Supramolecular Nanostructures Based on Perylene Diimide Bioconjugates: From Self-Assembly to Applications
Abstract
:1. Introduction
2. Synthesis and Properties of PDI Derivatives
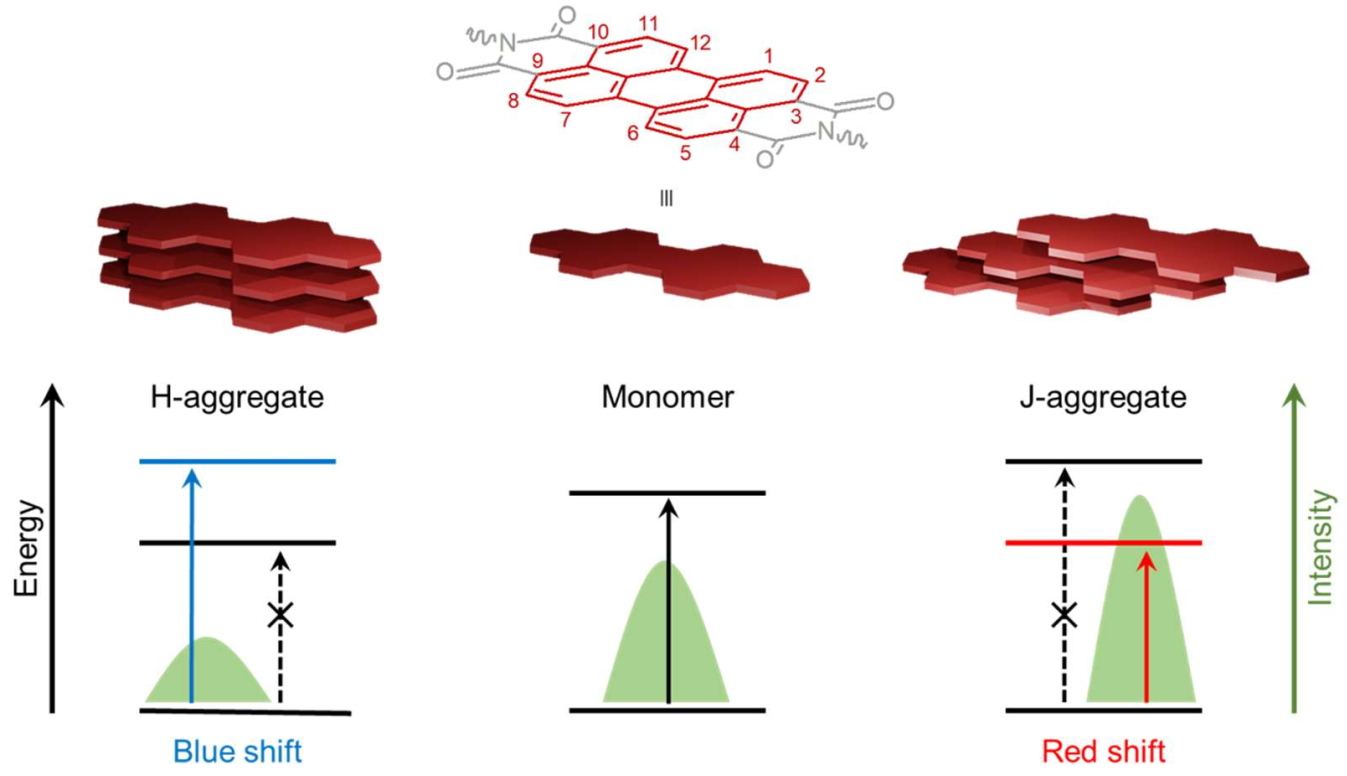
2.1. Physicochemical, Optical, and Self-Assembling Properties of PDIs
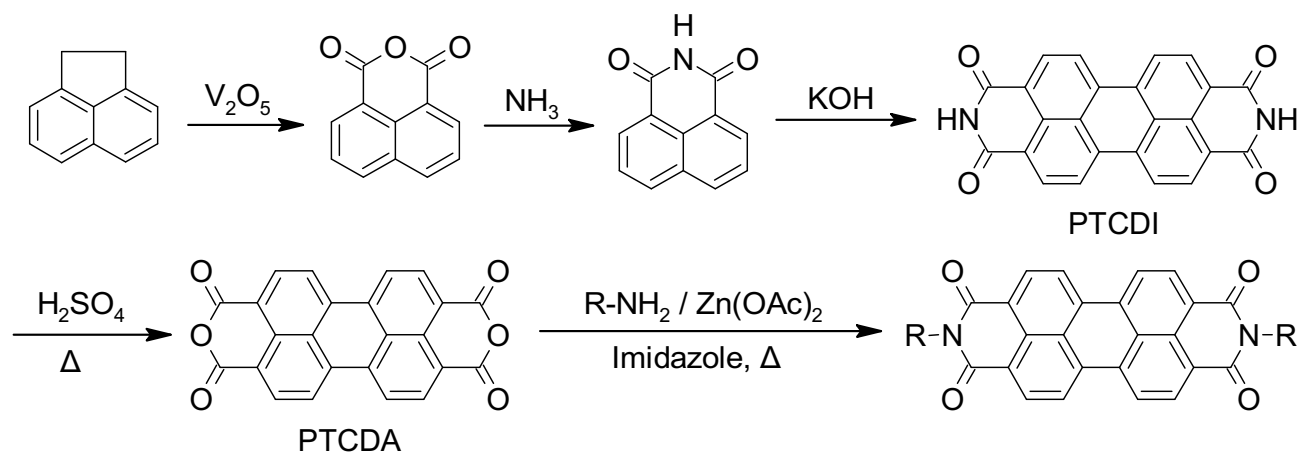
2.2. Synthetic Strategies to Access Symmetrical PDIs
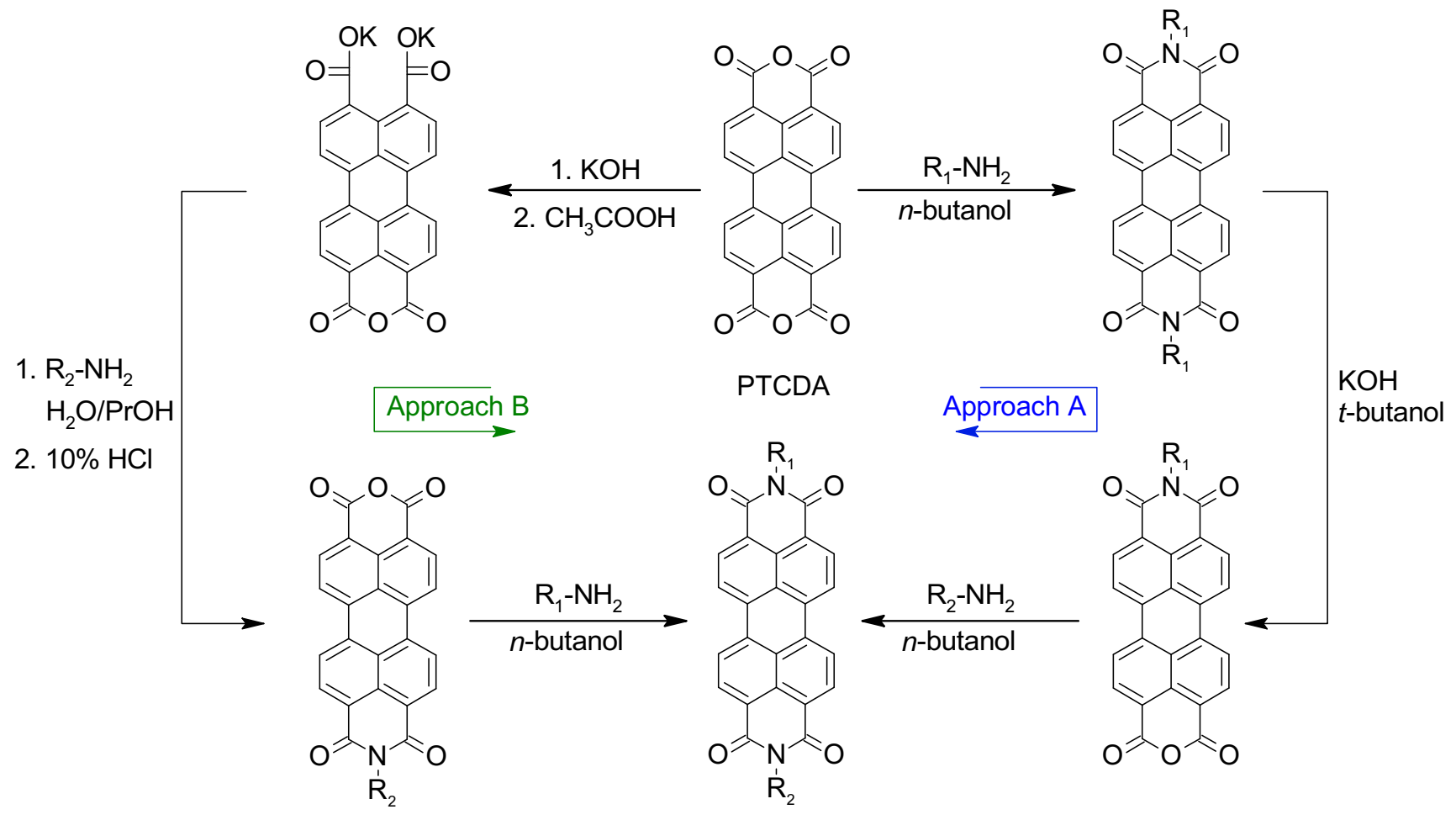
2.3. Synthetic Strategies to Access Asymmetrical PDIs
3. Peptide-PDI Bioconjugates
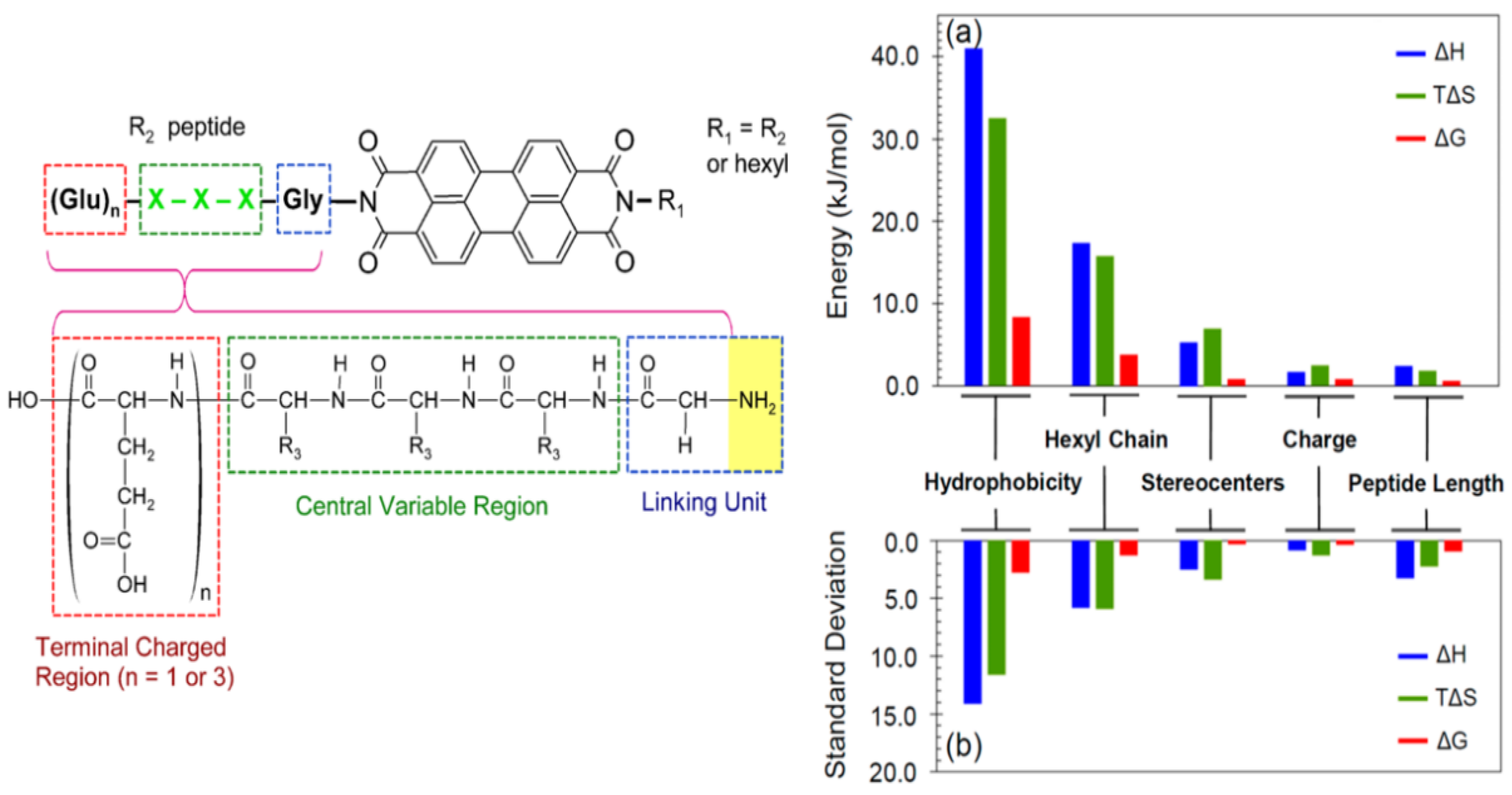
3.1. Synthesis of Peptide-Conjugated PDIs
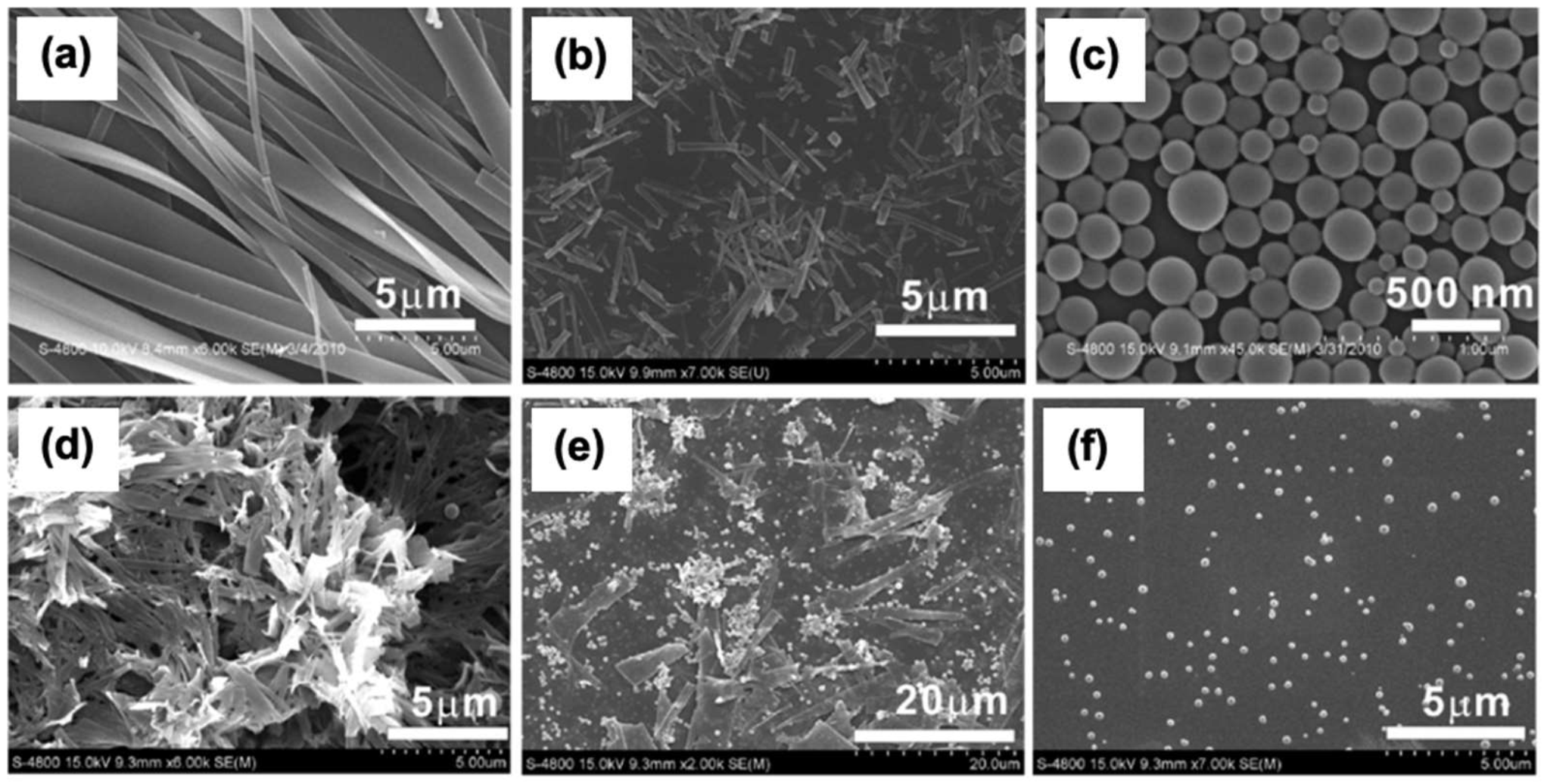
3.2. Sequence-Dependent Self-Assembly
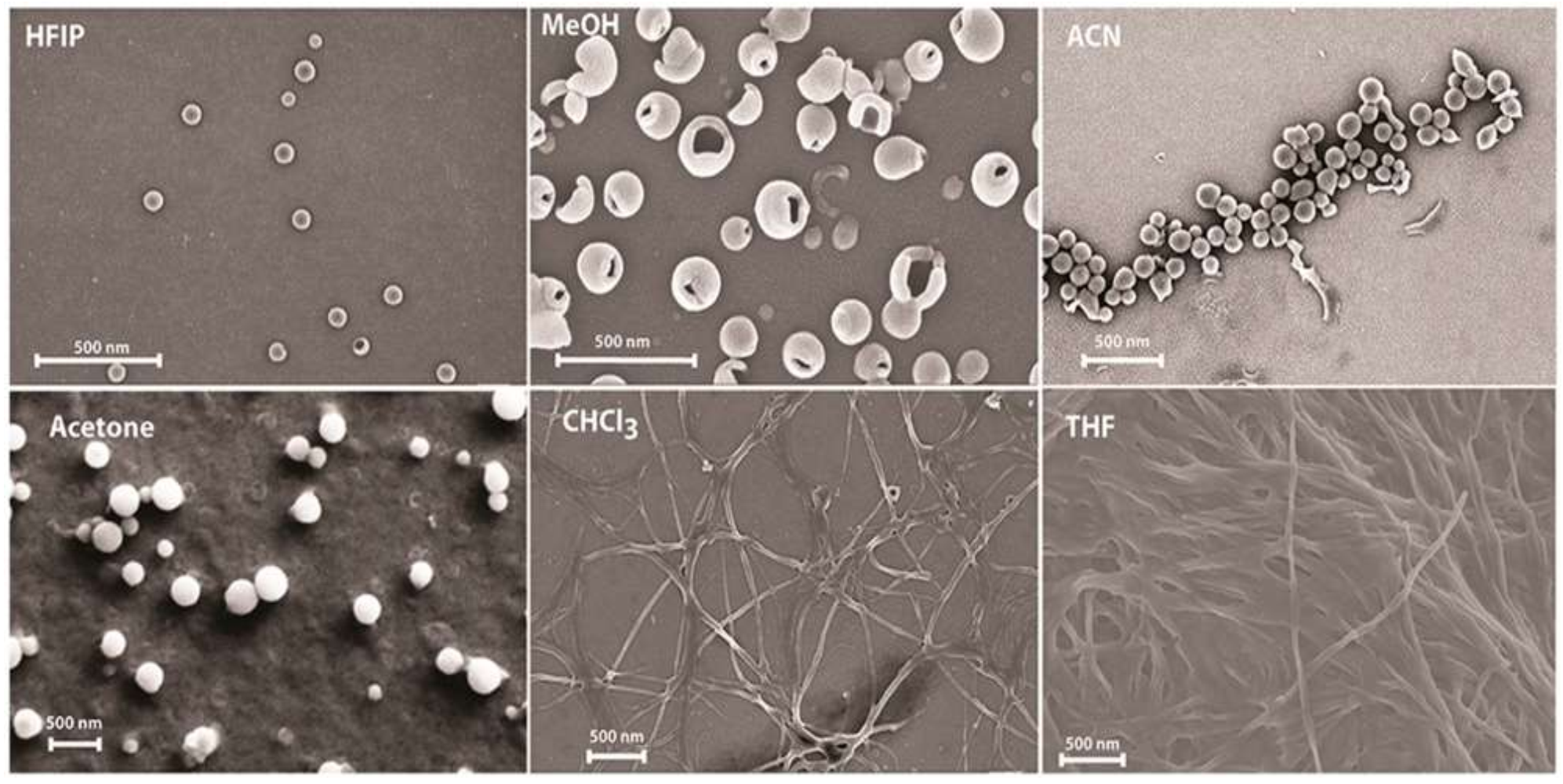
3.3. Solvent-Dependent Self-Assembly
3.4. PH-Dependent Self-Assembly
3.5. Concentration-Dependent Self-Assembly
4. Oligonucleotide–PDI Bioconjugates
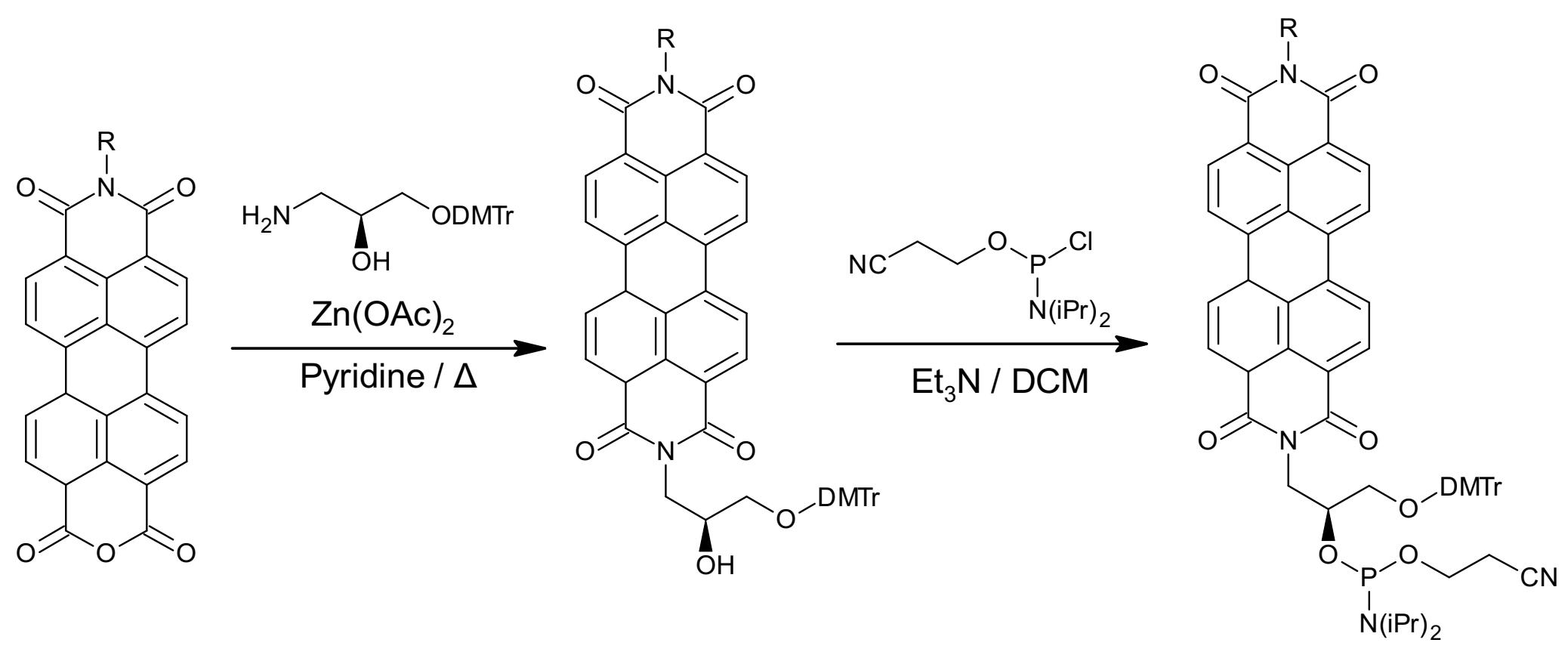
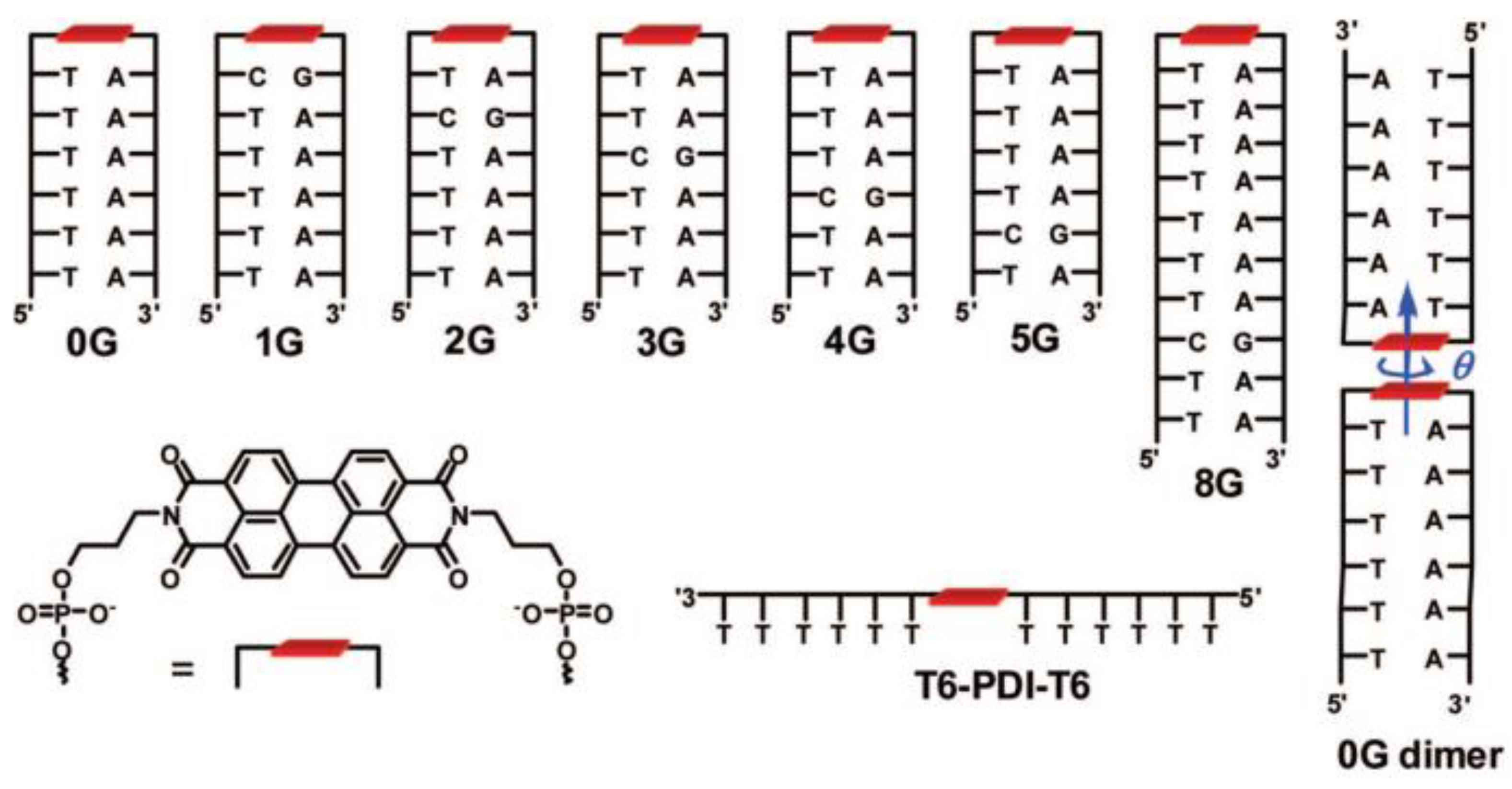
4.1. Synthesis of Oligonucleotide–PDI Bioconjugates
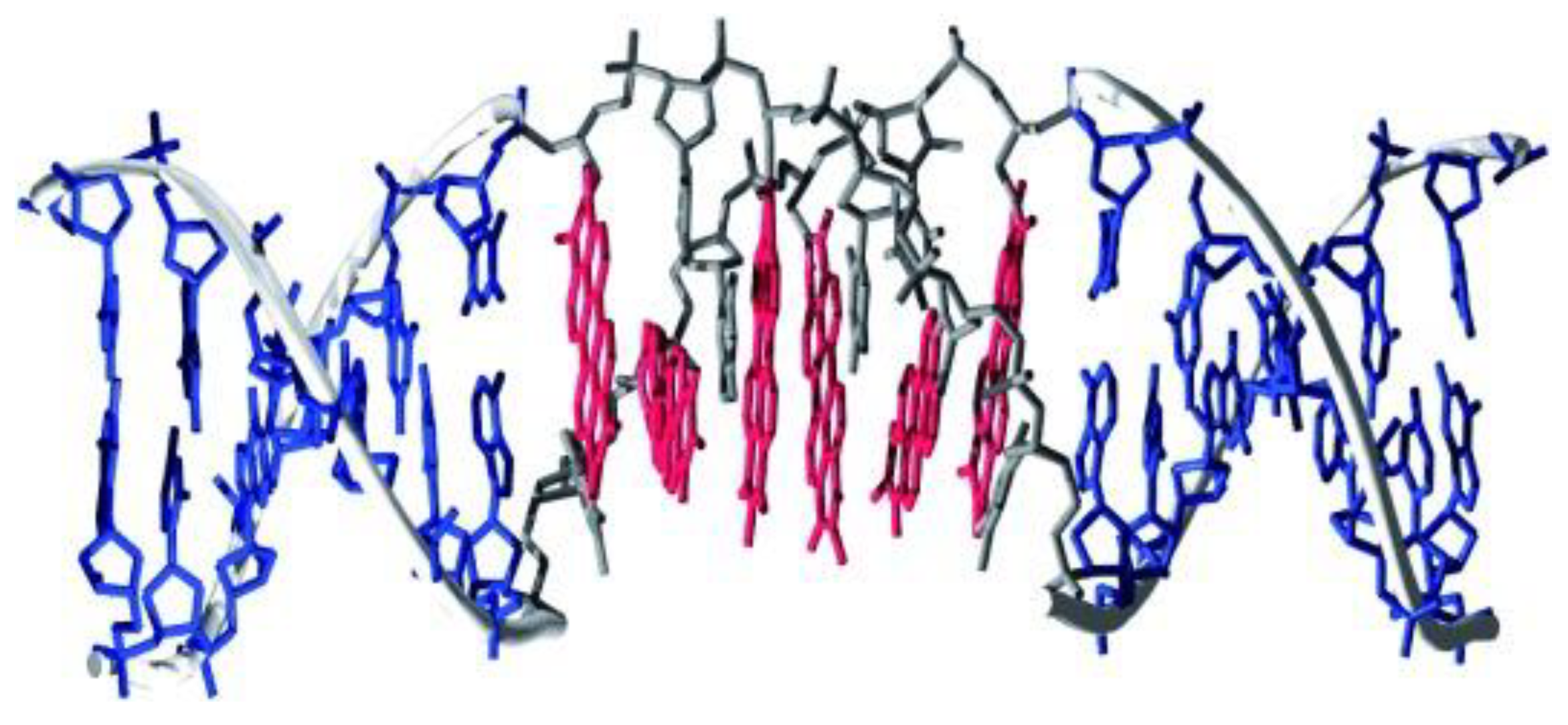
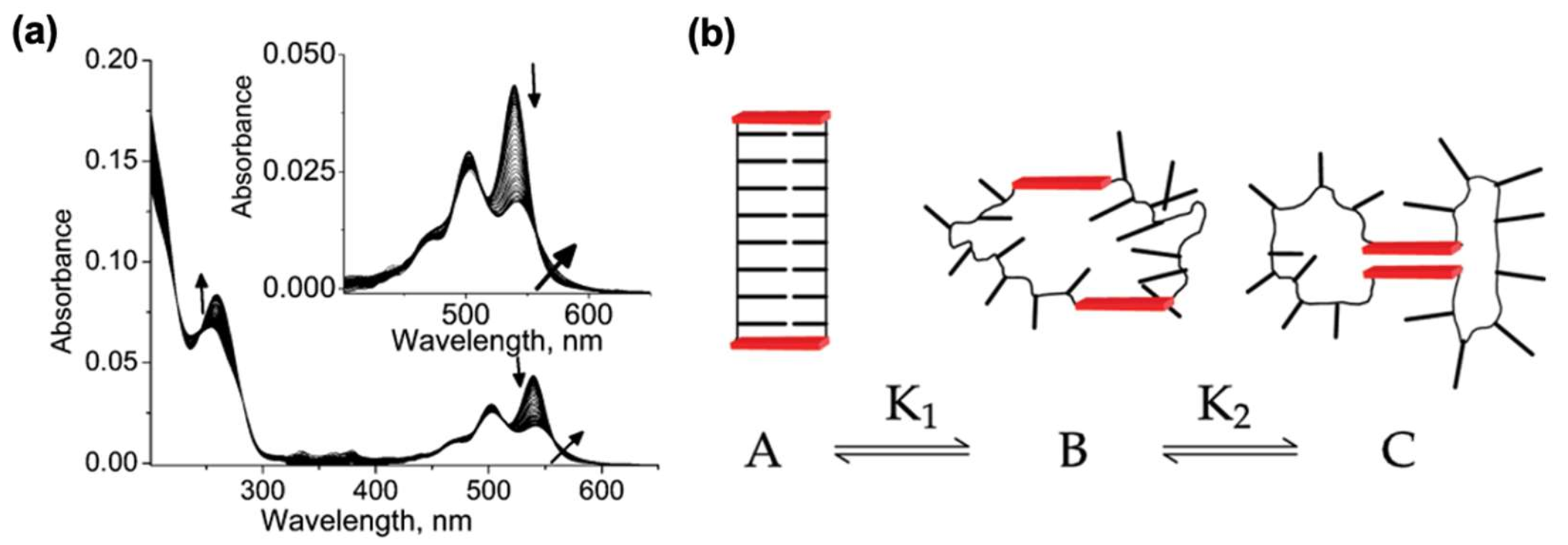
4.2. Structure-Dependent Self-Assembly of DNA–PDI Bioconjugates
4.3. pH-Dependent Self-Assembly of DNA–PDI Bioconjugates
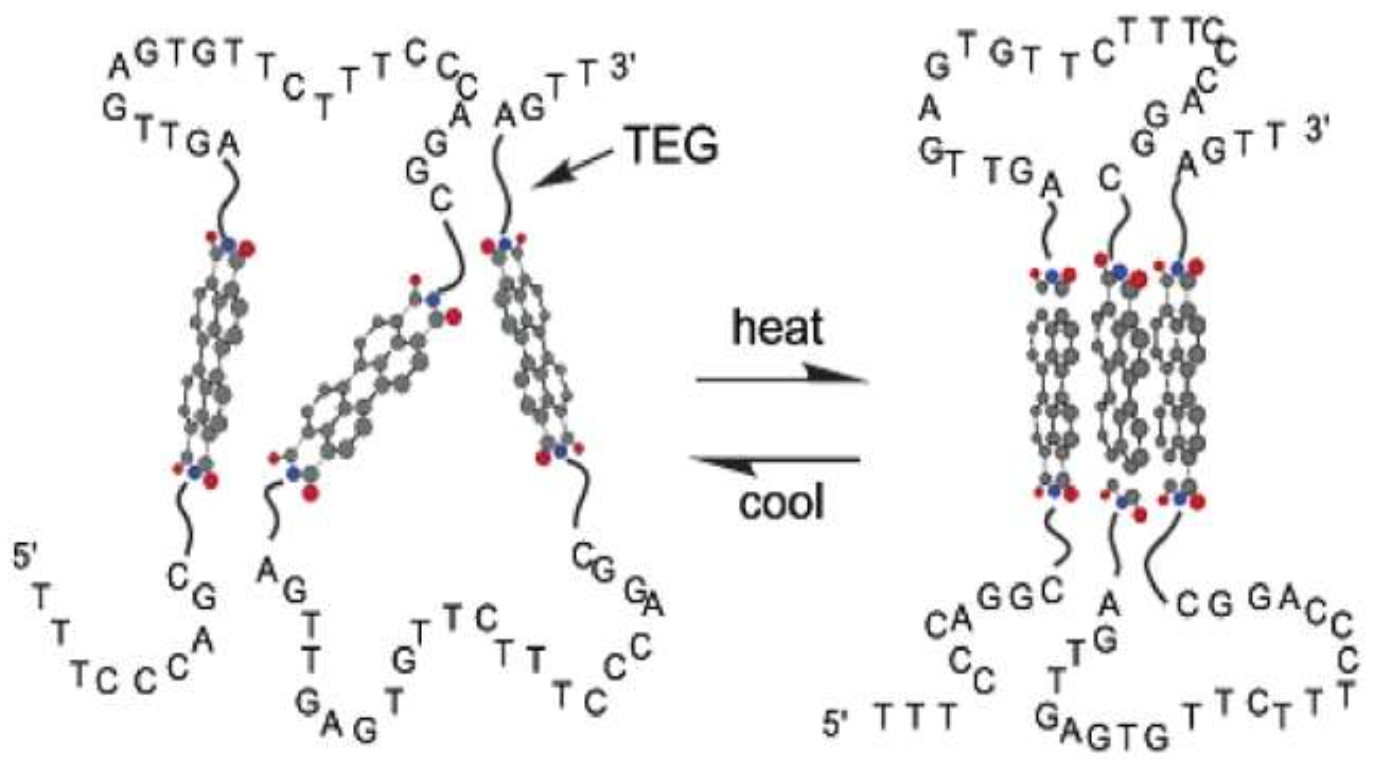
4.4. Temperature-Dependent Self-Assembly of DNA–PDIs
5. Potential Applications of PDI Bioconjugates
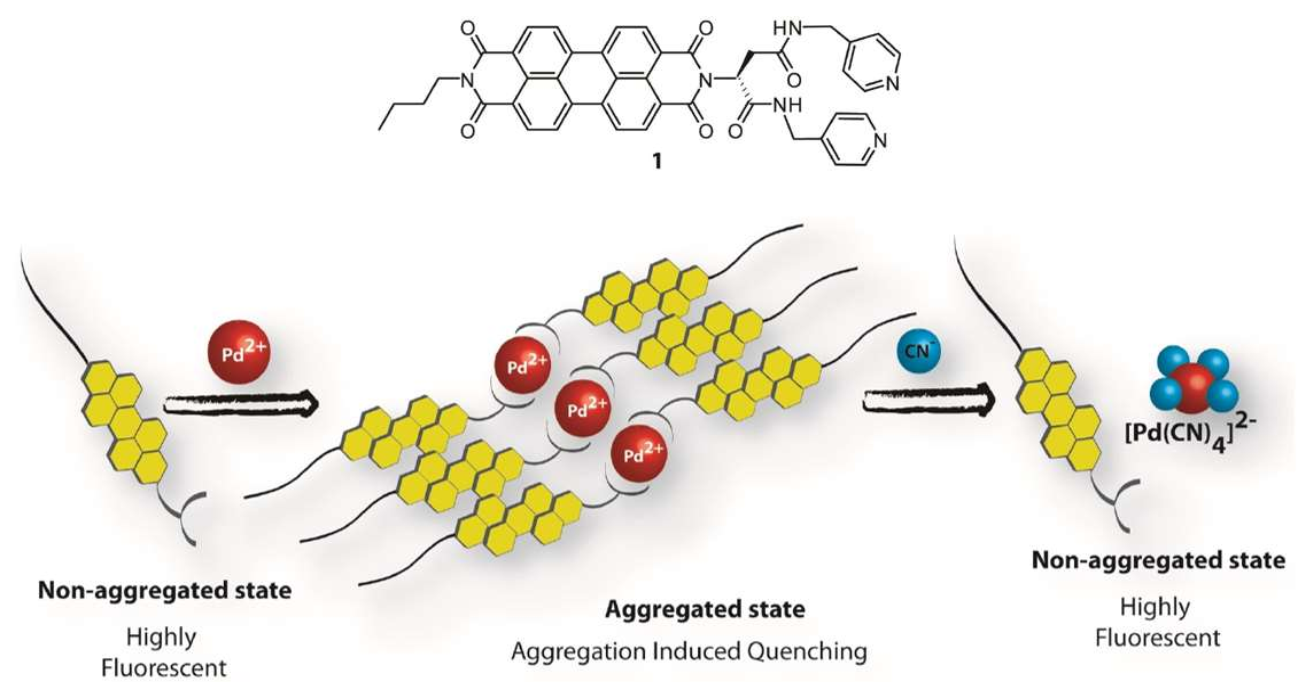
5.1. Applications of Peptide–PDI Bioconjugates
5.2. Applications of Oligonucleotide-Conjugated PDIs
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yadav, S.; Sharma, A.K.; Kumar, P. Nanoscale Self-Assembly for Therapeutic Delivery. Front. Bioeng. Biotechnol. 2020, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Ekiz, M.S.; Cinar, G.; Khalily, M.A.; Guler, M.O. Self-assembled peptide nanostructures for functional materials. Nanotechnology 2016, 27, 402002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkaš, P.; Bystrický, S. Chemical conjugation of biomacromolecules: A mini-review. Chem. Pap. 2010, 64, 683–695. [Google Scholar] [CrossRef]
- Cigánek, M.; Richtár, J.; Weiter, M.; Krajčovič, J. Organic π-Conjugated Molecules: From Nature to Artificial Applications. Where are the Boundaries? Isr. J. Chem. 2021, 61, 1–15. [Google Scholar] [CrossRef]
- Würthner, F.; Saha-Möller, C.R.; Fimmel, B.; Ogi, S.; Leowanawat, P.; Schmidt, D. Perylene Bisimide Dye Assemblies as Archetype Functional Supramolecular Materials. Chem. Rev. 2016, 116, 962–1052. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Wang, K.; Gu, L.-L.; Li, H. The study of perylene diimide–amino acid derivatives for the fluorescence detection of anions. RSC Adv. 2017, 7, 42685–42689. [Google Scholar] [CrossRef] [Green Version]
- Würthner, F.; Stepanenko, V.; Chen, Z.; Saha-Möller, C.R.; Kocher, N.; Stalke, D. Preparation and Characterization of Regioisomerically Pure 1,7-Disubstituted Perylene Bisimide Dyes. J. Org. Chem. 2004, 69, 7933–7939. [Google Scholar] [CrossRef]
