Artificial Lipid Droplets: Novel Effective Biomaterials to Protect Cells against Oxidative Stress and Lipotoxicity
Abstract
:1. Introduction
2. Material and Methods
2.1. Materials
2.2. Construction of Nanolipid Particles
2.3. Cell Culture
2.4. Cellular Uptake
2.5. Reactive Oxygen Species (ROS) Measure
2.6. In Vitro H2O2 Concentration Measurement
2.7. Cell Counting Kit-8 (CCK8)
2.8. Protein Expression and Purification
2.9. Antibodies
2.10. Isolation of Lipid Droplets (LDs)
2.11. Isolation of Mitochondria
2.12. Nanolipid Particle Lyophilization
2.13. Fluorescence Microscopy
2.14. Cryo-Scanning Electron Microscopy (SEM)
2.15. Lipid Analysis by Thin-Layer Chromatography
2.16. Recruitment of Proteins to NLPs
2.17. Silver Staining
2.18. Statistical Analyses
3. Results
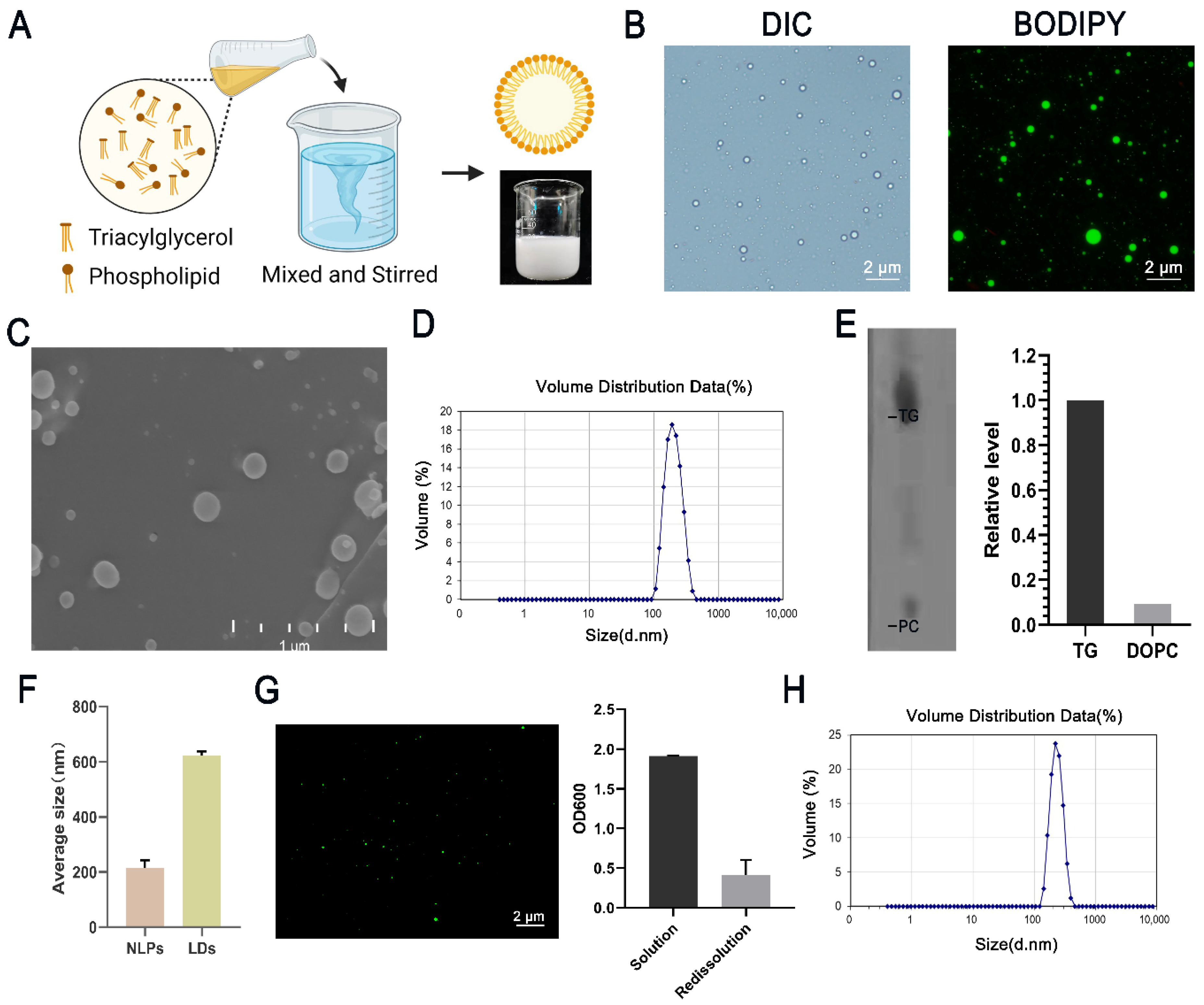
3.1. Formation of Nanolipid Particles and Their Characters
3.2. Stability and Utilization of Nanolipid Particles
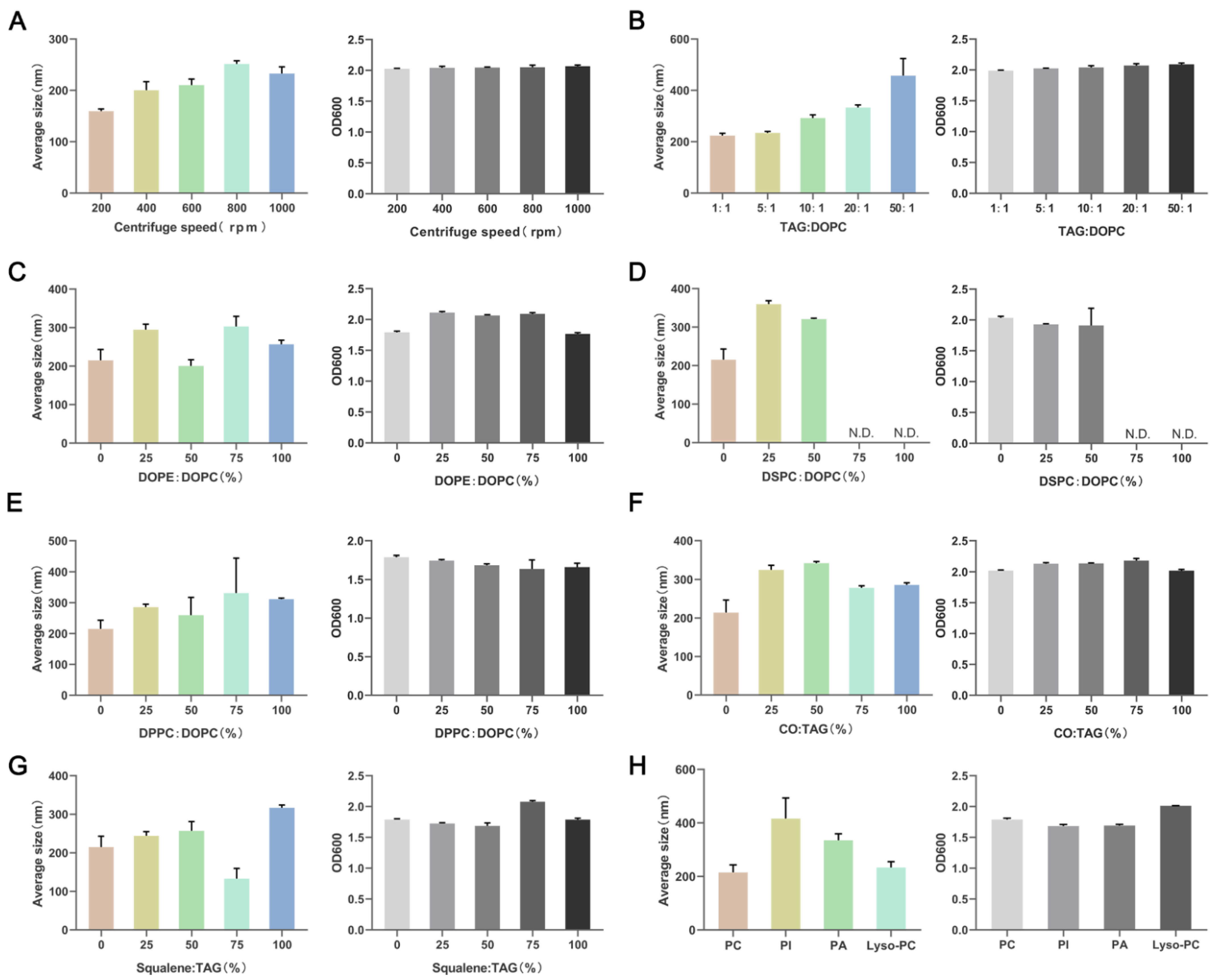
3.3. Lipid Composition Affects the Particle Size of NLPs
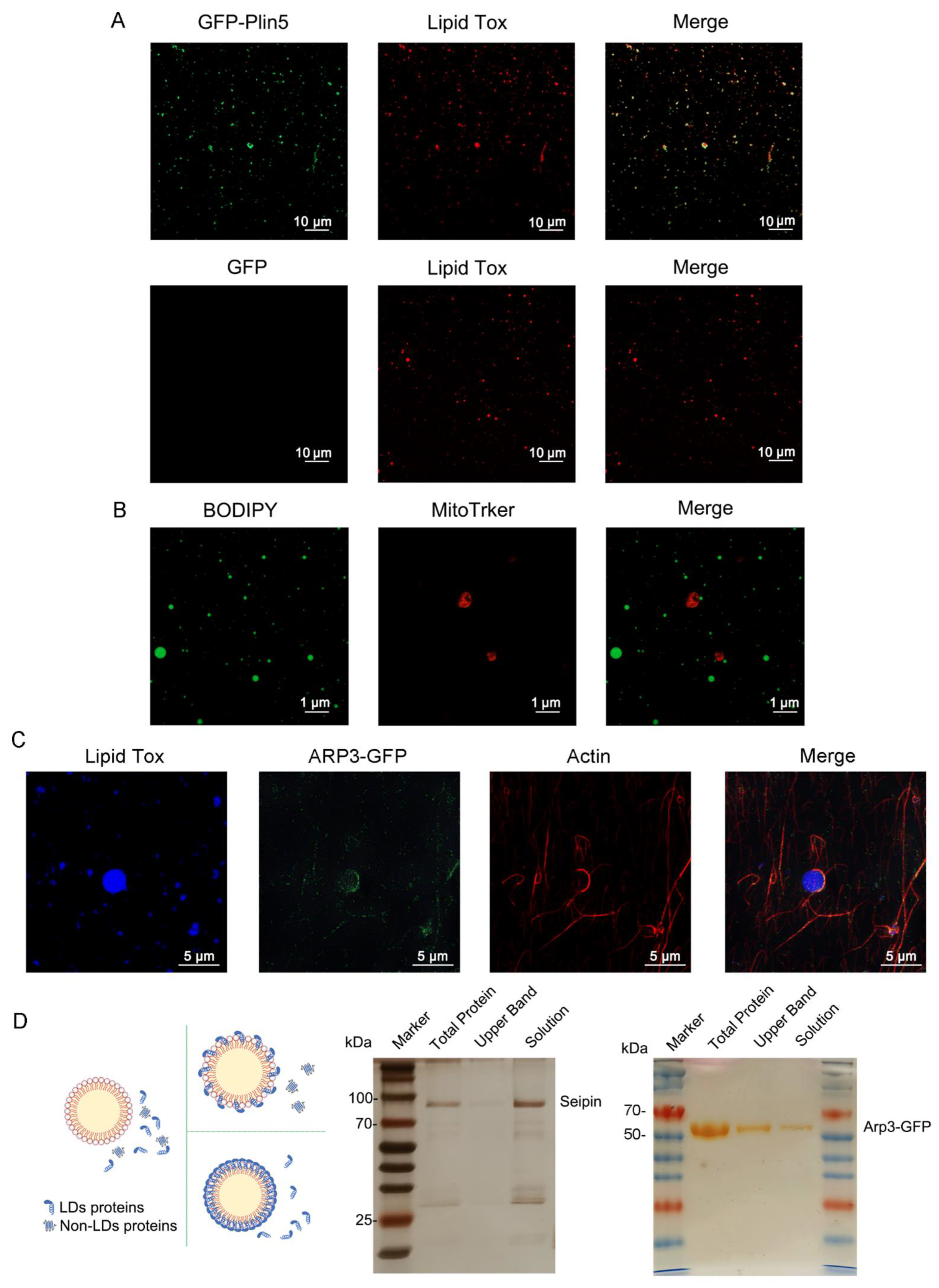
3.4. Organelles and LD-Resident Protein Recruitment of NLPs
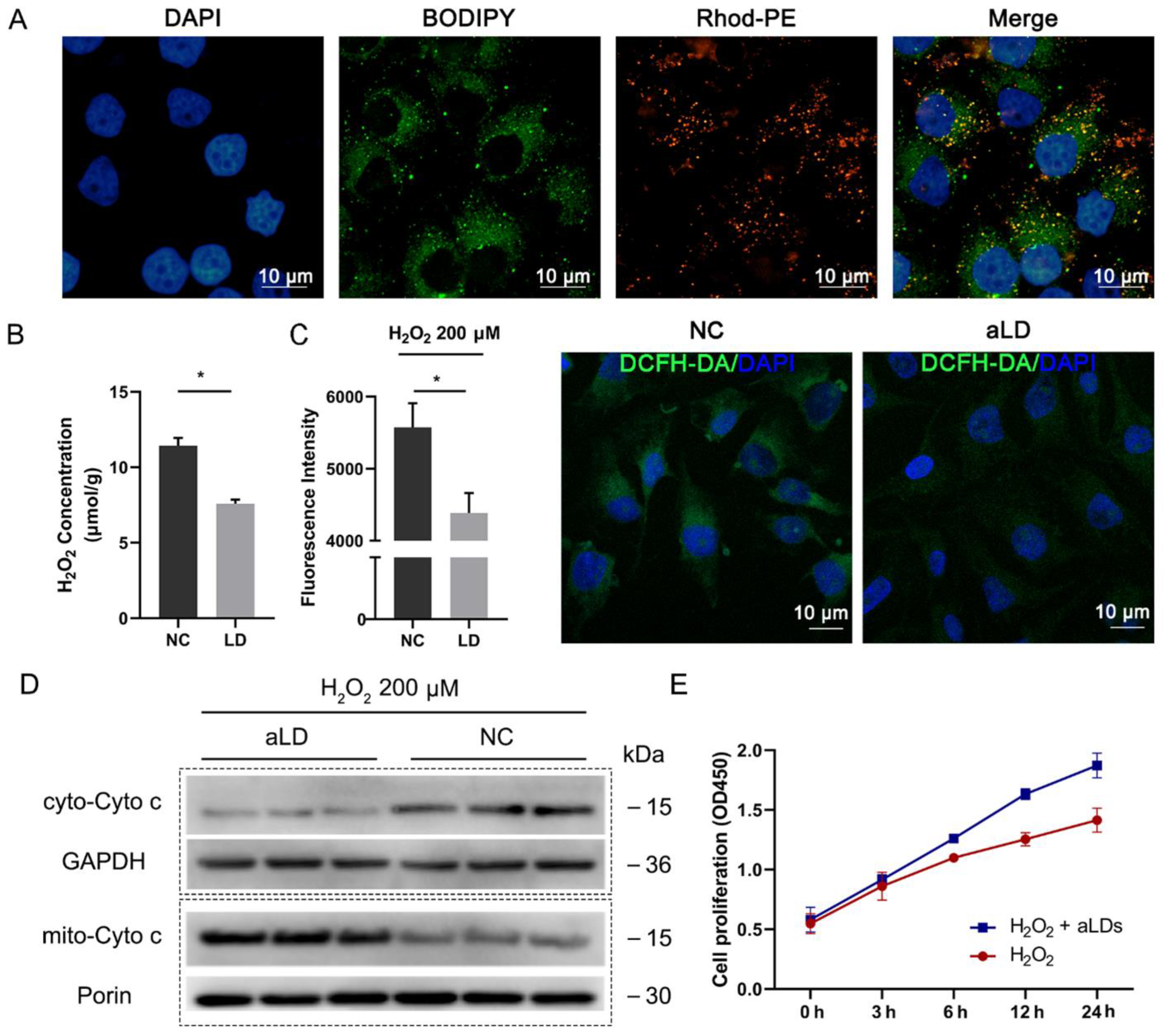
3.5. Perilipin-Coated aLDs Promote Cells to Protect against the ROS
3.6. Perilipin-Coated aLDs Promote Protection by Cells against Lipotoxicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, there, and everywhere: The importance of ER membrane contact sites. Science 2018, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, K. Lipid droplets from the inside. Nat. Rev. Mol. Cell Biol. 2018, 19, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herms, A.; Bosch, M.; Ariotti, N.; Reddy, B.J.; Fajardo, A.; Fernández-Vidal, A.; Alvarez-Guaita, A.; Fernandez-Rojo, M.A.; Rentero, C.; Tebar, F.; et al. Cell-to-Cell Heterogeneity in Lipid Droplets Suggests a Mechanism to Reduce Lipotoxicity. Curr. Biol. 2013, 23, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Cabodevilla, A.G.; Sánchez-Caballero, L.; Nintou, E.; Boiadjieva, V.G.; Picatoste, F.; Gubern, A.; Claro, E. Cell Survival during Complete Nutrient Deprivation Depends on Lipid Droplet-fueled β-Oxidation of Fatty Acids. J. Biol. Chem. 2013, 288, 27777–27788. [Google Scholar] [CrossRef] [Green Version]
- Qiu, B.; Ackerman, D.; Sanchez, D.J.; Li, B.; Ochocki, J.D.; Grazioli, A.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; Simon, M.C. HIF2α-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015, 5, 652–667. [Google Scholar] [CrossRef] [Green Version]
- Bailey, A.P.; Koster, G.; Guillermier, C.; Hirst, E.M.; MacRae, J.I.; Lechene, C.P.; Postle, A.D.; Gould, A.P. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell 2015, 163, 340–353. [Google Scholar] [CrossRef] [Green Version]
- Welte, M.A. How Brain Fat Conquers Stress. Cell 2015, 163, 269–270. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, J.E. Lipotoxicity: When tissues overeat. Curr. Opin. Lipidol. 2003, 14, 281–287. [Google Scholar] [CrossRef]
