect of AgNO3 and Silver Nanoparticles Synthesized from Penicillium fellutanum. Mat. Sci. Forum 2013, 760, 9–14. [Google Scholar] [CrossRef]
- Cervantes, C.; Gutierrez-Corona, F. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol. Rev. 1994, 14, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y.; Kitamoto, K.; Anraku, Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 1988, 170, 2676–2682. [Google Scholar] [CrossRef] [Green Version]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Rad. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Avery, S.V.; Howlett, N.G.; Radice, S. Copper toxicity towards Saccharomyces cerevisiae: Dependence on plasma membrane fatty acid composition. Appl. Environ. Microbiol. 1996, 62, 3960–3966. [Google Scholar] [CrossRef] [Green Version]
- Duffy, L.L.; Osmond-McLeod, M.J.; Judy, J.; King, T. Investigation into the antibacterial activity of silver, zinc oxide and copper oxide nanoparticles against poultry-relevant isolates of Salmonella and Campylobacter. Food Control 2018, 92, 293–300. [Google Scholar] [CrossRef]
- DeAlba-Montero, I.; Guajardo-Pacheco, J.; Morales-Sánchez, E.; Araujo-Martínez, R.; Loredo-Becerra, G.M.; Martínez-Castañón, G.A.; Ruiz, F.; Compeán Jasso, M.E. Antimicrobial properties of copper nanoparticles and amino acid chelated copper nanoparticles produced by using a soya extract. Bioinorg. Chem. Appl. 2017, 2017, 1064918. [Google Scholar] [CrossRef] [Green Version]
- Eckhardt, S.; Brunetto, P.S.; Gagnon, J.; Priebe, M.; Giese, B.; Fromm, K.M. Nanobio silver: Its interactions with peptides and bacteria, and its uses in medicine. Chem. Rev. 2013, 113, 4708–4754. [Google Scholar] [CrossRef] [Green Version]
- Sinha, R.; Karan, R.; Sinha, A.; Khare, S.K. Interaction and nanotoxic effect of ZnO and Ag nanoparticles on mesophilic and halophilic bacterial cells. Bioresour. Tech. 2011, 102, 1516–1520. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.; Dille, J.; Godet, S. Synthesis and antibacterial activity of silver nanoparticles against gram-positive and gram-negative bacteria. Nanomed. Nanotech. Biol. Med. 2012, 8, 37–45. [Google Scholar] [CrossRef]