- Abd-Ellah, M.; Cann, J.; Dayneko, S.V.; Laventure, A.; Cieplechowicz, E.; Welch, G.C. Interfacial ZnO Modification Using a Carboxylic Acid Functionalized N-Annulated Perylene Diimide for Inverted Type Organic Photovoltaics. ACS Appl. Electron. Mater. 2019, 1, 1590–1596. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, X.; Wang, Y.; Li, Z. Supramolecular Self-Assembly of Perylene Bisimide Derivatives Assisted by Various Groups. Langmuir 2019, 35, 342–358. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Król, A.; Würthner, F. Progress in the synthesis of perylene bisimide dyes. Org. Chem. Front. 2019, 6, 1272–1318. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Slattum, P.; Wang, C.; Zang, L. Self-Assembly of Perylene Imide Molecules into 1D Nanostructures: Methods, Morphologies, and Applications. Chem. Rev. 2015, 115, 11967–11998. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, P.; Kaur, N.; Mittal, L.S.; Kumar, K. Perylene diimides: Will they flourish as reaction-based probes? Anal. Methods 2020, 12, 3560–3574. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xue, Z.; Gao, N.; Yang, X.; Zang, L. Perylene Diimide-Based Fluorescent and Colorimetric Sensors for Environmental Detection. Sensors 2020, 20, 917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinbrück, N.; Kickelbick, G. Perylene polyphenylmethylsiloxanes for optoelectronic applications. J. Polym. Sci. Part. B Polym. Phys. 2019, 57, 1062–1073. [Google Scholar] [CrossRef] [Green Version]
- Yuen, J.D.; Pozdin, V.A.; Young, A.T.; Turner, B.L.; Giles, I.D.; Naciri, J.; Trammell, S.A.; Charles, P.T.; Stenger, D.A.; Daniele, M.A. Perylene-diimide-based n-type semiconductors with enhanced air and temperature stable photoconductor and transistor properties. Dyes Pigments 2020, 174, 108014. [Google Scholar] [CrossRef]
- Song, C.; Liu, X.; Li, X.; Wang, Y.-C.; Wan, L.; Sun, X.; Zhang, W.; Fang, J. Perylene Diimide-Based Zwitterion as the Cathode Interlayer for High-Performance Nonfullerene Polymer Solar Cells. ACS Appl. Mater. Interfaces 2018, 10, 14986–14992. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Chen, X. Semiconducting Perylene Diimide Nanostructure: Multifunctional Phototheranostic Nanoplatform. Accounts Chem. Res. 2019, 52, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xue, K.-F.; Yang, Y.; Hu, H.; Xu, J.-F.; Zhang, X. In Situ Hypoxia-Induced Supramolecular Perylene Diimide Radical Anions in Tumors for Photothermal Therapy with Improved Specificity. J. Am. Chem. Soc. 2022, 144, 2360–2367. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Müllen, K.; Yin, M. Water-soluble perylenediimides: Design concepts and biological applications. Chem. Soc. Rev. 2016, 45, 1513–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langhals, H. Cyclic Carboxylic Imide Structures as Structure Elements of High Stability. Novel Developments in Perylene Dye Chemistry. Heterocycles 1995, 40, 477. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Barlow, S.; Marder, S.R. Perylene-3,4,9,10-tetracarboxylic Acid Diimides: Synthesis, Physical Properties, and Use in Organic Electronics. J. Org. Chem. 2011, 76, 2386–2407. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Böckmann, M.; Jiang, W.; Doltsinis, N.L.; Wang, Z. Perylene Diimide-Embedded Double [8]Helicenes. J. Am. Chem. Soc. 2020, 142, 7092–7099. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Wu, K.; Zhou, Z.; Wang, G.; Zhao, B.; Tan, S. Alkynyl-Functionalized Pyrene-Cored Perylene Diimide Electron Acceptors for Efficient Nonfullerene Organic Solar Cells. ACS Appl. Energy Mater. 2019, 2, 3918–3926. [Google Scholar] [CrossRef]
