Nanoparticle-Induced m6A RNA Modification: Detection Methods, Mechanisms and Applications
Abstract
:1. Introduction
2. m6A RNA Modification
2.1. Dynamic Regulation of m6A RNA Modification
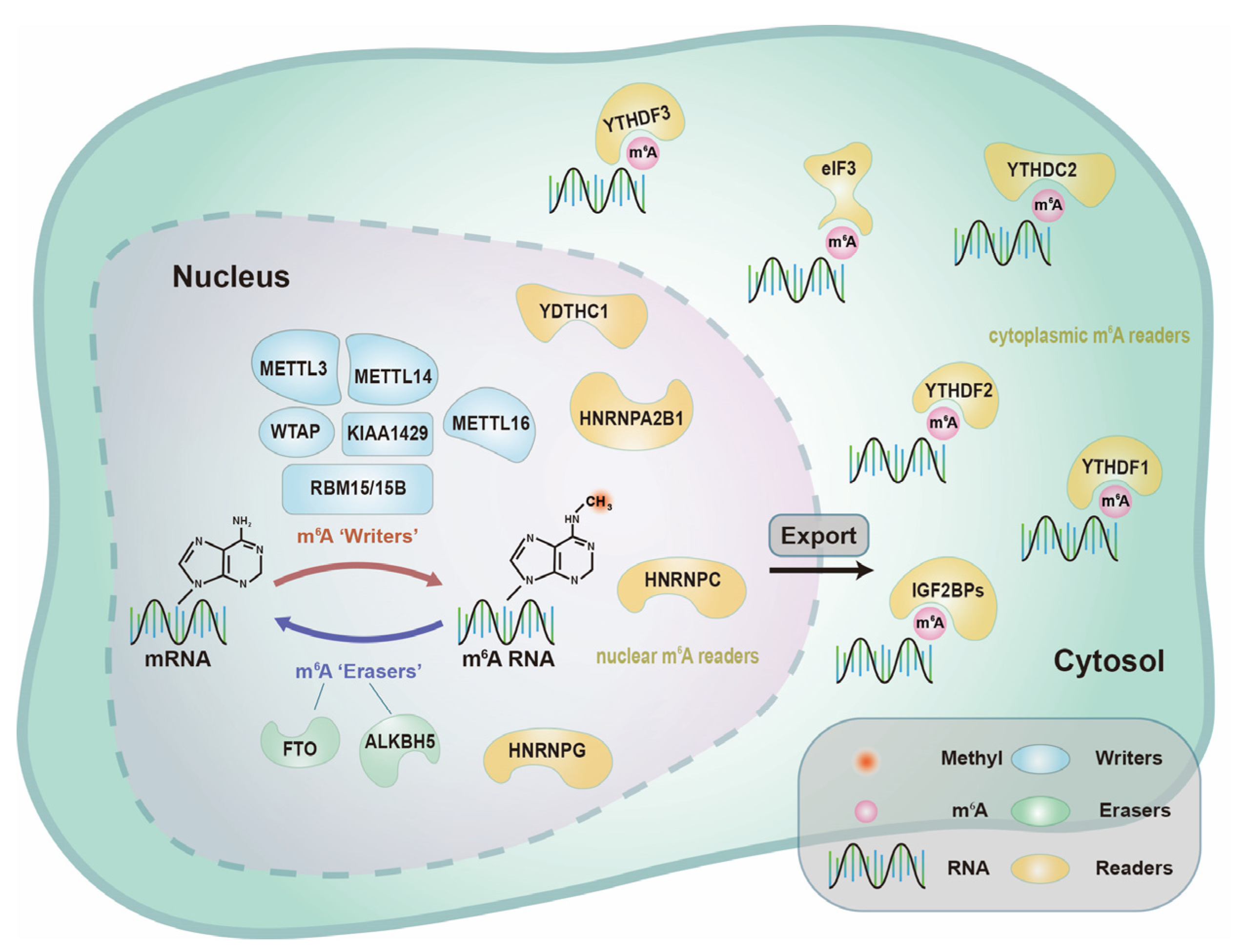
2.1.1. m6A “Writers”—Adenosine Methyltransferases
2.1.2. m6A “Erasers”—Demethylases
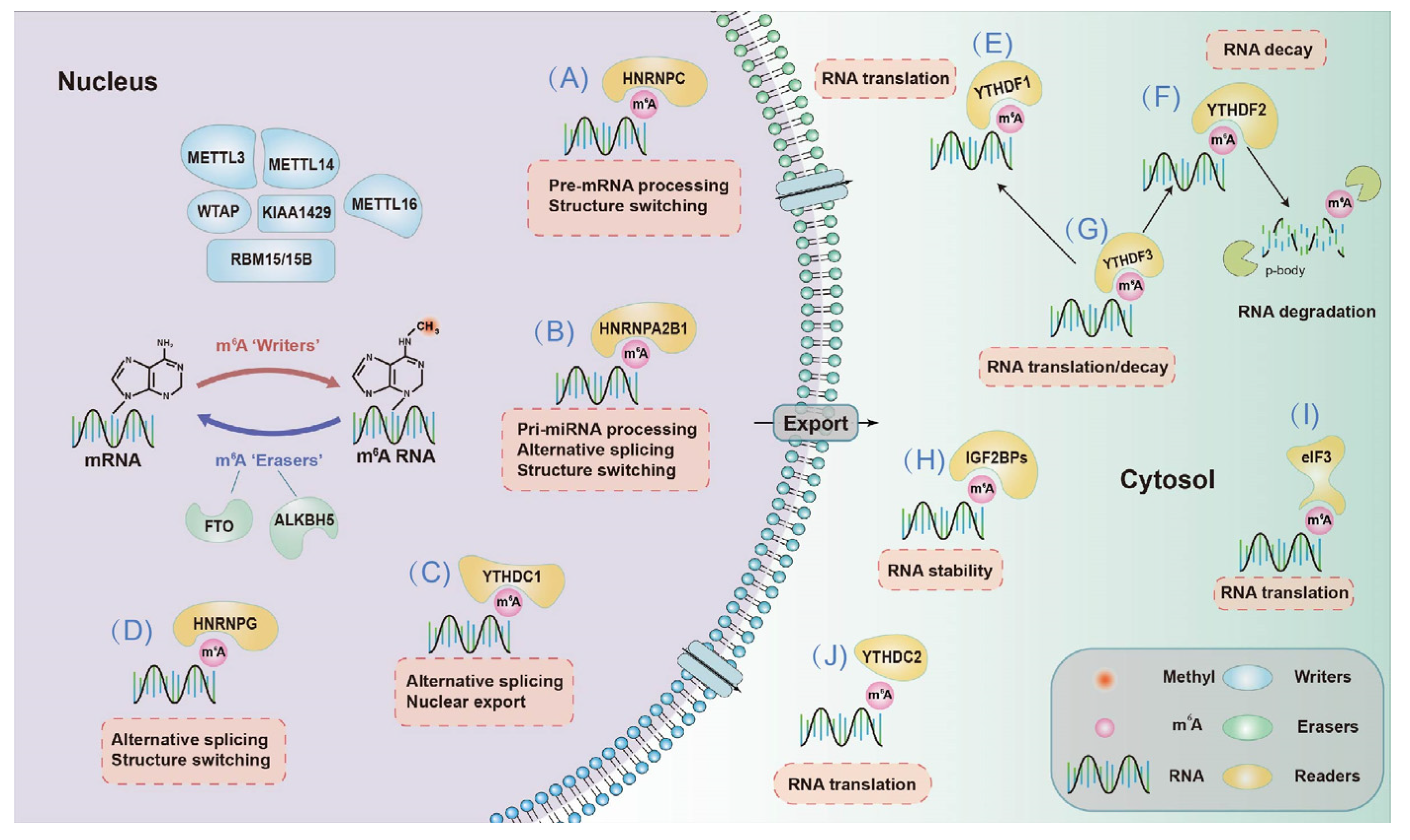
2.1.3. m6A “Readers”—Binding Proteins
2.2. Biofunctions of m6A RNA Modification
2.2.1. Effects on mRNA Fate—Splicing, Processing, Translation and Degradation
2.2.2. Biological Consequences of m6A—Dysregulation in Cellular Processes and Diseases
Cellular Stress
Hematopoietic Development
Neurogenesis
Fertilization
Immune Response
Cancer
Diabetes
| Processes /Diseases | m6A Regulator | Cells/ Organisms | Effect of Gene Loss/Gain of Function | Mechanism | Ref. |
|---|---|---|---|---|---|
| METTL3 | mRTECs | ↑ROS | ↑METTL3/Keap1/Nrf2 | [54] | |
| FTO | L02 cells | ↓ROS | ↓FTO/↑PGC-1α | [55] | |
| YTHDF1 | Beas-2B cells | ↓Hypoxia adaptation | ↓YTHDF1/Keap1/Nrf2-AKR1C1 | [56] | |
| Cell stress | METTL3 | HCCs | ↓Glycolytic capacity | ↓METTL3/mTORC | [57] |
| YTHDF2 | MEFs | ↑Heat shock stress | ↓YTHDF2/↑HSP90, HSP60, HSPB1 | [58,59] | |
| METTL16 | MEFs | ↑DNA damage | ↑METTL16/γH2AX | [60,61] | |
| FTO | Mice | ↑ER stress | ↓FTO/↓HSP70/↑NF-κB | [62] | |
| FTO | Mice | ↑autophagy | ↓FTO/↓Atg5, Atg7 | [63] | |
| FTO | 293T cells | ↑autophagy | ↓FTO/↑ULK1 | [64] | |
| Haematopoietic | METTL3 | HSCs | ↓Proliferation, ↓differentiation | ↓METTL3/↑MDA5/RIG-I | [65] |
| development | METTL3 | HSPCs | ↑Differentiation, ↓cell proliferation | ↑METTL3/c-MYC/BCL2/PTEN | [66] |
| YTHDF2 | HSCs | ↑Regeneration | ↓YTHDF2/↑Wnt target genes | [67] | |
| METTL3 | HSPCs | ↑Endothelial to haematopoietic transition | ↓METTL3/↑YTHDF2/↓Notch1a | [68] | |
| Neurogenesis | YTHDF2 | NSPCs | ↓Self-renewal | ↓YTHDF1/JAK–STAT | [69] |
| FTO | NSCs | ↓Proliferation, ↓differentiation | ↓FTO/BDNF/PI3K/Akt2/Akt3 | [70] | |
| METTL3/14 | RGCs | ↑Neurogenesis, ↑cell cycle | ↓METTL3/14/↑Neurog2/Neurod1 | [71] | |
| YTHDF1 | Mice | ↓Learning, memory defects | ↓YTHDF1/Camk2a | [72] | |
| Fertilization | METTL3 | Zebrafish | ↓Sperm motility | ↓METTL14/11-KT/17β-E2 | [73] |
| ALKBH5 | Mice | ↓Fertility | ↓ALKBH5/↑Dnmt1 | [37,74] | |
| YTHDC1 | Germ cells | ↓Oocyte growth, maturation | ↓YTHDC1/CPSF6/SRSF3 | [75] | |
| YTHDF2 | Mice | ↓Oocyte maturation | ↓YTHDF2/Trpc5 | [76] | |
| FTO | SA patients | ↑Spontaneous abortion | ↓FTO/VEGFA, VEGFR | [77] | |
| ALKBH5 | SA patients | ↑Spontaneous abortion | ↓FTO/↑CYR61 | [78] | |
| Immune | METTL3 | Mice | ↓T cell proliferation | ↓METTL3/IL-7/STAT5/SOCS | [82] |
| response | METTL14 | DCs | ↓B cell development | ↓METTL14/TLR4/NF-κB | [83] |
| Cancer | METTL3/14 | GBM | ↑Proliferation and self-renewal of GSCs | ↓METTL3/14/↑ADAM19 | [87] |
| METTL3 | AML | ↓Cell cycle and differentiation of leukaemic cells | ↓METTL3/↑c-MYC | [91] | |
| ALKBH5 | AML | ↓Prognosis of AML patients | ↓ALKBH5/↑TP53 | [92] | |
| Diabetes | METTL14 | Mice | ↓Insulin secretion | ↓METTL14/IGF1–AKT–PDX1 | [93] |
| FTO | HepG2 cells | ↓Glucose metabolism | ↓FTO/FOXO1/G6PC/DGAT2 | [94] |
2.3. Detection
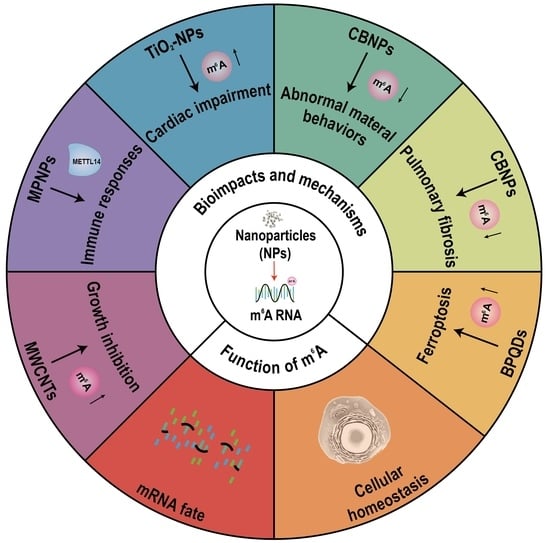
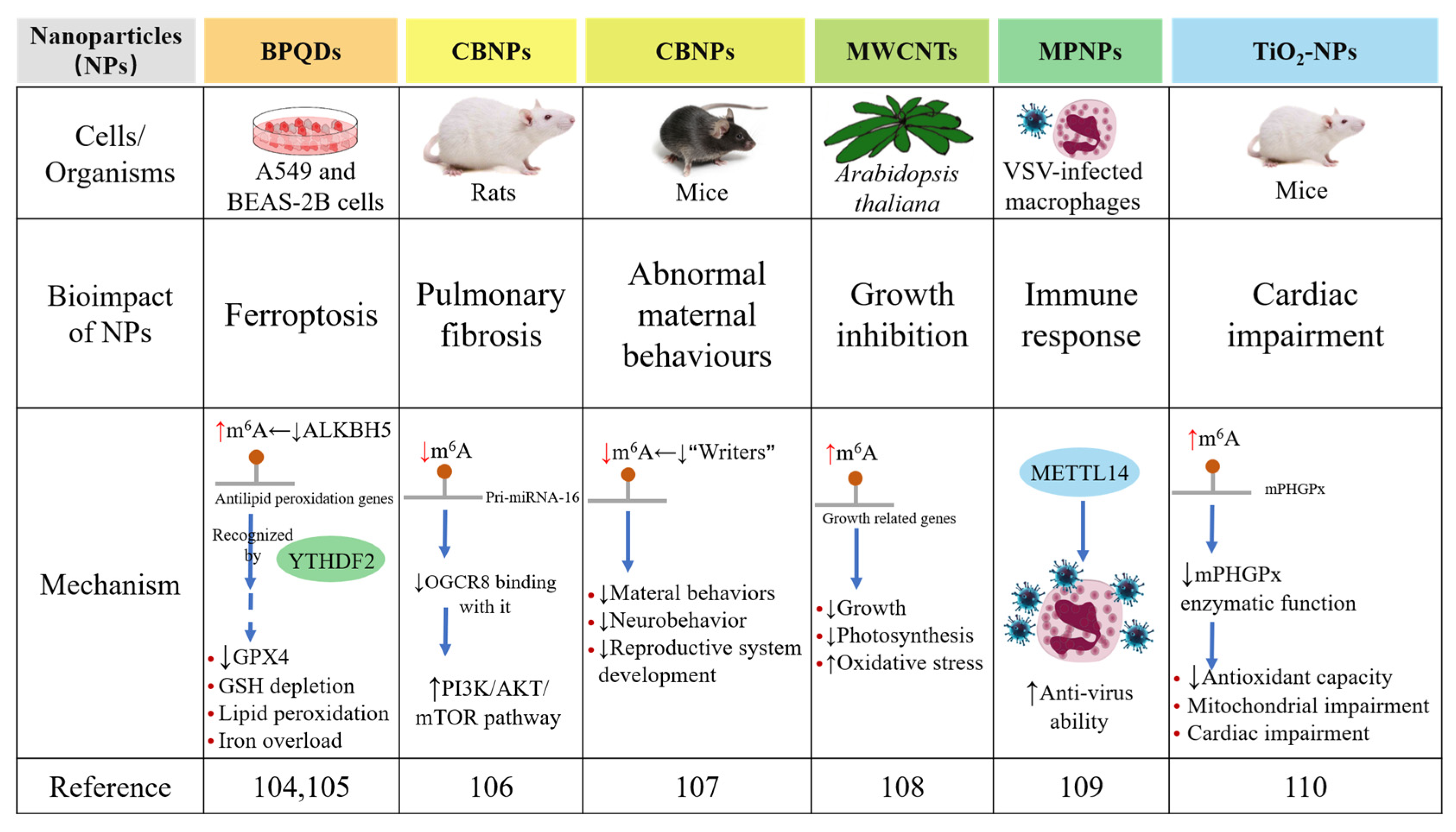
3. m6A RNA Modification Modulates the Bioimpacts of Nanoparticles
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NPs | nanoparticles |
| m6A | N6-methyladenosine |
| SiO2-NPs | silica dioxide nanoparticles |
| TiO2-NPs | titanium dioxide nanoparticles |