- Matai, I.; Sachdev, A.; Dubey, P.; Kumar, S.U.; Bhushan, B.; Gopinath, P. Antibacterial activity and mechanism of Ag–ZnO nanocomposite on S. aureus and GFP-expressing antibiotic resistant E. coli. Coll. Surf. B Biointerfaces 2014, 115, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Pramanik, A.; Sikdar, N.; Ghosh, S.K.; Pramanik, P. Amelioration studies on optimization of low molecular weight chitosan nanoparticle preparation, characterization with potassium per sulfate and silver nitrate combined action with aid of drug delivery to tetracycline resistant bacteria. Int. J. Pharm. Sci. Drug Res. 2010, 2, 247–253. [Google Scholar]
- Ghosh, D.; Pramanik, A.; Sikdar, N.; Pramanik, P. Synthesis of low molecular weight alginic acid nanoparticles through persulfate treatment as effective drug delivery system to manage drug resistant bacteria. Biotechnol. Bioprocess Eng. 2011, 16, 383–392. [Google Scholar] [CrossRef]
- Wenning, M.; Breitenwieser, F.; Konrad, R.; Huber, I.; Busch, U.; Scherer, S. Identification and differentiation of food-related bacteria: A comparison of FTIR spectroscopy and MALDI-TOF mass spectrometry. J. Microbiol. Methods 2014, 103, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Mariey, L.; Signolle, J.P.; Amiel, C.; Travert, J. Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics. Vib. Spectrosc. 2001, 26, 151–159. [Google Scholar] [CrossRef]
- Zarnowiec, P.; Lechowicz, L.; Czerwonka, G.; Kaca, W. Fourier transform infrared spectroscopy (FTIR) as a tool for the identification and differentiation of pathogenic bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Tang, M.; McEwen, G.D.; Wu, Y.; Miller, C.D.; Zhou, A. Characterization and analysis of mycobacteria and Gram-negative bacteria and co-culture mixtures by Raman microspectroscopy, FTIR, and atomic force microscopy. Anal. Bioanal. Chem. 2013, 405, 1577–1591. [Google Scholar] [CrossRef]
- Huleihel, M.; Pavlov, V.; Erukhimovitch, V. The use of FTIR microscopy for the evaluation of anti-bacterial agents activity. J. Photochem. Photobiol. B Biol. 2009, 96, 17–23. [Google Scholar] [CrossRef]
- Kudryashov, S.I.; Nastulyavichus, A.A.; Tolordava, E.R.; Kirichenko, A.N.; Saraeva, I.N.; Rudenko, A.A.; Romanova, Y.M.; Panarin, A.Y.; Ionin, A.A.; Itina, T.E. Surface-Enhanced IR-Absorption Microscopy of Staphylococcus aureus Bacteria on Bactericidal Nanostructured Si Surfaces. Molecules 2019, 24, 4488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Los, D.A.; Murata, N. Membrane fluidity and its roles in the perception of Env. signals. Biochim. Biophys. Acta (BBA)-Biomembr. 