- Zhang, L.; Song, I.; Ahn, J.; Han, M.; Linares, M.; Surin, M.; Zhang, H.-J.; Oh, J.H.; Lin, J. π-Extended perylene diimide double-heterohelicenes as ambipolar organic semiconductors for broadband circularly polarized light detection. Nat. Commun. 2021, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- Dayneko, S.V.; Rahmati, M.; Pahlevani, M.; Welch, G.C. Solution processed red organic light-emitting-diodes using an N-annulated perylene diimide fluorophore. J. Mater. Chem. C 2020, 8, 2314–2319. [Google Scholar] [CrossRef]
- Wang, J.; He, E.; Liu, X.; Yu, L.; Wang, H.; Zhang, R.; Zhang, H. High performance hydrazine vapor sensor based on redox mechanism of twisted perylene diimide derivative with lower reduction potential. Sens. Actuators B Chem. 2017, 239, 898–905. [Google Scholar] [CrossRef]
- Seo, J.; Khazi, M.I.; Kim, J.-M. Highly responsive triethylamine vapor sensor based on a perylene diimide-polydiacetylene system via heat-induced tuning of the molecular packing approach. Sens. Actuators B Chem. 2021, 334, 129660. [Google Scholar] [CrossRef]
- Yang, Z.; Song, J.; Tang, W.; Fan, W.; Dai, Y.; Shen, Z.; Lin, L.; Cheng, S.; Liu, Y.; Niu, G.; et al. Stimuli-Responsive Nanotheranostics for Real-Time Monitoring Drug Release by Photoacoustic Imaging. Theranostics 2019, 9, 526–536. [Google Scholar] [CrossRef]
- Gong, Q.; Xing, J.; Huang, Y.; Wu, A.; Yu, J.; Zhang, Q. Perylene Diimide Oligomer Nanoparticles with Ultrahigh Photothermal Conversion Efficiency for Cancer Theranostics. ACS Appl. Bio Mater. 2020, 3, 1607–1615. [Google Scholar] [CrossRef]
- Würthner, F. Perylene bisimide dyes as versatile building blocks for functional supramolecular architectures. Chem. Commun. 2004, 35, 1564–1579. [Google Scholar] [CrossRef]
- Arantes, J.T.; Lima, M.P.; Fazzio, A.; Xiang, H.; Wei, S.-H.; Dalpian, G.M. Effects of Side-Chain and Electron Exchange Correlation on the Band Structure of Perylene Diimide Liquid Crystals: A Density Functional Study. J. Phys. Chem. B 2009, 113, 5376–5380. [Google Scholar] [CrossRef]
- Polkehn, M.; Tamura, H.; Eisenbrandt, P.; Haacke, S.; Méry, S.; Burghardt, I. Molecular Packing Determines Charge Separation in a Liquid Crystalline Bisthiophene–Perylene Diimide Donor–Acceptor Material. J. Phys. Chem. Lett. 2016, 7, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Spano, F.C.; Silva, C. H- and J-Aggregate Behavior in Polymeric Semiconductors. Annu. Rev. Phys. Chem. 2014, 65, 477–500. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Li, X.-Q.; Stepanenko, V.; Würthner, F. Control of H- and J-Type π Stacking by Peripheral Alkyl Chains and Self-Sorting Phenomena in Perylene Bisimide Homo- and Heteroaggregates. Chem. A Eur. J. 2008, 14, 11343–11357. [Google Scholar] [CrossRef]