| PEG | polyethylene glycol |
| QDs | quantum dots |
| FTO | fat mass and obesity-associated protein |
| METTL3 | methyltransferase-like 3 |
| METTL14 | methyltransferase-like 14 |
| WTAP | Wilms tumour 1-associated protein |
| RBM15/RBM15B | RNA-binding motif protein 15/15B |
| ALKBH5 | ALKB homologue 5 |
| YTH | YT521-B homology |
| YTHDF1 | YTH domain-containing 1 |
| YTHDF2 | YTH domain-containing 2 |
| YTHDF3 | YTH domain-containing 3 |
| YTHDC1 | YTH domain-containing 1 |
| YTHDC2 | YTH domain-containing 2 |
| eIF3 | eukaryotic initiation factor 3 |
| IGF2BPs | insulin-like growth factor 2 mRNA-binding proteins |
| HNRNPA2B1 | heterogeneous nuclear ribonucleoprotein A2B1 |
| HNRNPC | heterogeneous nuclear ribonucleoprotein C |
| HNRNPG | heterogeneous nuclear ribonucleoprotein G |
| pre-mRNA | precursor mRNA |
| SRSF2 | serine and arginine rich splicing factor 2 |
| mESCs | mouse embryonic stem cells |
| EBs | embryoid bodies |
| mRTECs | mouse renal tubular epithelial cells |
| ROS | reactive oxygen species |
| mRTECs | mouse renal tubular epithelial cells |
| PGC1α | peroxisome proliferator-activated receptor-γ coactivator-1α |
| Beas-2B cells | human bronchial epithelium cells |
| HCCs | hepatocellular carcinoma cells |
| NER | nucleotide excision repair |
| dsRNAs | double-stranded RNAs |
| HSCs | haematopoietic stem cells |
| NSCs | neural stem/progenitor cells |
| RGCs | radial glial cells |
| DCs | dendritic cells |
| GBM | glioblastoma |
| AML | acute myeloid leukaemia |
| GSCs | glioblastoma stem cells |
| MeRIP-seq | methylated RNA immunoprecipitation followed by high-throughput sequencing |
| m6A-seq | m6A RNA immunoprecipitation sequencing |
| PA-m6A-seq | photocrosslinking-assisted m6A sequencing |
| miCLIP-seq | m6A individual nucleotide resolution cross-linking and immunoprecipitation sequencing |
| SCARLET | site-specific cleavage and radioactive labelling followed by ligation-assisted extraction and thin-layer chromatography |
| m6A-REF-seq | m6A-sensitive RNA endoribonuclease-facilitated sequencing |
| HPLC–MS | high-performance liquid chromatography-tandem mass spectrometry |
| hESCs | human embryonic stem cells |
| BPQDs | black phosphorus quantum dots |
| ER | endoplasmic reticulum |
| GPX4 | glutathione peroxidase 4 |
| GSH | glutathione |
| RIP | RNA immunoprecipitation |
| CBNPs | carbon black nanoparticles |
| MWCNTs | multiwalled carbon nanotubes |
| VSV | vesicular stomatitis virus |
| MPNPs | metal–protein nanoparticles |
| IFN-β | interferon-beta |
| mPHGPx | mitochondrial phospholipid hydroperoxide glutathione peroxidase |
References
- Gupta, R.; Xie, H. Nanoparticles in daily life: Applications, toxicity and regulations. J. Environ. Pathol. Toxicol. Oncol. 2018, 37, 209–230. [Google Scholar] [CrossRef]
- Hoseinnejad, M.; Jafari, S.M.; Katouzian, I. Inorganic and metal nanoparticles and their antimicrobial activity in food packaging applications. Crit. Rev. Microbiol. 2018, 44, 161–181. [Google Scholar] [CrossRef]
- Lu, P.J.; Cheng, W.L.; Huang, S.C.; Chen, Y.P.; Chou, H.K.; Cheng, H.F. Characterizing titanium dioxide and zinc oxide nanoparticles in sunscreen spray. Int. J. Cosmet. Sci. 2015, 37, 620–626. [Google Scholar] [CrossRef]
- Knowles, B.; Wagner, P.; MacLaughlin, S.; Higgins, M.; Molino, P.J. Silica nanoparticles functionalized with zwitterionic sulfobetaine siloxane for application as a versatile antifouling coating System. ACS Appl. Mater. Interfaces 2017, 9, 18584–18594. [Google Scholar] [CrossRef]