2004, 1666, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Wharton, C.W. Infrared spectroscopy of enzyme reaction intermediates. Nat. Prod. Rep. 2000, 17, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Barth, A.; Zscherp, C. What vibrations tell about proteins. Q. Rev. Biophys. 2002, 35, 369–430. [Google Scholar] [CrossRef]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Coll. Int. Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef]
- Kreibig, U.; Vollmer, M. Optical Properties of Metal Clusters; Springer: Berlin, Germany, 1995. [Google Scholar] [CrossRef]
- Szymańska-Chargot, M.; Gruszecka, A.; Smolira, A.; Bederski, K.; Głuch, K.; Cytawa, J.; Michalak, L. Formation of nanoparticles and nanorods via UV irradiation of AgNO3 solutions. J. Alloys Compd. 2009, 486, 66–69. [Google Scholar] [CrossRef]
- Henglein, A.; Meisel, D. Spectrophotometric observations of the adsorption of organosulfur compounds on Coll.al silver nanoparticles. J. Phys. Chem. B 1998, 102, 8364–8366. [Google Scholar] [CrossRef]
- Wu, S.H.; Chen, D.H. Synthesis of high-concentration Cu nanoparticles in aqueous CTAB solutions. J. Coll. Int. Sci. 2004, 273, 165–169. [Google Scholar] [CrossRef]
- Higuchi, M.; Imoda, D.; Hirao, T. Redox Behavior of Polyaniline− Transition Metal Complexes in Solution. Macromolecules 1996, 29, 8277–8279. [Google Scholar] [CrossRef]
- Salavati-Niasari, M.; Davar, F. Synthesis of copper and copper (I) oxide nanoparticles by thermal decomposition of a new precursor. Mat. Lett. 2009, 63, 441–443. [Google Scholar] [CrossRef]
- Babu, S.A.; Prabu, H.G. Synthesis of AgNPs using the extract of Calotropis procera flower at room temperature. Mat. Lett. 2011, 65, 1675–1677. [Google Scholar] [CrossRef]
- Shamaila, S.; Wali, H.; Sharif, R.; Nazir, J.; Zafar, N.; Rafique, M.S. Antibacterial effects of laser ablated Ni nanoparticles. Appl. Phys. Lett. 2013, 103, 153701. [Google Scholar] [CrossRef]
- Romaniuk, J.A.; Cegelski, L. Peptidoglycan and teichoic acid levels and alterations in Staphylococcus aureus by cell-wall and whole-cell nuclear magnetic resonance. Biochemistry 2018, 57, 3966–3975. [Google Scholar] [CrossRef] [PubMed]
- Vernon-Parry, K.D. Scanning electron microscopy: An introduction. III-Vs Rev. 2000, 13, 40–44. [Google Scholar] [CrossRef] [Green Version]