- Pasaogullari, N.; Icil, H.; Demuth, M. Symmetrical and unsymmetrical perylene diimides: Their synthesis, photophysical and electrochemical properties. Dyes Pigments 2006, 69, 118–127. [Google Scholar] [CrossRef]
- Nagao, Y.; Misono, T. Synthesis and Reactions of Perylenecarboxylic Acid Derivatives. VII. Hydrolysis ofN,N′-Dialkyl-3,4: 9,10-Perylenebis(dicarboximide) with Sulfuric Acid. Bull. Chem. Soc. Jpn. 1981, 54, 1269–1270. [Google Scholar] [CrossRef] [Green Version]
- Nagao, Y. Synthesis and properties of perylene pigments. Prog. Org. Coat. 1997, 31, 43–49. [Google Scholar] [CrossRef]
- Tröster, H. Untersuchungen zur Protonierung von Perylen-3,4,9,10-tetracarbonsäurealkalisalzen. Dyes Pigments 1983, 4, 171–177. [Google Scholar] [CrossRef]
- Eakins, G.L.; Gallaher, J.K.; Keyzers, R.A.; Falber, A.; Webb, J.E.A.; Laos, A.; Tidhar, Y.; Weissman, H.; Rybtchinski, B.; Thordarson, P.; et al. Thermodynamic Factors Impacting the Peptide-Driven Self-Assembly of Perylene Diimide Nanofibers. J. Phys. Chem. B 2014, 118, 8642–8651. [Google Scholar] [CrossRef]
- Zottig, X.; Côté-Cyr, M.; Arpin, D.; Archambault, D.; Bourgault, S. Protein Supramolecular Structures: From Self-Assembly to Nanovaccine Design. Nanomaterials 2020, 10, 1008. [Google Scholar] [CrossRef] [PubMed]
- Makishima, A. (Ed.) Fundamental Knowledges and Techniques in Biochemistry. In Biochemistry for Materials; Elsevier Science: Amsterdam, The Netherlands, 2019; pp. 35–51. [Google Scholar] [CrossRef]
- Ouellette, R.J.; Rawn, J.D. Amino Acids, Peptides, and Proteins. In Organic Chemistry Study Guide; Elsevier: Amsterdam, The Netherlands, 2015; p. 569. [Google Scholar] [CrossRef]
- Vadehra, G.S.; Wall, B.D.; Diegelmann, S.R.; Tovar, J.D. On-resin dimerization incorporates a diverse array of π-conjugated functionality within aqueous self-assembling peptide backbones. Chem. Commun. 2010, 46, 3947–3949. [Google Scholar] [CrossRef]
- Godin, E.; Nguyen, P.T.; Zottig, X.; Bourgault, S. Identification of a hinge residue controlling islet amyloid polypeptide self-assembly and cytotoxicity. J. Biol. Chem. 2019, 294, 8452–8463. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Kim, Y.-O.; Park, S.-J.; Jung, B.Y.; Jang, H.-S.; Choi, S.K.; Kim, J.; Kim, S.; Jung, Y.C.; Shin, D.-S.; Lee, Y.-S. Solid-Phase Synthesis of Peptide-Conjugated Perylene Diimide Bolaamphiphile and Its Application in Photodynamic Therapy. ACS Omega 2018, 3, 5896–5902. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Debnath, S.; Javid, N.; Frederix, P.W.J.M.; Fleming, S.; Pappas, C.; Ulijn, R.V. Differential Self-Assembly and Tunable Emission of Aromatic Peptide Bola-Amphiphiles Containing Perylene Bisimide in Polar Solvents Including Water. Langmuir 2014, 30, 7576–7584. [Google Scholar] [CrossRef] [PubMed]
- Eakins, G.L.; Pandey, R.; Wojciechowski, J.P.; Zheng, H.Y.; Webb, J.E.A.; Valéry, C.; Thordarson, P.; Plank, N.O.V.; Gerrard, J.A.; Hodgkiss, J.M. Functional Organic Semiconductors Assembled via Natural Aggregating Peptides. Adv. Funct. Mater. 2015, 25, 5640–5649. [Google Scholar] [CrossRef]