- Petan, T.; Jarc, E.; Jusovic, M. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sreenivasan, U.; Hu, H.; Saladino, A.; Polster, B.M.; Lund, L.M.; Gong, D.-W.; Stanley, W.C.; Sztalryd, C. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 2011, 52, 2159–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granneman, J.G.; Moore, H.-P.H.; Krishnamoorthy, R.; Rathod, M. Perilipin Controls Lipolysis by Regulating the Interactions of AB-hydrolase Containing 5 (Abhd5) and Adipose Triglyceride Lipase (Atgl). J. Biol. Chem. 2009, 284, 34538–34544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granneman, J.G.; Moore, H.-P.H.; Mottillo, E.P.; Zhu, Z.; Zhou, L. Interactions of Perilipin-5 (Plin5) with Adipose Triglyceride Lipase. J. Biol. Chem. 2011, 286, 5126–5135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najt, C.P.; Khan, S.A.; Heden, T.D.; Witthuhn, B.A.; Perez, M.; Heier, J.L.; Mead, L.E.; Franklin, M.P.; Karanja, K.K.; Graham, M.J.; et al. Lipid Droplet-Derived Monounsaturated Fatty Acids Traffic via PLIN5 to Allosterically Activate SIRT1. Mol. Cell 2020, 77, 810–824.e8. [Google Scholar] [CrossRef]
- Greenwood, D.J.; Dos Santos, M.S.; Huang, S.; Russell, M.R.G.; Collinson, L.M.; MacRae, J.I.; West, A.; Jiang, H.; Gutierrez, M.G. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science 2019, 364, 1279–1282. [Google Scholar] [CrossRef] [Green Version]
- Dubey, R.; Stivala, C.E.; Nguyen, H.Q.; Goo, Y.-H.; Paul, A.; Carette, J.E.; Trost, B.M.; Rohatgi, R. Lipid droplets can promote drug accumulation and activation. Nat. Chem. Biol. 2020, 16, 206–213. [Google Scholar] [CrossRef]
- Liang, T.; Wen, D.; Chen, G.; Chan, A.; Chen, Z.; Li, H.; Wang, Z.; Han, X.; Jiang, L.; Zhu, J.J.; et al. Adipocyte-Derived Anticancer Lipid Droplets. Adv. Mater. 2021, 33, 2100629. [Google Scholar] [CrossRef]