- Newbury, D.E.; Ritchie, N.W. Elemental mapping of microstructures by scanning electron microscopy-energy dispersive X-ray spectrometry (SEM-EDS): Extraordinary Adv. with the silicon drift detector (SDD). J. Anal. At. Spectrom. 2013, 28, 973–988. [Google Scholar] [CrossRef]
- Jabbari, E.; Peppas, N.A. Quantitative measurement of interdiffusion at polymer–polymer interfaces with TEM/EDS and EELS. J. Appl. Polym. Sci. 1995, 57, 775–779. [Google Scholar] [CrossRef]
- Stevie, F.A.; Vartuli, C.B.; Giannuzzi, L.A.; Shofner, T.L.; Brown, S.R.; Rossie, B.; Hillion, F.; Mills, R.H.; Antonell, M.; Irwin, R.B.; et al. Application of focused ion beam lift-out specimen preparation to TEM, SEM, STEM, AES and SIMS analysis. Surf. Interface Anal. 2001, 31, 345–351. [Google Scholar] [CrossRef]
- Davis, R.; Mauer, L.J. Fourier transform infrared (FT-IR) spectroscopy: A rapid tool for detection and analysis of foodborne pathogenic bacteria. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Formatex Research Center: Badajoz, Spain, 2010; Volume 2, pp. 1582–1594. Available online: https://d1wqtxts1xzle7.cloudfront.net/61513069/Davis___Mauer__201020191214-15260-1c3stns-with-cover-page-v2.pdf?Expires=1667211919&Signature=OyHGC9yeSIZ2xAY~U6Bp9tyI9p4cu039Ba7yEsDOyXAGI-~qfWGvgpfI~9Bbc5r~kGiBg4sSzHyethXnWiYyECu6EnsxkR4-~KQSyzApAH3d4m6CoVe9VmlxKiH-q6JIOOClmuoYajUgKC6o70oUnYq0UvnMYaq3rkmRC0d~Ei06haIKPTmK9DdP2Ltl0uEpHscZTMEmLEF5VsmmEEh2y82Ik9p7ZMJgIpmIFOgLDNjExPf3imWFx0BxJOxnYMW5M9~1NKcIGvMwDQTsc5MuxA7GpfF3PrIH4z1jo4MU3j3x-GOO3wfecvuLzljlVxuTaJTgXY166oAnlypqT~wCEQ__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA (accessed on 24 October 2022).
- Melin, A.M.; Perromat, A.; Déléris, G. Pharmacologic application of Fourier transform IR spectroscopy: In vivo toxicity of carbon tetrachloride on rat liver. Biopolym. Orig. Res. Biomol. 2000, 57, 160–168. [Google Scholar] [CrossRef]
- Gorgulu, S.T.; Dogan, M.; Severcan, F. The characterization and differentiation of higher plants by Fourier transform infrared spectroscopy. Appl. Spectrosc. 2007, 61, 300–308. [Google Scholar] [CrossRef]
- Garip, S.; Gozen, A.C.; Severcan, F. Use of Fourier transform infrared spectroscopy for rapid comparative analysis of Bacillus and Micrococcus isolates. Food Chem. 2009, 113, 1301–1307. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; van den Braak, N.; Endtz, H.P.; Naumann, D.; Puppels, G.J. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 2002, 51, 255–271. [Google Scholar] [CrossRef]