- Wei, D.; Ge, L.; Wang, Z.; Wang, Y.; Guo, R. Self-Assembled Dual Helical Nanofibers of Amphiphilic Perylene Diimides with Oligopeptide Substitution. Langmuir 2019, 35, 11745–11754. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.S.; Shmilovich, K.; Herringer, N.S.M.; Marin, N.; Ferguson, A.L.; Tovar, J.D. Computationally Guided Tuning of Peptide-Conjugated Perylene Diimide Self-Assembly. Langmuir 2021, 37, 8594–8606. [Google Scholar] [CrossRef] [PubMed]
- Marty, R.; Nigon, R.; Leite, D.; Frauenrath, H. Two-Fold Odd–Even Effect in Self-Assembled Nanowires from Oligopeptide-Polymer-Substituted Perylene Bisimides. J. Am. Chem. Soc. 2014, 136, 3919–3927. [Google Scholar] [CrossRef]
- Ke, D.; Tang, A.; Zhan, C.; Yao, J. Conformation-variable PDI@β-sheet nanohelices show stimulus-responsive supramolecular chirality. Chem. Commun. 2013, 49, 4914–4916. [Google Scholar] [CrossRef]
- Eakins, G.L.; Wojciechowski, J.P.; Martin, A.D.; Webb, J.E.A.; Thordarson, P.; Hodgkiss, J.M. Chiral effects in peptide-substituted perylene imide nanofibres. Supramol. Chem. 2015, 27, 746–756. [Google Scholar] [CrossRef]
- Ahmed, S.; Pramanik, B.; Sankar, K.N.A.; Srivastava, A.; Singha, N.; Dowari, P.; Srivastava, A.; Mohanta, K.; Debnath, A.; Das, D. Solvent Assisted Tuning of Morphology of a Peptide-Perylenediimide Conjugate: Helical Fibers to Nano-Rings and Their Differential Semiconductivity. Sci. Rep. 2017, 7, 9485. [Google Scholar] [CrossRef]
- Ahmed, S.; Sankar, K.N.A.; Pramanik, B.; Mohanta, K.; Das, D. Solvent Directed Morphogenesis and Electrical Properties of a Peptide–Perylenediimide Conjugate. Langmuir 2018, 34, 8355–8364. [Google Scholar] [CrossRef]
- Sun, Y.; He, C.; Sun, K.; Li, Y.; Dong, H.; Wang, Z.; Li, Z. Fine-Tuned Nanostructures Assembled from l-Lysine-Functionalized Perylene Bisimides. Langmuir 2011, 27, 11364–11371. [Google Scholar] [CrossRef]
- Chen, Z.; Lohr, A.; Saha-Möller, C.R.; Würthner, F. Self-assembled π-stacks of functional dyes in solution: Structural and thermodynamic features. Chem. Soc. Rev. 2009, 38, 564–584. [Google Scholar] [CrossRef]
- Farooqi, M.J.; Penick, M.A.; Burch, J.; Negrete, G.R.; Brancaleon, L. Characterization of novel perylene diimides containing aromatic amino acid side chains. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 153, 124–131. [Google Scholar] [CrossRef] [Green Version]
- Chal, P.; Shit, A.; Nandi, A.K. Optoelectronic Properties of Supramolecular of Aggregates Phenylalanine Conjugated Perylene Bisimide. ChemistrySelect 2018, 3, 3993–4003. [Google Scholar] [CrossRef]
- Draper, E.R.; Walsh, J.J.; McDonald, T.O.; Zwijnenburg, M.A.; Cameron, P.J.; Cowan, A.J.; Adams, D.J. Air-stable photoconductive films formed from perylene bisimide gelators. J. Mater. Chem. C 2014, 2, 5570–5575. [Google Scholar] [CrossRef] [Green Version]
- Kozma, E.; Grisci, G.; Mróz, W.; Catellani, M.; Eckstein-Andicsovà, A.; Pagano, K.; Galeotti, F. Water-soluble aminoacid functionalized perylene diimides: The effect of aggregation on the optical properties in organic and aqueous media. Dyes Pigments 2016, 125, 201–209. [Google Scholar] [CrossRef]