- Martin, S.; Parton, R.G. Lipid droplets: A unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006, 7, 373–378. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, S.; Yang, L.; Na, H.; Zhang, P.; Zhang, H.; Wang, Y.; Chen, Y.; Yu, J.; Huo, C.; et al. Isolating lipid droplets from multiple species. Nat. Protoc. 2013, 8, 43–51. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Li, J.; Yu, J.; Pu, J.; Li, L.; Zhang, H.; Zhang, S.; Peng, G.; Yang, F.; et al. Proteome of Skeletal Muscle Lipid Droplet Reveals Association with Mitochondria and Apolipoprotein A-I. J. Proteome Res. 2011, 10, 4757–4768. [Google Scholar] [CrossRef]
- Weiss, M.; Frohnmayer, J.P.; Benk, L.T.; Haller, B.; Janiesch, J.-W.; Heitkamp, T.; Börsch, M.; Lira, R.B.; Dimova, R.; Lipowsky, R.; et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 2018, 17, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, X.M.; Ma, X.J.; Du, Y.L.; Zheng, L.M.; Liu, P.S. Construction of Nanodroplet/Adiposome and Artificial Lipid Droplets. ACS Nano 2016, 10, 3312–3322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, L.; Ding, Y.; Wang, Y.; Lan, L.; Ma, Q.; Chi, X.; Wei, P.; Zhao, Y.; Steinbüchel, A.; et al. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 2017, 8, 15979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhi, Z.; Zhang, S.; Zhou, C.; Mechler, A.; Liu, P. Validating an artificial organelle: Studies of lipid droplet-specific proteins on adiposome platform. IScience 2021, 24, 102834. [Google Scholar] [CrossRef] [PubMed]