- Naumann, D. FT-infrared and FT-Raman spectroscopy in Biomed. Res. Appl. Spectrosc. Rev. 2001, 36, 239–298. [Google Scholar] [CrossRef]
- Ozek, N.S.; Bal, I.B.; Sara, Y.; Onur, R.; Severcan, F. Structural and functional characterization of simvastatin-induced myotoxicity in different skeletal muscles. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2014, 1840, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Kamnev, A.A. FTIR spectroscopic studies of bacterial cellular responses to Env. factors, plant-bacterial interactions and signalling. Spectroscopy 2008, 22, 83–95. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Jackson, M.; Sowa, M.G.; Mantsch, H.H. Infrared spectroscopy: A new frontier in medicine. Biophys. Chem. 1997, 68, 109–125. [Google Scholar] [CrossRef]
- Szalontai, B.; Nishiyama, Y.; Gombos, Z.; Murata, N. Membrane dynamics as seen by Fourier transform infrared spectroscopy in a cyanobacterium, Synechocystis PCC 6803: The effects of lipid unsaturation and the protein-to-lipid ratio. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1509, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Severcan, F.; Cannistraro, S. A spin label ESR and saturation transfer ESR study of alpha-tocopherol containing model membranes. Chem. Phys. Lipids 1990, 53, 17–26. [Google Scholar] [CrossRef]
- Boyar, H.; Boyar, F. Oestrogen-phospholipid membrane interactions: An FTIR study. J. Mol. Struct. 1997, 408/409, 269–272. [Google Scholar] [CrossRef]
- Cakmak, G.; Togan, I.; Severcan, F. 17-Estradiol induced compositional, structural and functional changes in rainbow trout liver, revealed by FT-IR spectroscopy: A comparative study with nonylphenol. Aquat. Toxicol. 2006, 77, 53–63. [Google Scholar] [CrossRef]
- Kardas, M.; Gozen, A.G.; Severcan, F. FTIR spectroscopy offers hints towards widespread molecular changes in cobalt-acclimated fresh water. bacteria. Aquat. Toxicol. 2014, 155, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Lovrić, J.; Cho, S.J.; Winnik, F.M.; Maysinger, D. Unmodified cadmium telluride quantum dots induce reactive oxygen species formation leading to multiple organelle damage and cell death. Chem. Biol. 2005, 12, 1227–1234. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Pinault, M.; Pfefferle, L.D.; Elimelech, M. Single-walled carbon nanotubes exhibit strong antimicrobial activity. Langmuir 2007, 23, 8670–8673. [Google Scholar] [CrossRef] [PubMed]