- Nolan, M.C.; Walsh, J.J.; Mears, L.L.E.; Draper, E.R.; Wallace, M.; Barrow, M.; Dietrich, B.; King, S.M.; Cowan, A.J.; Adams, D.J. pH dependent photocatalytic hydrogen evolution by self-assembled perylene bisimides. J. Mater. Chem. A 2017, 5, 7555–7563. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Basu, K.; Gayen, K.; Panigrahi, S.; Mondal, S.; Basak, D.; Banerjee, A. TiO2 Nanoparticles Incorporated Peptide Appended Perylene Bisimide-Based Nanohybrid System: Enhancement of Photo-Switching Behavior. J. Phys. Chem. C 2017, 121, 5428–5435. [Google Scholar] [CrossRef]
- Pandeeswar, M.; Govindaraju, T. Engineering molecular self-assembly of perylene diimide through pH-responsive chiroptical switching. Mol. Syst. Des. Eng. 2016, 1, 202–207. [Google Scholar] [CrossRef]
- Li, J.; Pei, H.; Zhu, B.; Liang, L.; Wei, M.; He, Y.; Chen, N.; Li, D.; Huang, Q.; Fan, C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. [Google Scholar] [CrossRef]
- Oliviero, G.; D’Errico, S.; Pinto, B.; Nici, F.; Dardano, P.; Rea, I.; De Stefano, L.; Mayol, L.; Piccialli, G.; Borbone, N. Self-Assembly of G-Rich Oligonucleotides Incorporating a 3′-3′ Inversion of Polarity Site: A New Route Towards G-Wire DNA Nanostructures. ChemistryOpen 2017, 6, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Campolongo, M.J.; Nhi Tran, T.N.; Ruiz, R.C.H.; Kahn, J.S.; Luo, D. Novel DNA materials and their applications. WIREs Nanomed. Nanobiotechnol. 2010, 2, 648–669. [Google Scholar] [CrossRef]
- Marras, A.E.; Zhou, L.; Su, H.-J.; Castro, C.E. Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. USA 2015, 112, 713–718. [Google Scholar] [CrossRef] [Green Version]
- Wagner, C.; Wagenknecht, H.-A. Perylene-3,4:9,10-tetracarboxylic Acid Bisimide Dye as an Artificial DNA Base Surrogate. Org. Lett. 2006, 8, 4191–4194. [Google Scholar] [CrossRef]
- Ustinov, A.V.; Dubnyakova, V.V.; Korshun, V.A. A convenient ‘click chemistry’ approach to perylene diimide–oligonucleotide conjugates. Tetrahedron 2008, 64, 1467–1473. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, H.-H.; Niu, A.S.; Li, A.D.Q. Alternating DNA and π-Conjugated Sequences. Thermophilic Foldable Polymers. J. Am. Chem. Soc. 2003, 125, 5248–5249. [Google Scholar] [CrossRef]
- Takada, T.; Ishino, S.; Takata, A.; Nakamura, M.; Fujitsuka, M.; Majima, T.; Yamana, K. Rapid Electron Transfer of Stacked Heterodimers of Perylene Diimide Derivatives in a DNA Duplex. Chem. A Eur. J. 2018, 24, 8228–8232. [Google Scholar] [CrossRef]
- Mishra, A.K.; Weissman, H.; Krieg, E.; Votaw, K.A.; Mccullagh, M.; Rybtchinski, B.; Lewis, F.D. Self-Assembly of Perylenediimide–Single-Strand-DNA Conjugates: Employing Hydrophobic Interactions and DNA Base-Pairing To Create a Diverse Structural Space. Chem. A Eur. J. 2017, 23, 10328–10337. [Google Scholar] [CrossRef] [PubMed]