- Bogdan, J.; Jackowska-Tracz, A.; Zarzynska, J.; Pławińska-Czarnak, J. Chances and limitations of nanosized titanium dioxide practical application in view of its physicochemical properties. Nanoscale Res. Lett. 2016, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Ashfaq, U.A.; Riaz, M.; Yasmeen, E.; Yousaf, M.Z. Recent advances in nanoparticle-based targeted drug-delivery systems against cancer and role of tumor microenvironment. Crit. Rev. Ther. Drug Carr. Syst. 2017, 34, 317–353. [Google Scholar] [CrossRef]
- Gill, K.K.; Kaddoumi, A.; Nazzal, S. PEG–lipid micelles as drug carriers: Physiochemical attributes, formulation principles and biological implication. J. Drug Target. 2015, 23, 222–231. [Google Scholar] [CrossRef]
- Mansuriya, B.D.; Altintas, Z. Applications of graphene quantum dots in biomedical sensors. Sensors 2020, 20, 1072. [Google Scholar] [CrossRef] [Green Version]
- Barani, M.; Reza Hajinezhad, M.; Sargazi, S.; Zeeshan, M.; Rahdar, A.; Pandey, S.; Khatami, M.; Zargari, F. Simulation, in vitro, and in vivo cytotoxicity assessments of methotrexate-loaded pH-responsive nanocarriers. Polymers 2021, 13, 3153. [Google Scholar] [CrossRef]
- Barani, M.; Rahdar, A.; Sargazi, S.; Amiri, M.S.; Sharma, P.K.; Bhalla, N. Nanotechnology for inflammatory bowel disease management: Detection, imaging and treatment. Sens. Bio-Sens. Res. 2021, 32, 100417. [Google Scholar] [CrossRef]
- Arora, S.; Rajwade, J.M.; Paknikar, K. Nanotoxicology and in vitro studies: The need of the hour. Toxicol. Appl. Pharmacol. 2012, 258, 151–165. [Google Scholar] [CrossRef]
- Cao, Y.; Li, S.; Chen, J. Modeling better in vitro models for the prediction of nanoparticle toxicity: A review. Toxicol. Mech. Methods 2020, 31, 1–17. [Google Scholar] [CrossRef]
- Lewinski, N.; Colvin, V.; Drezek, R. Cytotoxicity of nanoparticles. Small 2008, 4, 26–49. [Google Scholar] [CrossRef]
- Li, B.; Tang, M. Research progress of nanoparticle toxicity signaling pathway. Life Sci. 2020, 263, 118542. [Google Scholar] [CrossRef]
- Manohar, A.; Vijayakanth, V.; Kim, K.H. Influence of Ca doping on ZnFe2O4 nanoparticles magnetic hyperthermia and cytotoxicity study. J. Alloy. Compd. 2021, 886, 161276. [Google Scholar] [CrossRef]
- Kononenko, V.; Narat, M.; Drobne, D. Nanoparticle interaction with the immune system. Arch. Ind. Hyg. Toxicol. 2015, 66, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Wu, R.; Ming, L. The role of m6A RNA methylation in cancer. Biomed. Pharmacother. 2019, 112, 108613. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z. Nanoparticles induced embryo–fetal toxicity. Toxicol. Ind. Health 2020, 36, 181–213. [Google Scholar] [CrossRef]
- Ahsan, S.M.; Rao, C.M.; Ahmad, M.F. Nanoparticle-protein interaction: The significance and role of protein corona. Adv. Exp. Med. Biol. 2018, 1048, 175–198. [Google Scholar] [CrossRef]
- Dinischiotu, A.; Stanca, L.; Gradinaru, D.; Petrache, S.N.; Radu, M.; Serban, A.I. Lipid peroxidation due to in vitro and in vivo exposure of biological samples to nanoparticles. Methods Mol. Biol. 2013, 1028, 155–164. [Google Scholar] [CrossRef]
- Fu, P.P.; Xia, Q.; Hwang, H.-M.; Ray, P.C.; Yu, H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014, 22, 64–75. [Google Scholar] [CrossRef] [Green Version]
- Manshian, B.B.; Poelmans, J.; Saini, S.; Pokhrel, S.; Grez, J.J.; Himmelreich, U.; Mädler, L.; Soenen, S.J. Nanoparticle-induced inflammation can increase tumor malignancy. Acta Biomater. 2018, 68, 99–112. [Google Scholar] [CrossRef]
