Zeolitic Imidazolate Framework-8 (ZIF-8) as a Drug Delivery Vehicle for the Transport and Release of Telomerase Inhibitor BIBR 1532
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
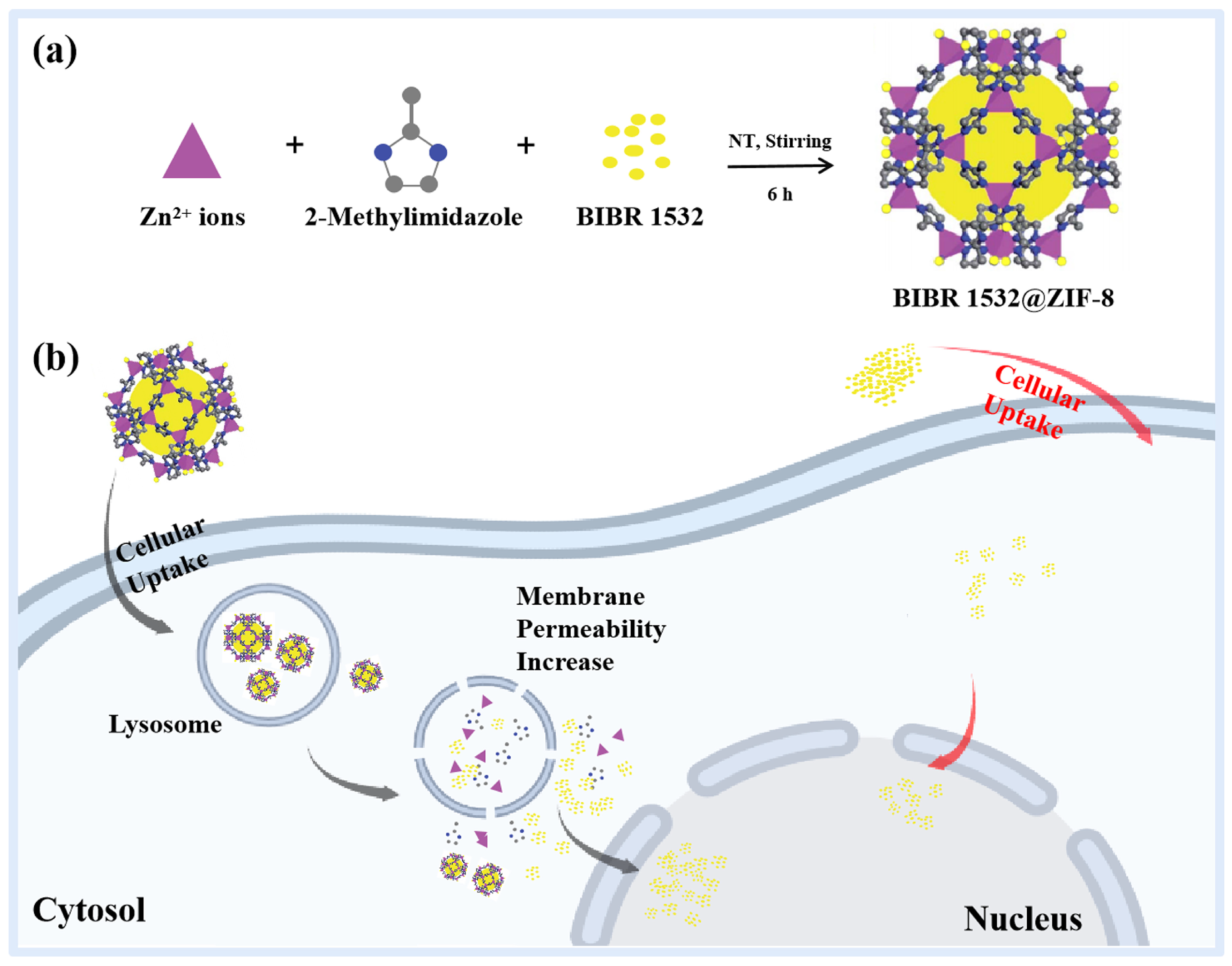