- Markegard, C.B.; Mazaheripour, A.; Jocson, J.-M.; Burke, A.M.; Dickson, M.N.; Gorodetsky, A.A.; Nguyen, H.D. Molecular Dynamics Simulations of Perylenediimide DNA Base Surrogates. J. Phys. Chem. B 2015, 119, 11459–11465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumstark, D.; Wagenknecht, H.-A. Fluorescent Hydrophobic Zippers inside Duplex DNA: Interstrand Stacking of Perylene-3,4:9,10-tetracarboxylic Acid Bisimides as Artificial DNA Base Dyes. Chem. A Eur. J. 2008, 14, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Franceschin, M.; Borbone, N.; Oliviero, G.; Casagrande, V.; Scuotto, M.; Coppola, T.; Borioni, S.; Mayol, L.; Ortaggi, G.; Bianco, A.; et al. Synthesis of a Dibromoperylene Phosphoramidite Building Block and Its Incorporation at the 5′ End of a G-Quadruplex Forming Oligonucleotide: Spectroscopic Properties and Structural Studies of the Resulting Dibromoperylene Conjugate. Bioconj. Chem. 2011, 22, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Carmieli, R.; Zeidan, T.A.; Kelley, R.F.; Mi, Q.; Lewis, F.D.; Wasielewski, M.R. Excited State, Charge Transfer, and Spin Dynamics in DNA Hairpin Conjugates with Perylenediimide Hairpin Linkers. J. Phys. Chem. A 2009, 113, 4691–4700. [Google Scholar] [CrossRef] [PubMed]
- Kerwin, S.M.; Chen, G.; Kern, J.T.; Thomas, P.W. Perylene Diimide G-Quadruplex DNA Binding Selectivity is Mediated by Ligand Aggregation. Bioorg. Med. Chem. Lett. 2001, 12, 447–450. [Google Scholar] [CrossRef]
- Hariharan, M.; Siegmund, K.; Zheng, Y.; Long, H.; Schatz, G.C.; Lewis, F.D. Perylenediimide-Linked DNA Dumbbells: Long-Distance Electronic Interactions and Hydrophobic Assistance of Base-Pair Melting. J. Phys. Chem. C 2010, 114, 20466–20471. [Google Scholar] [CrossRef]
- Hariharan, M.; Zheng, Y.; Long, H.; Zeidan, T.A.; Schatz, G.C.; Vura-Weis, J.; Wasielewski, M.R.; Zuo, X.; Tiede, D.M.; Lewis, F.D. Hydrophobic Dimerization and Thermal Dissociation of Perylenediimide-Linked DNA Hairpins. J. Am. Chem. Soc. 2009, 131, 5920–5929. [Google Scholar] [CrossRef]
- Draper, E.R.; Schweins, R.; Akhtar, R.; Groves, P.; Chechik, V.; Zwijnenburg, M.A.; Adams, D.J. Reversible Photoreduction as a Trigger for Photoresponsive Gels. Chem. Mater. 2016, 28, 6336–6341. [Google Scholar] [CrossRef] [Green Version]
- Pramanik, B.; Ahmed, S.; Singha, N.; Das, D. Self-Assembly Assisted Tandem Sensing of Pd2+ and CN− by a Perylenediimide-Peptide Conjugate. ChemistrySelect 2017, 2, 10061–10066. [Google Scholar] [CrossRef]
- Heinonen, J.K. Regulatory Roles of PPi. In Biological Role of Inorganic Pyrophosphate; Springer: Boston, MA, USA, 2001; p. 123. [Google Scholar] [CrossRef]
- Feng, X.; An, Y.; Yao, Z.; Li, C.; Shi, G. A Turn-on Fluorescent Sensor for Pyrophosphate Based on the Disassembly of Cu2+-Mediated Perylene Diimide Aggregates. ACS Appl. Mater. Interfaces 2012, 4, 614–618. [Google Scholar] [CrossRef]
- Grisci, G.; Mróz, W.; Catellani, M.; Kozma, E.; Galeotti, F. Off-On Fluorescence Response of a Cysteine-based Perylene Diimide for Mercury Detection in Water. ChemistrySelect 2016, 1, 3033–3037. [Google Scholar] [CrossRef]
- Kalita, A.; Hussain, S.; Malik, A.H.; Subbarao, N.V.V.; Iyer, P.K. Vapor phase sensing of ammonia at the sub-ppm level using a perylene diimide thin film device. J. Mater. Chem. C 2015, 3, 10767–10774. [Google Scholar] [CrossRef]