- Pogribna, M.; Hammons, G. Epigenetic effects of nanomaterials and nanoparticles. J. Nanobiotechnol. 2021, 19, 2. [Google Scholar] [CrossRef]
- Sierra, M.I.; Valdés, A.; Fernández, A.; Torrecillas, R.; Fraga, M.F. The effect of exposure to nanoparticles and nanomaterials on the mammalian epigenome. Int. J. Nanomed. 2016, 11, 6297–6306. [Google Scholar] [CrossRef] [Green Version]
- Wong, B.S.E.; Hu, Q.; Baeg, G.H. Epigenetic modulations in nanoparticle-mediated toxicity. Food Chem. Toxicol. 2017, 109, 746–752. [Google Scholar] [CrossRef]
- Cayir, A.; Byun, H.-M.; Barrow, T.M. Environmental epitranscriptomics. Environ. Res. 2020, 189, 109885. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Li, H.-B.; Yin, Z.; Flavell, R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016, 6, 160003. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.; Mumbach, M.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Boissel, S.; Reish, O.; Proulx, K.; Kawagoe-Takaki, H.; Sedgwick, B.; Yeo, G.S.H.; Meyre, D.; Golzio, C.; Molinari, F.; Kadhom, N.; et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009, 85, 106–111. [Google Scholar] [CrossRef] [Green Version]
- Dina, C.; Meyre, D.; Gallina, S.; Durand, E.; Körner, A.; Jacobson, P.; Carlsson, L.M.S.; Kiess, W.; Vatin, V.; Lecoeur, C.; et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007, 39, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef] [Green Version]
- Scuteri, A.; Sanna, S.; Chen, W.-M.; Uda, M.; Albai, G.; Strait, J.; Najjar, S.; Nagaraja, R.; Orrú, M.; Usala, G.; et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007, 3, e115. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fu, Y.; Klungland, A.; Yang, Y.-G.; He, C. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013, 10, 915–918. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, B.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2013, 505, 117–120. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Dong, L.; Liu, X.-M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.-B. M6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Patil, D.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A promotes cap-independent translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zou, Q.; Ding, J.; Ling, M.; Wang, W.; Li, H.; Huang, B. Increased N6-methyladenosine causes infertility is associated with FTO expression. J. Cell. Physiol. 2018, 233, 7055–7066. [Google Scholar] [CrossRef] [PubMed]
- Lan, N.; Lu, Y.; Zhang, Y.; Pu, S.; Xi, H.; Nie, X.; Liu, J.; Yuan, W. FTO—A common genetic basis for obesity and cancer. Front. Genet. 2020, 11, 559138. [Google Scholar] [CrossRef]
- Meng, T.-G.; Lu, X.; Guo, L.; Hou, G.-M.; Ma, X.-S.; Li, Q.-N.; Huang, L.; Fan, L.-H.; Zhao, Z.-H.; Ou, X.-H.; et al. Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. FASEB J. 2018, 33, 1179–1187. [Google Scholar] [CrossRef]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA 2018, 24, 499–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Coker, H.; Wei, G.; Brockdorff, N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta 2018, 1862, 310–318. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, E.; Cui, Y.-H.; He, Y.-Y. Context-dependent roles of RNA modifications in stress responses and diseases. Int. J. Mol. Sci. 2021, 22, 1949. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ishfaq, M.; Xu, L.; Xia, C.; Chen, C.; Li, J. METTL3/m6A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front. Pharmacol. 2019, 10, 517. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, C.; Zhuang, C.; Luo, X.; Huang, X.; Yao, L.; Li, J.; Li, Y.; Xiong, T.; Ye, J.; Zhang, F.; et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1α signalling axis. J. Cell. Mol. Med. 2019, 23, 2163–2173. [Google Scholar] [CrossRef]