- Thiam, A.R.; Ikonen, E. Lipid Droplet Nucleation. Trends Cell Biol. 2021, 31, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Thiam, A.R.; Antonny, B.; Wang, J.; Delacotte, J.; Wilfling, F.; Walther, T.C.; Beck, R.; Rothman, J.E.; Pincet, F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. USA 2013, 110, 13244–13249. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Jin, Y.; Li, M.; Wang, H.; Xiong, M.; Zeng, W.; Yuan, H.; Liu, C.; Ren, Z.; Liu, C. Faster and More Specific: Excited-State Intramolecular Proton Transfer-Based Dyes for High-Fidelity Dynamic Imaging of Lipid Droplets within Cells and Tissues. Anal. Chem. 2020, 92, 10342–10349. [Google Scholar] [CrossRef]
- Long, T.; Sun, Y.; Hassan, A.; Qi, X.; Li, X. Structure of nevanimibe-bound tetrameric human ACAT1. Nature 2020, 581, 339–343. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Benador, I.Y.; Veliova, M.; Mahdaviani, K.; Petcherski, A.; Wikstrom, J.D.; Assali, E.A.; Acin-Perez, R.; Shum, M.; Oliveira, M.F.; Cinti, S.; et al. Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab. 2018, 27, 869–885.e6. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Mirza, A.H.; Zhang, S.; Liang, B.; Liu, P. Lipid droplets and mitochondria are anchored during brown adipocyte differentiation. Protein Cell 2019, 10, 921–926. [Google Scholar] [CrossRef] [Green Version]