- Thill, A.; Zeyons, O.; Spalla, O.; Chauvat, F.; Rose, J.; Auffan, M.; Flank, A.M. Cytotoxicity of CeO2 nanoparticles for Escherichia coli. Physico-Chemical. insight of the cytotoxicity mechanism. Env. Sci. Tech. 2006, 40, 6151–6156. [Google Scholar] [CrossRef] [PubMed]
- Stoimenov, P.K.; Klinger, R.L.; Marchin, G.L.; Klabunde, K.J. Metal oxide nanoparticles as bactericidal agents. Langmuir 2002, 18, 6679–6686. [Google Scholar] [CrossRef]
- Thayer, M.M.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J. Biol. Chem. 1991, 266, 2864–2871. [Google Scholar] [CrossRef]
- Bitter, W.; Koster, M.; Latijnhouwers, M.; De Cock, H.; Tommassen, J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 1998, 27, 209–219. [Google Scholar] [CrossRef]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Cho, M.H. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotech. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Lok, C.N.; Ho, C.M.; Chen, R.; He, Q.Y.; Yu, W.Y.; Sun, H.; Che, C.M. Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J. Proteome Res. 2006, 5, 916–924. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346. [Google Scholar] [CrossRef]



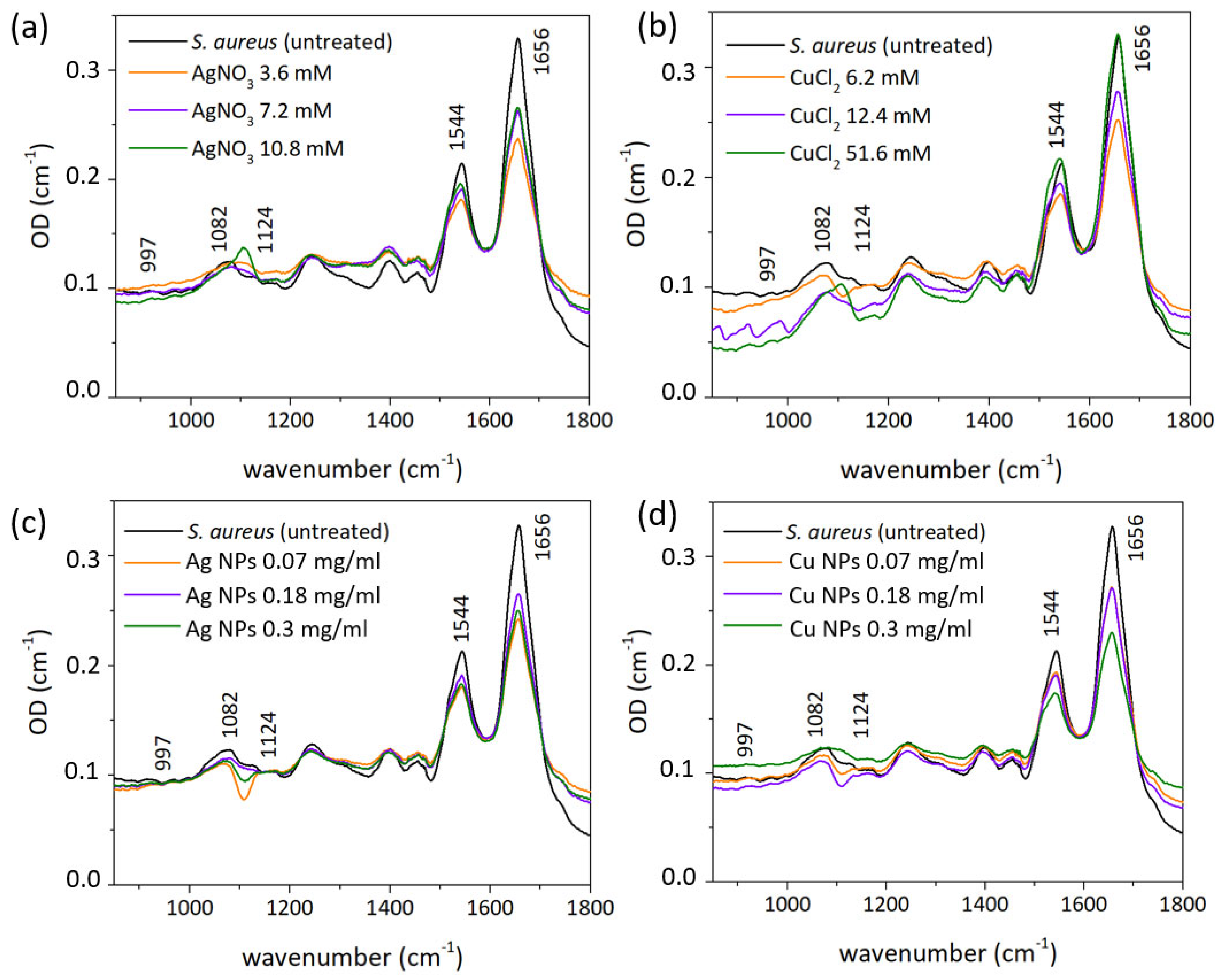
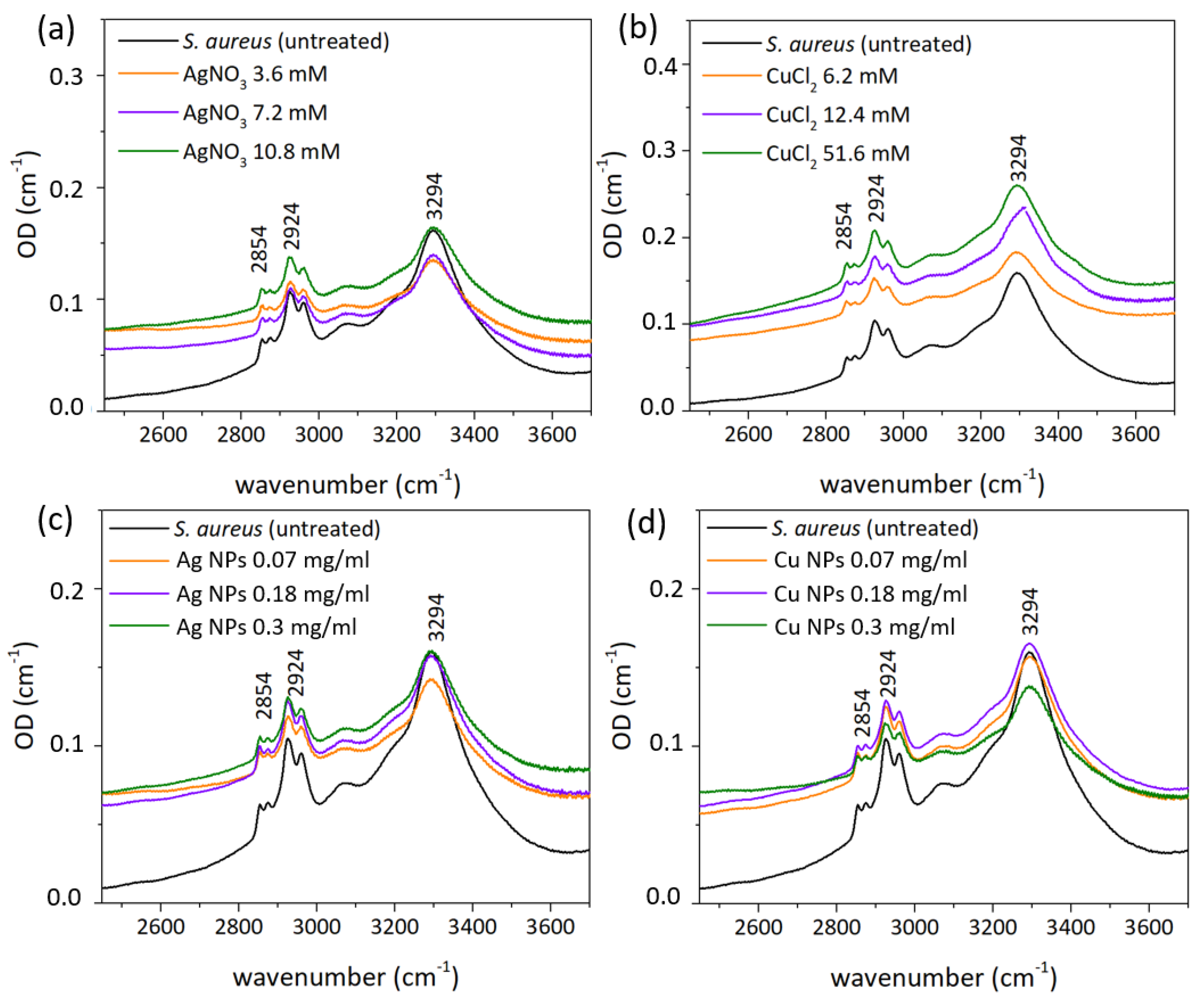
| Functional Groups | Frequency (cm−1) | ||||
|---|---|---|---|---|---|
| Control | AgNO3-Treated | Ag NP-Treated | CuCl2-Treated | Cu NP-Treated | |
| Amide A | 3294 | 3294 | 3294 | 3294 | 3294 |
| CH2 asym str | 2924 | 2922 | 2925 | 2925 | 2925 |
| CH2 sym str | 2854 | 2852 | 2854 | 2854 | 2852 |
| Amide I | 1656 | 1656 | 1655 | 1655 | 1655 |
| Amide II | 1544 | 1541 | 1541 | 1543 | 1541 |
| C-C | 1124 | 1104 | - | 1106 | - |
| PO2 | 1082 | - | 1082 | - | 1085 |
| ribose skeleton | 997 | 997 | 997 | 997 | 997 |
| Functional Groups | Bandwidth (cm−1) | %Change | %Change | %Change | %Change | ||||
|---|---|---|---|---|---|---|---|---|---|
| Control | AgNO3-Treated | Ag NP-Treated | CuCl2-Treated | Cu NP-Treated | |||||
| Amide I | 53.5 ± 2.35 | 61.39 ± 1.83 | +14.75 | 63.28 ± 1.67 | +18.28 | 55.69 ± 1.28 | +4.09 | 64.17 ± 1.64 | +19.94 |
| Amide II | 57.19 ± 2.74 | 64.59 ± 1.83 | +12.94 | 68.54 ± 1.34 | +19.85 | 52.71 ± 1.53 | −7.83 | 70.30 ± 1.45 | +22.92 |