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 4892. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Wei, X.; Jian, Z.; Zhang, X. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 2020, 9, 2859–2867. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Li, Y.; Wang, T.; Zhong, X. Modification of N6-methyladenosine RNA methylation on heat shock protein expression. PLoS ONE 2018, 13, e0198604. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Kovaříková, A.S.; Stixová, L.; Kovařík, A.; Komůrková, D.; Legartová, S.; Fagherazzi, P.; Bártová, E. N6-adenosine methylation in RNA and a reduced m3G/TMG level in non-coding RNAs appear at microirradiation-induced DNA lesions. Cells 2020, 9, 360. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Riddle, R.C.; Yang, Q.; Rosen, C.R.; Guttridge, D.C.; Dirckx, N.; Faugere, M.-C.; Farber, C.R.; Clemens, T.L. The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc. Natl. Acad. Sci. USA 2019, 116, 17980–17989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. M6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2019, 16, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.; Miao, Y.; Liang, P.; Zhu, K.; She, Y.; Wu, Y.; Liu, D.-A.; Huang, J.; Ren, J.; et al. M6A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018, 28, 955–957. [Google Scholar] [CrossRef]
- Gao, Y.; Vasic, R.; Song, Y.; Teng, R.; Liu, C.; Gbyli, R.; Biancon, G.; Nelakanti, R.; Lobben, K.; Kudo, E.; et al. M6A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity 2020, 52, 1007.e8–1021.e8. [Google Scholar] [CrossRef]
- Vu, L.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; Mackay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Wang, H.; Zuo, H.; Liu, J.; Wen, F.; Gao, Y.; Zhu, X.; Liu, B.; Xiao, F.; Wang, W.; Huang, G.; et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018, 28, 1035–1038. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Yusheng, C.; Ding, Y.; Xue, Y.; et al. M6A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef]
- Li, M.; Zhao, X.; Wang, W.; Shi, H.; Pan, Q.; Lu, Z.; Perez, S.P.; Suganthan, R.; He, C.; Bjørås, M.; et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018, 19, 69. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411. [Google Scholar] [CrossRef]
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 2017, 171, 877–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Zhang, X.; Weng, Y.-L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. M6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Zhong, C.; Wu, X.; Chen, J.; Tao, B.; Xia, X.; Shi, M.; Zhu, Z.; Trudeau, V.L.; Hu, W. Mettl3 mutation disrupts gamete maturation and reduces fertility in zebrafish. Genetics 2018, 208, 729–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landfors, M.; Nakken, S.; Fusser, M.; Dahl, J.-A.; Klungland, A.; Fedorcsak, P. Sequencing of FTO and ALKBH5 in men undergoing infertility work-up identifies an infertility-associated variant and two missense mutations. Fertil. Steril. 2016, 105, 1170–1179.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasowitz, S.; Ma, J.; Anderson, S.J.; Leu, N.A.; Xu, Y.; Gregory, B.D.; Schultz, R.M.; Wang, P.J. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018, 14, e1007412. [Google Scholar] [CrossRef]