2.2. Synthesis of Nanoparticles
2.3. Characterization
2.4. BIBR 1532 Loading
2.5. Cell Viability Assay
2.6. Inductively Coupled Plasma-Optical Emission Spectrometer (ICP-OES)
2.7. Confocal Laser Scanning Microscopy
2.8. Acridine Orange (AO) Staining
2.9. Western Blot Analysis
2.10. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Analysis
2.11. Cytokinesis-Block Micronucleus (CBMN) Assay
2.12. Cell Cycle Analysis
2.13. Senescence-Associated β-Galactosidase Detection
2.14. Statistical Analysis
3. Results
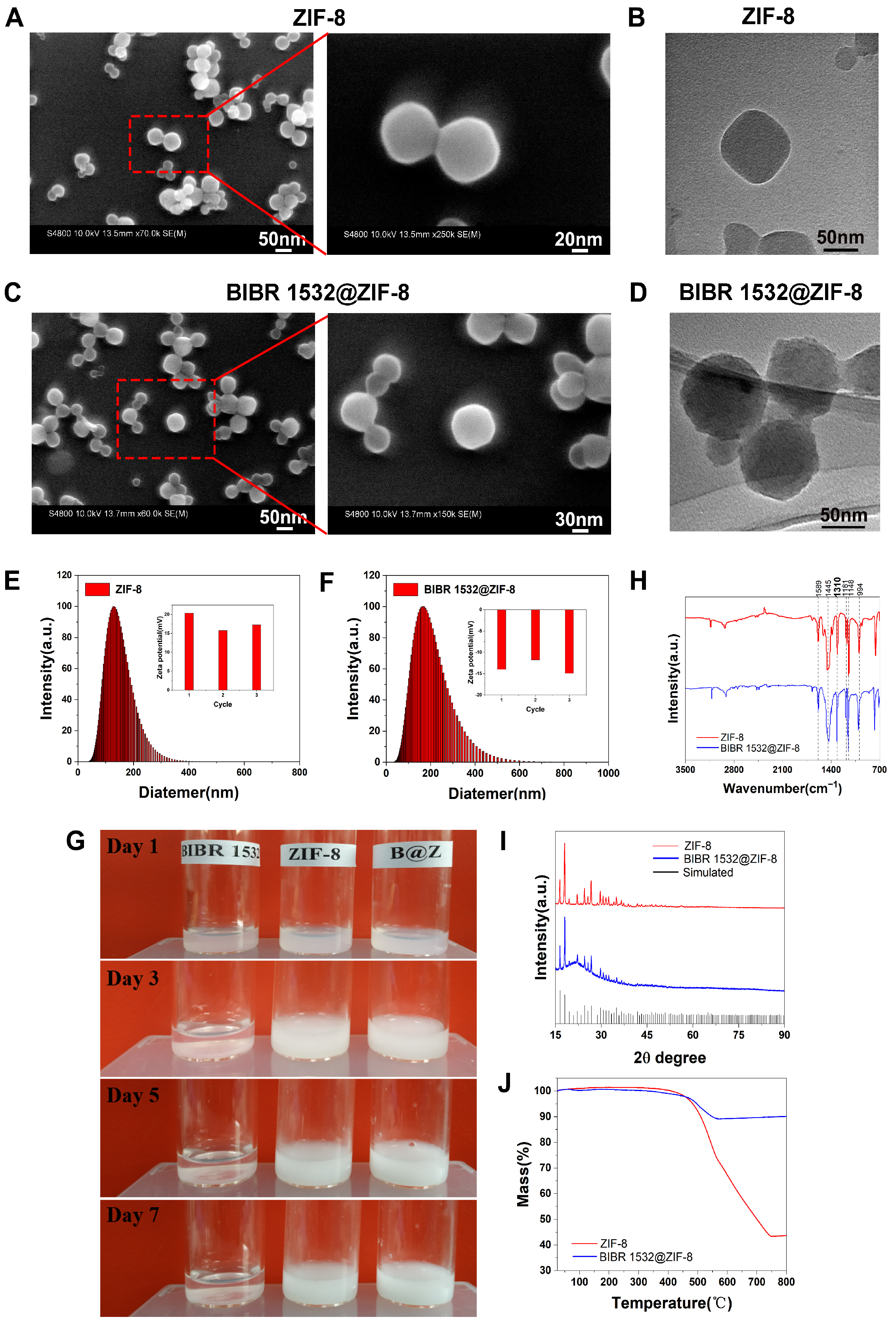
3.1. Physicochemical Characterizations of ZIF-8 and BIBR 1532@ZIF-8