- Liu, F.; Mu, J.; Wu, X.; Bhattacharjya, S.; Yeow, E.K.L.; Xing, B. Peptide–perylene diimide functionalized magnetic nano-platforms for fluorescence turn-on detection and clearance of bacterial lipopolysaccharides. Chem. Commun. 2014, 50, 6200–6203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthuraj, B.; Chowdhury, S.R.; Iyer, P.K. Modulation of Amyloid-β Fibrils into Mature Microrod-Shaped Structure by Histidine Functionalized Water-Soluble Perylene Diimide. ACS Appl. Mater. Interfaces 2015, 7, 21226–21234. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Zottig, X.; Sebastiao, M.; Arnold, A.A.; Marcotte, I.; Bourgault, S. Identification of transmissible proteotoxic oligomer-like fibrils that expand conformational diversity of amyloid assemblies. Commun. Biol. 2021, 4, 939. [Google Scholar] [CrossRef] [PubMed]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Takada, T.; Ashida, A.; Nakamura, M.; Yamana, K. Cationic perylenediimide as a specific fluorescent binder to mismatch containing DNA. Bioorg. Med. Chem. 2013, 21, 6011–6014. [Google Scholar] [CrossRef]
- Takada, T.; Yamaguchi, K.; Tsukamoto, S.; Nakamura, M.; Yamana, K. Light-up fluorescent probes utilizing binding behavior of perylenediimide derivatives to a hydrophobic pocket within DNA. Analyst 2014, 139, 4016–4021. [Google Scholar] [CrossRef] [PubMed]
- Bevers, S.; Schutte, S.; McLaughlin, L.W. Naphthalene- and Perylene-Based Linkers for the Stabilization of Hairpin Triplexes. J. Am. Chem. Soc. 2000, 122, 5905–5915. [Google Scholar] [CrossRef]
- Wohlgamuth, C.H.; McWilliams, M.A.; Mazaheripour, A.; Burke, A.M.; Lin, K.-Y.; Doan, L.; Slinker, J.D.; Gorodetsky, A.A. Electrochemistry of DNA Monolayers Modified With a Perylenediimide Base Surrogate. J. Phys. Chem. C 2014, 118, 29084–29090. [Google Scholar] [CrossRef]
- Vasimalla, S.; Sato, S.; Takenaka, F.; Kurose, Y.; Takenaka, S. Cyclic perylene diimide: Selective ligand for tetraplex DNA binding over double stranded DNA. Bioorg. Med. Chem. 2017, 25, 6404–6411. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kihal, N.; Nazemi, A.; Bourgault, S. Supramolecular Nanostructures Based on Perylene Diimide Bioconjugates: From Self-Assembly to Applications. Nanomaterials 2022, 12, 1223. https://doi.org/10.3390/nano12071223
Kihal N, Nazemi A, Bourgault S. Supramolecular Nanostructures Based on Perylene Diimide Bioconjugates: From Self-Assembly to Applications. Nanomaterials. 2022; 12(7):1223. https://doi.org/10.3390/nano12071223
Chicago/Turabian StyleKihal, Nadjib, Ali Nazemi, and Steve Bourgault. 2022. "Supramolecular Nanostructures Based on Perylene Diimide Bioconjugates: From Self-Assembly to Applications" Nanomaterials 12, no. 7: 1223. https://doi.org/10.3390/nano12071223
APA StyleKihal, N., Nazemi, A., & Bourgault, S. (2022). Supramolecular Nanostructures Based on Perylene Diimide Bioconjugates: From Self-Assembly to Applications. Nanomaterials, 12(7), 1223. https://doi.org/10.3390/nano12071223