- Pfisterer, S.G.; Gateva, G.; Horvath, P.; Pirhonen, J.; Salo, V.T.; Karhinen, L.; Varjosalo, M.; Ryhänen, S.J.; Lappalainen, P.; Ikonen, E. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat. Commun. 2017, 8, 14858. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Schuster, M.; Guimarães, S.C.; Ashwin, P.; Schrader, M.; Metz, J.; Hacker, C.; Gurr, S.J.; Steinberg, G. Active diffusion and microtubule-based transport oppose myosin forces to position organelles in cells. Nat. Commun. 2016, 7, 11814. [Google Scholar] [CrossRef]
- Salo, V.T.; Li, S.Q.; Vihinen, H.; Holtta-Vuori, M.; Szkalisity, A.; Horvath, P.; Belevich, I.; Peranen, J.; Thiele, C.; Somerharju, P.; et al. Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev. Cell 2019, 50, 478–493.e9. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Tan, Y.; Zhao, P.; Ren, Z. SEIPIN: A Key Factor for Nuclear Lipid Droplet Generation and Lipid Homeostasis. Int. J. Mol. Sci. 2020, 21, 8208. [Google Scholar] [CrossRef]
- Jarc, E.; Petan, T. Lipid Droplets and the Management of Cellular Stress. Yale J. Biol. Med. 2019, 92, 435–452. [Google Scholar]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.B.; Louie, S.M.; Daniele, J.R.; Tran, Q.; Dillin, A.; Zoncu, R.; Nomura, D.K.; Olzmann, J.A. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell 2017, 42, 9–21.e5. [Google Scholar] [CrossRef] [Green Version]
- Bischof, J.; Salzmann, M.; Streubel, M.K.; Hasek, J.; Geltinger, F.; Duschl, J.; Bresgen, N.; Briza, P.; Haskova, D.; Lejskova, R.; et al. Clearing the outer mitochondrial membrane from harmful proteins via lipid droplets. Cell Death Discov. 2017, 3, 17016. [Google Scholar] [CrossRef]