| CH2 sym str | 8.82 ± 2.04 | 10.18 ± 1.81 | +15.4 | 8.82 ± 1.79 | 0 | 11.54 ± 2.63 | +30.84 | 8.56 ± 1.75 | −2.95 |
| CH2 asym str | 33.5 ± 1.75 | 33.76 ± 1.43 | +0.8 | 33.81 ± 1.64 | +0.93 | 30.54 ± 2.22 | −8.84 | 34.46 ± 1.52 | +2.09 |
| Amide A | 156.24 ± 2.42 | 164.21 ± 2.33 | +5.11 | 157.32 ± 1.96 | +0.69 | 147.59 ± 2.2 | −5.54 | 173.07 ± 2.19 | +10.77 |
| Functional Groups | Area (a.u.) | %Change | %Change | %Change | %Change | ||||
|---|---|---|---|---|---|---|---|---|---|
| Control | AgNO3-Treated | Ag NP-Treated | CuCl2-Treated | Cu NP-Treated | |||||
| Amide I | 21.51 ± 0.36 | 17.48 ± 0.17 | −18.74 | 17.12 ± 0.15 | −20.41 | 22.43 ± 0.4 | +4.28 | 14.47 ± 0.12 | −32.73 |
| Amide II | 11.6 ± 0.38 | 10.57 ± 0.1 | −8.88 | 10.66 ± 0.17 | −8.1 | 10.95 ± 0.4 | −5.6 | 9.48 ± 0.14 | −18.28 |
| CH2 sym str | 0.18 ± 0.04 | 0.18 ± 0.04 | 0 | 0.13 ± 0.03 | −27.8 | 0.24 ± 0.06 | +33.34 | 0.1 ± 0.02 | −44.4 |
| CH2 asym str | 2.51 ± 0.18 | 1.96 ± 0.1 | −21.91 | 1.63 ± 0.1 | −35.06 | 2.04 ± 0.24 | +18.73 | 1.42 ± 0.08 | −43.43 |
| Amide A | 29.35 ± 0.71 | 20.68 ± 0.48 | −29.54 | 17.92 ± 0.33 | −38.94 | 25.52 ± 0.58 | −13.05 | 18.01 ± 0.35 | −38.64 |
| S. aureus (CFU) | |
|---|---|
| control | 108 |
| AgNO3 | 0 |
| CuCl2 | 0 |
| Ag NPs | 0 |
| Cu NPs | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saraeva, I.; Tolordava, E.; Yushina, Y.; Sozaev, I.; Sokolova, V.; Khmelnitskiy, R.; Sheligyna, S.; Pallaeva, T.; Pokryshkin, N.; Khmelenin, D.; et al. Direct Bactericidal Comparison of Metal Nanoparticles and Their Salts against S. aureus Culture by TEM and FT-IR Spectroscopy. Nanomaterials 2022, 12, 3857. https://doi.org/10.3390/nano12213857
Saraeva I, Tolordava E, Yushina Y, Sozaev I, Sokolova V, Khmelnitskiy R, Sheligyna S, Pallaeva T, Pokryshkin N, Khmelenin D, et al. Direct Bactericidal Comparison of Metal Nanoparticles and Their Salts against S. aureus Culture by TEM and FT-IR Spectroscopy. Nanomaterials. 2022; 12(21):3857. https://doi.org/10.3390/nano12213857
Chicago/Turabian StyleSaraeva, Irina, Eteri Tolordava, Yulia Yushina, Islam Sozaev, Vera Sokolova, Roman Khmelnitskiy, Svetlana Sheligyna, Tatiana Pallaeva, Nikolay Pokryshkin, Dmitry Khmelenin, and et al. 2022. "Direct Bactericidal Comparison of Metal Nanoparticles and Their Salts against S. aureus Culture by TEM and FT-IR Spectroscopy" Nanomaterials 12, no. 21: 3857. https://doi.org/10.3390/nano12213857