- Qi, S.; Ma, J.-Y.; Wang, Z.-B.; Guo, L.; Hou, Y.; Sun, Q.-Y. N6-methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J. Biol. Chem. 2016, 291, 23020–23026. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Zhou, Y.; Wu, H.; Lv, X.; Yang, L.; Ren, Z.; Tian, H.; Yu, Q.; Li, J.; Lin, W.; et al. RNA demethylase FTO mediated RNA m6A modification is involved in maintaining maternal-fetal interface in spontaneous abortion. Front. Cell Dev. Biol. 2021, 9, 617172. [Google Scholar] [CrossRef]
- Li, X.-C.; Jin, F.; Wang, B.-Y.; Yin, X.-J.; Hong, W.; Tian, F.-J. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics 2019, 9, 3853–3865. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Akira, S. Toll-like receptor and RIG-1-like receptor signaling. Ann. N. Y. Acad. Sci. 2008, 1143, 1–20. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Lina, K.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Hu, X.; Huang, M.; Liu, J.; Gu, Y.; Ma, L.; Zhou, Q.; Cao, X. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019, 10, 1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Zhang, M.; Wu, J.; Wang, S.; Yang, X.; Yi, M.; Zhang, X.; Fang, X. Exploring diagnostic m6A regulators in endometriosis. Aging 2020, 12, 25916–25938. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Zhang, C.; Zhang, G. M6A RNA methylation controls proliferation of human glioma cells by influencing cell apoptosis. Cytogenet. Genome Res. 2019, 159, 119–125. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. M6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Paris, J.; Morgan, M.; Campos, J.; Spencer, G.J.; Shmakova, A.; Ivanova, I.; Mapperley, C.; Lawson, H.; Wotherspoon, D.A.; Sepulveda, C.; et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019, 25, 137.e6–148.e6. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell 2020, 27, 64.e9–80.e9. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Wang, P.; Han, G.; Zhang, T.; Chang, J.; Yin, R.; Shan, Y.; Wen, J.; Xie, X.; et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 2020, 27, 81.e8–97.e8. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Namshik, H.; Millan-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Hannes, P.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef]
- Kwok, C.-T.; Marshall, A.; Rasko, J.E.J.; Wong, J.J.L. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol. 2017, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- De Jesus, D.F.; Zhang, Z.; Brown, N.K.; Hu, J.; Ahriman, S.; Mathews, C.E.; Powers, A.C.; Atkinson, M.A.; Eizirik, D.L.; He, C.; et al. 287-OR: M6A mRNA methylation regulates the innate immune response in human β-Cells. Diabetes 2021, 70. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, F.; Huang, W.; Qin, S.; Huang, J.-T.; Sergi, C.; Yuan, B.-F.; Liu, S.-M. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2018, 104, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.-Z.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-resolution N6-methyladenosine (m6A) map using photo-crosslinking-assisted m6A sequencing. Angew. Chem. Int. Ed. 2014, 54, 1587–1590. [Google Scholar] [CrossRef] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Liu, N.; Pan, T. Probing RNA modification status at single-nucleotide resolution in total RNA. Methods Enzymol. 2015, 560, 149–159. [Google Scholar] [CrossRef]