- Ben M’barek, K.; Ajjaji, D.; Chorlay, A.; Vanni, S.; Foret, L.; Thiam, A.R. ER Membrane Phospholipids and Surface Tension Control Cellular Lipid Droplet Formation. Dev. Cell 2017, 41, 591–604.e7. [Google Scholar] [CrossRef] [Green Version]
- Chorlay, A.; Monticelli, L.; Ferreira, J.V.; Ben M’barek, K.; Ajjaji, D.; Wang, S.H.; Johnson, E.; Beck, R.; Omrane, M.; Beller, M.; et al. Membrane Asymmetry Imposes Directionality on Lipid Droplet Emergence from the ER. Dev. Cell 2019, 50, 25–42.e7. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, P.; Jin, Y.; Wu, X.; Huang, J.; Chen, L.; Tan, Y.; Yuan, H.; Wu, J.; Ren, Z. Artificial Lipid Droplets: Novel Effective Biomaterials to Protect Cells against Oxidative Stress and Lipotoxicity. Nanomaterials 2022, 12, 672. https://doi.org/10.3390/nano12040672
Zhao P, Jin Y, Wu X, Huang J, Chen L, Tan Y, Yuan H, Wu J, Ren Z. Artificial Lipid Droplets: Novel Effective Biomaterials to Protect Cells against Oxidative Stress and Lipotoxicity. Nanomaterials. 2022; 12(4):672. https://doi.org/10.3390/nano12040672
Chicago/Turabian StyleZhao, Pengxiang, Yi Jin, Xiang Wu, Jin Huang, Lupeng Chen, Yanjie Tan, Hong Yuan, Jian Wu, and Zhuqing Ren. 2022. "Artificial Lipid Droplets: Novel Effective Biomaterials to Protect Cells against Oxidative Stress and Lipotoxicity" Nanomaterials 12, no. 4: 672. https://doi.org/10.3390/nano12040672
APA StyleZhao, P., Jin, Y., Wu, X., Huang, J., Chen, L., Tan, Y., Yuan, H., Wu, J., & Ren, Z. (2022). Artificial Lipid Droplets: Novel Effective Biomaterials to Protect Cells against Oxidative Stress and Lipotoxicity. Nanomaterials, 12(4), 672. https://doi.org/10.3390/nano12040672