- Shu, X.; Cao, J.; Cheng, M.; Xiang, S.; Gao, M.; Li, T.; Ying, X.; Wang, F.; Yue, Y.; Lu, Z.; et al. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 2020, 16, 887–895. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.; Xu, Z.; Cao, M.; Hu, Q.; Pan, C.; Guo, M.; Wei, J.; Yang, H. Detection of N6-methyladenosine modification residues (Review). Int. J. Mol. Med. 2019, 43, 2267–2278. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Wang, L.; Mettenbrink, E.M.; DeAngelis, P.L.; Wilhelm, S. Nanoparticle toxicology. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Ordaz, J.; Lo, C.-L.; Damayanti, N.P.; Zhou, F.; Irudayaraj, J. ZnO nanoparticles induced reactive oxygen species promotes multimodal cyto- and epigenetic toxicity. Toxicol. Sci. 2017, 156, 261–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, C.; Lee, Q.Y.; Cai, Y.; Liu, X.; Ding, J.; Yung, L.-Y.L.; Bay, B.-H.; Baeg, G.-H. Silver nanoparticles disrupt germline stem cell maintenance in the Drosophila testis. Sci. Rep. 2016, 6, 20632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Ruan, F.; Jiang, S.; Zeng, J.; Yin, H.; Liu, R.; Zhang, Y.; Huang, L.; Wang, C.; Ma, S.; et al. Black phosphorus quantum dots cause nephrotoxicity in organoids, mice, and human cells. Small 2020, 16, e2001371. [Google Scholar] [CrossRef]
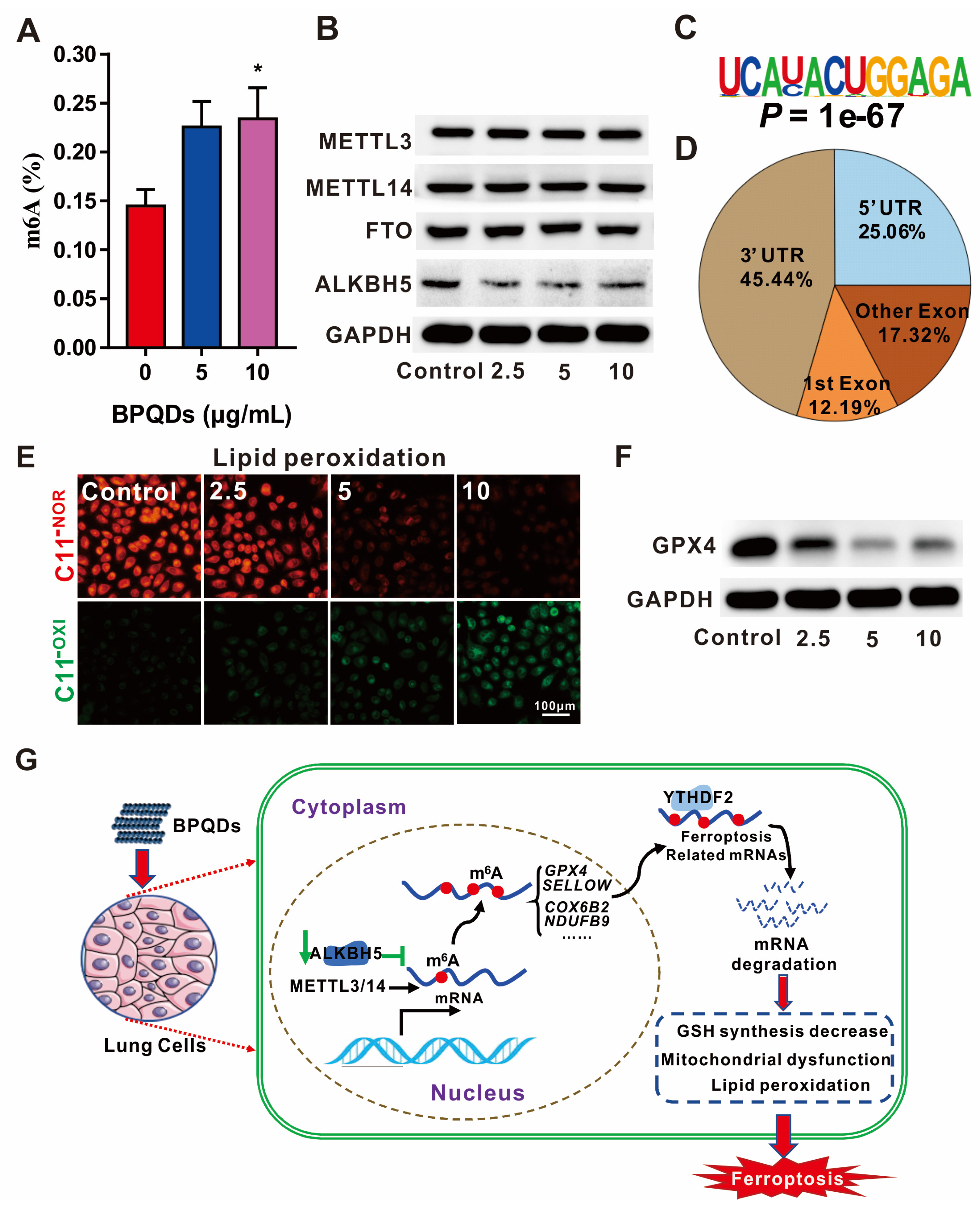
- Ruan, F.; Zeng, J.; Yin, H.; Jiang, S.; Cao, X.; Zheng, N.; Han, C.; Zhang, C.; Zuo, Z.; He, C. RNA m6A modification alteration by black phosphorus quantum dots regulates cell ferroptosis: Implications for nanotoxicological assessment. Small Methods 2021, 5, 2001045. [Google Scholar] [CrossRef]
- Han, B.; Chu, C.; Su, X.; Zhang, N.; Zhou, L.; Zhang, M.; Yang, S.; Shi, L.; Zhao, B.; Niu, Y.; et al. N6-methyladenosine-dependent primary microRNA-126 processing activated PI3K-AKT-mTOR pathway drove the development of pulmonary fibrosis induced by nanoscale carbon black particles in rats. Nanotoxicology 2019, 14, 1–20. [Google Scholar] [CrossRef]
- Zhang, S.; Meng, P.; Cheng, S.; Jiang, X.; Zhang, J.; Qin, X.; Tang, Q.; Bai, L.; Zou, Z.; Chen, C. Pregnancy exposure to carbon black nanoparticles induced neurobehavioral deficits that are associated with altered m6A modification in offspring. NeuroToxicology 2020, 81, 40–50. [Google Scholar] [CrossRef]
- Yang, Z.; Deng, C.; Wu, Y.; Dai, Z.; Tang, Q.; Cheng, C.; Xu, Y.; Hu, R.; Liu, C.; Chen, X.; et al. Insights into the mechanism of multi-walled carbon nanotubes phytotoxicity in Arabidopsis through transcriptome and m6A methylome analysis. Sci. Total Environ. 2021, 787, 147510. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, J.; Zheng, M.; Yang, Z.; Zhao, L.; Zhang, W.; Zhong, W.; Chen, Y.; Lin, J. Metal–protein nanoparticles facilitate anti-VSV and H1N1 viruses through the coordinative actions on innate immune responses and METTL14. Macromol. Biosci. 2021, 21, 2000382. [Google Scholar] [CrossRef]
- Kunovac, A.; Hathaway, Q.A.; Pinti, M.V.; Durr, A.J.; Taylor, A.D.; Goldsmith, W.T.; Garner, K.L.; Nurkiewicz, T.R.; Hollander, J.M. Enhanced antioxidant capacity prevents epitranscriptomic and cardiac alterations in adult offspring gestationally-exposed to ENM. Nanotoxicology 2021, 15, 812–831. [Google Scholar] [CrossRef]
- Yin, H.; Wang, H.; Jiang, W.; Zhou, Y.; Ai, S. Electrochemical immunosensor for N6-methyladenosine detection in human cell lines based on biotin-streptavidin system and silver-SiO2 signal amplification. Biosens. Bioelectron. 2017, 90, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, S.; Gunasekaran, K.; Sasidharan, S.; Mathan, V.J.; Perumal, E. MicroRNAs and xenobiotic toxicity: An overview. Toxicol. Rep. 2020, 7, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Pogribna, M.; Koonce, N.A.; Mathew, A.; Word, B.; Patri, A.K.; Lyn-Cook, B.; Hammons, G. Effect of titanium dioxide nanoparticles on DNA methylation in multiple human cell lines. Nanotoxicology 2020, 14, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Rossner, J.P.; Vrbova, K.; Rossnerova, A.; Zavodna, T.; Milcova, A.; Klema, J.; Vecera, Z.; Mikuska, P.; Coufalik, P.; Capka, L.; et al. Gene expression and epigenetic changes in mice following inhalation of copper (II) oxide nanoparticles. Nanomaterials 2020, 10, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Yang, D.; Ma, X.-X. Immune infiltration-related N6-methyladenosine RNA methylation regulators influence the malignancy and prognosis of endometrial cancer. Aging 2021, 13, 16287–16315. [Google Scholar] [CrossRef]
- Rossnerova, A.; Honkova, K.; Pelclova, D.; Zdimal, V.; Hubacek, J.A.; Chvojkova, I.; Vrbova, K.; Rossner, J.P.; Topinka, J.; Vlckova, S.; et al. DNA methylation profiles in a group of workers occupationally exposed to nanoparticles. Int. J. Mol. Sci. 2020, 21, 2420. [Google Scholar] [CrossRef] [Green Version]
- Poma, A.; Colafarina, S.; Fontecchio, G.; Chichiriccò, G. Transgenerational Effects of NMs; Springer: Berlin/Heidelberg, Germany, 2014; Volume 811, pp. 235–254. [Google Scholar] [CrossRef]
- Bhagat, J.; Zang, L.; Nishimura, N.; Shimada, Y. Zebrafish: An emerging model to study microplastic and nanoplastic toxicity. Sci. Total Environ. 2020, 728, 138707. [Google Scholar] [CrossRef]
- Hu, J.; Lin, W.; Lin, B.; Wu, K.; Fan, H.; Yu, Y. Persistent DNA methylation changes in zebrafish following graphene quantum dots exposure in surface chemistry-dependent manner. Ecotoxicol. Environ. Saf. 2018, 169, 370–375. [Google Scholar] [CrossRef]
- Yee, M.; Hii, L.-W.; Looi, C.; Lim, W.-M.; Wong, S.-F.; Kok, Y.-Y.; Tan, B.-K.; Wong, C.-Y.; Leong, C.-O. Impact of microplastics and nanoplastics on human health. Nanomaterials 2021, 11, 496. [Google Scholar] [CrossRef]
- Guo, P.X. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [Google Scholar] [CrossRef]
| Methods | Resolution | Sample RNA Demand (n) | Need for Antibodies | References |
|---|---|---|---|---|
| MeRIP-seq | 100–200 nt | Extremely large | Yes | [95] |
| PA-m6A-seq | 20–30 nt | Large | Yes | [96] |
| miCLIP-seq | Single base | Large | Yes | [97] |
| SCARLET | Single base | Large | No | [98] |
| m6A-REF-seq | Single base | Little | No | [99] |
| Colorimetry | Total amount of m6A | Little | Yes | [100] |
| m6A dot-blot | Large | Yes | ||
| HPLC–MS/MS | Large | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ruan, F.; Zuo, Z.; He, C. Nanoparticle-Induced m6A RNA Modification: Detection Methods, Mechanisms and Applications. Nanomaterials 2022, 12, 389. https://doi.org/10.3390/nano12030389
Wang Y, Ruan F, Zuo Z, He C. Nanoparticle-Induced m6A RNA Modification: Detection Methods, Mechanisms and Applications. Nanomaterials. 2022; 12(3):389. https://doi.org/10.3390/nano12030389
Chicago/Turabian StyleWang, Yi, Fengkai Ruan, Zhenghong Zuo, and Chengyong He. 2022. "Nanoparticle-Induced m6A RNA Modification: Detection Methods, Mechanisms and Applications" Nanomaterials 12, no. 3: 389. https://doi.org/10.3390/nano12030389
APA StyleWang, Y., Ruan, F., Zuo, Z., & He, C. (2022). Nanoparticle-Induced m6A RNA Modification: Detection Methods, Mechanisms and Applications. Nanomaterials, 12(3), 389. https://doi.org/10.3390/nano12030389