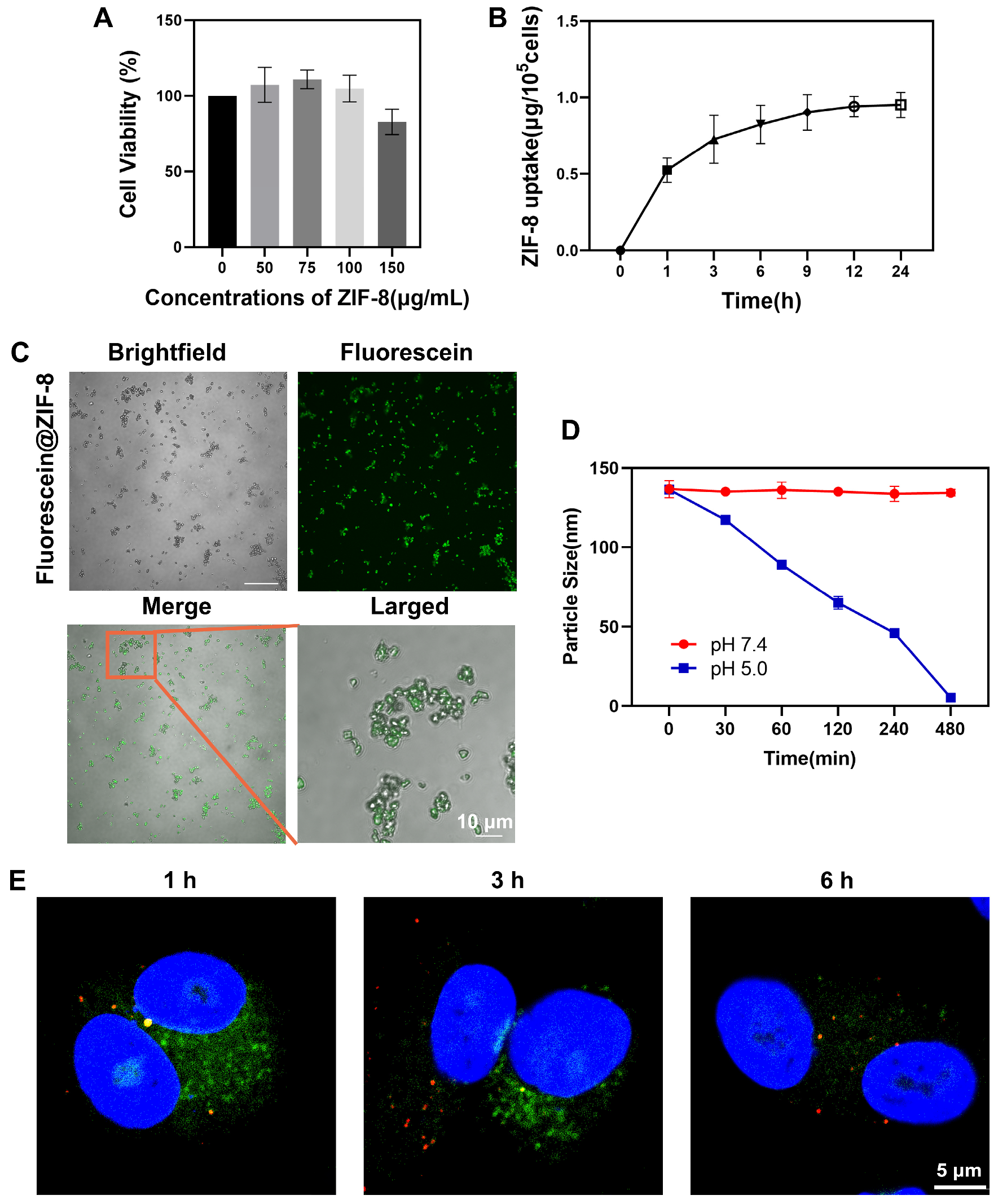
3.2. ZIF-8 Localized in Lysosomes
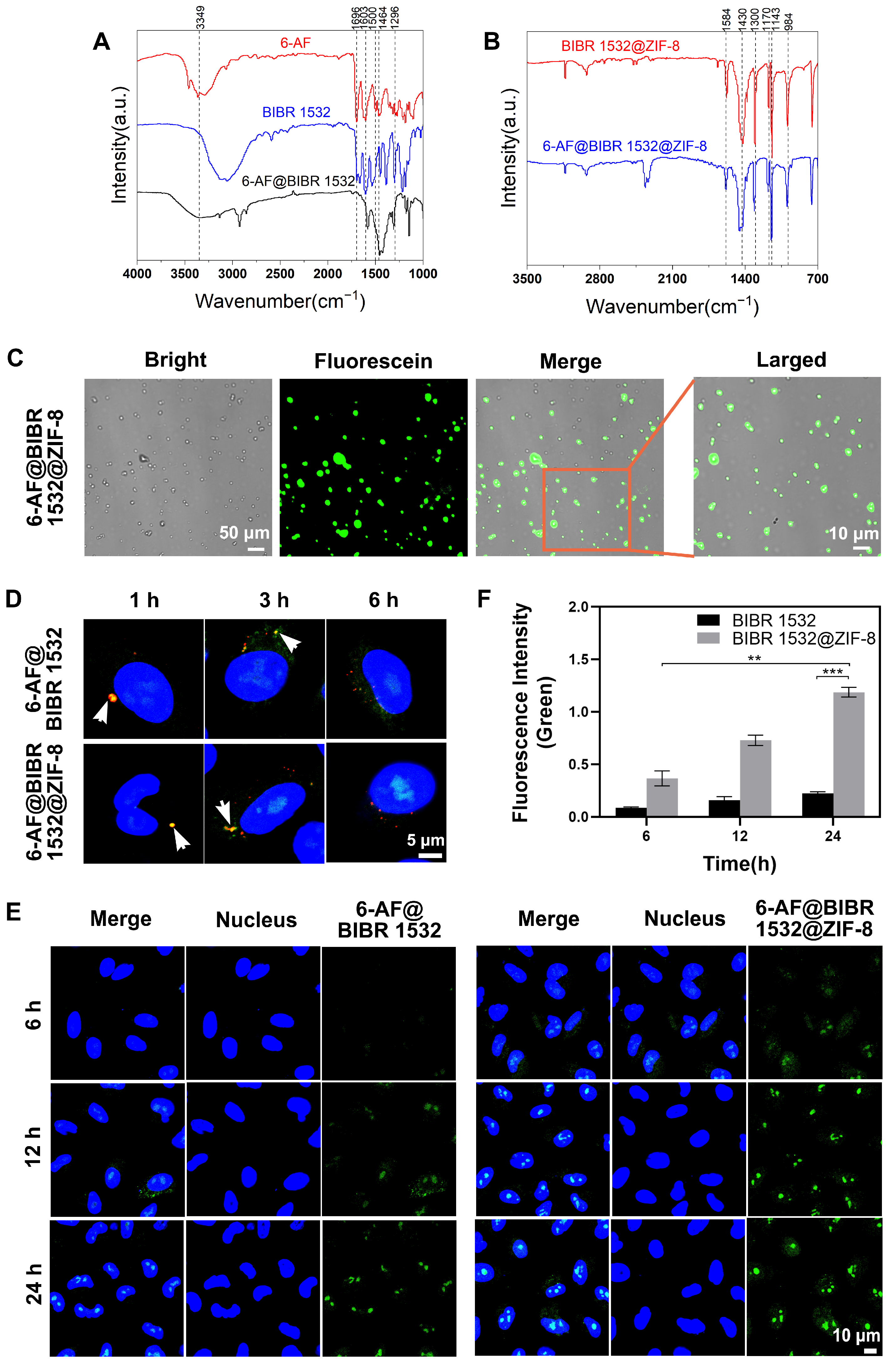
3.3. BIBR 1532 Encapsulation by ZIF-8 Increased Delivery Efficacy
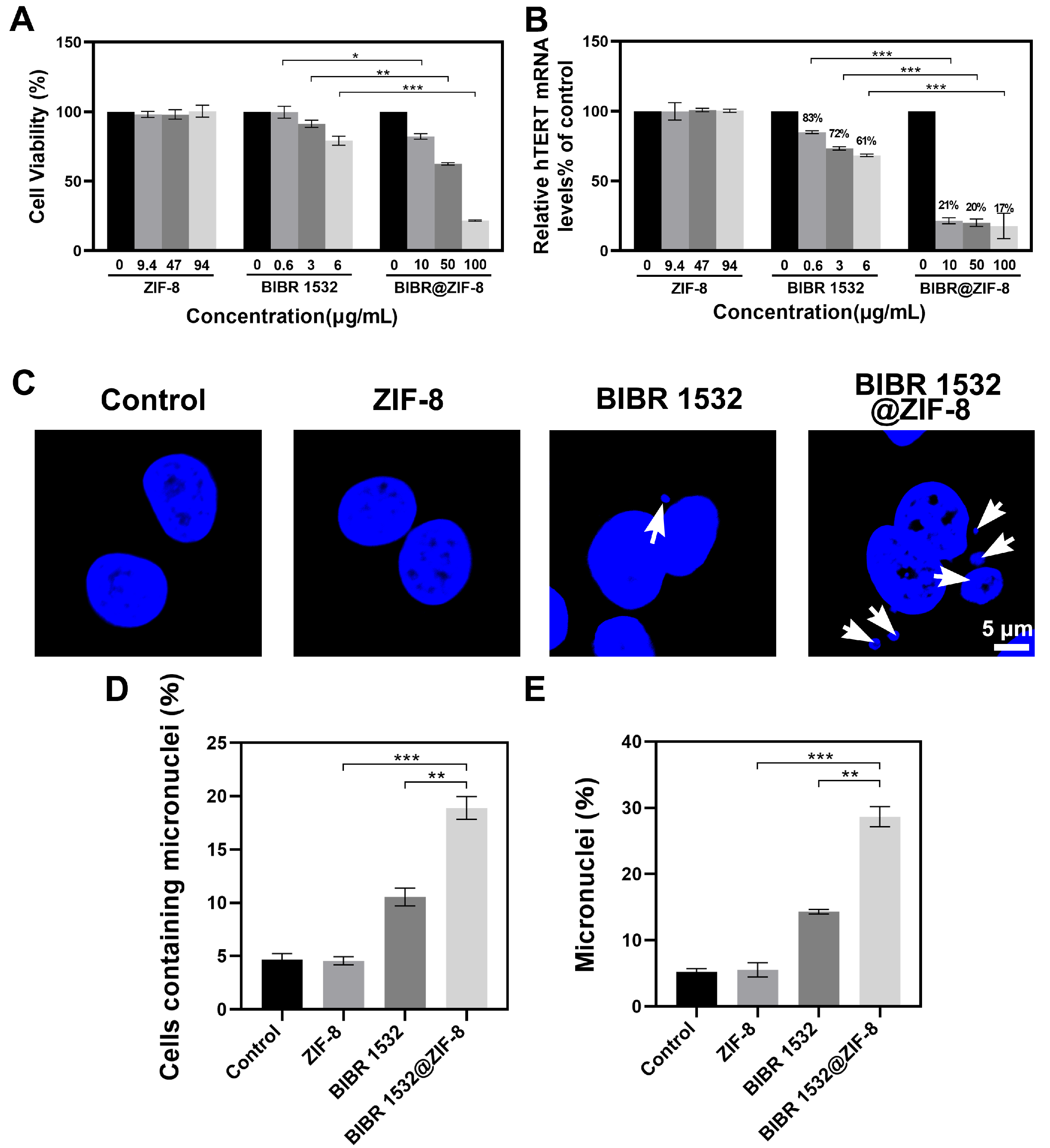
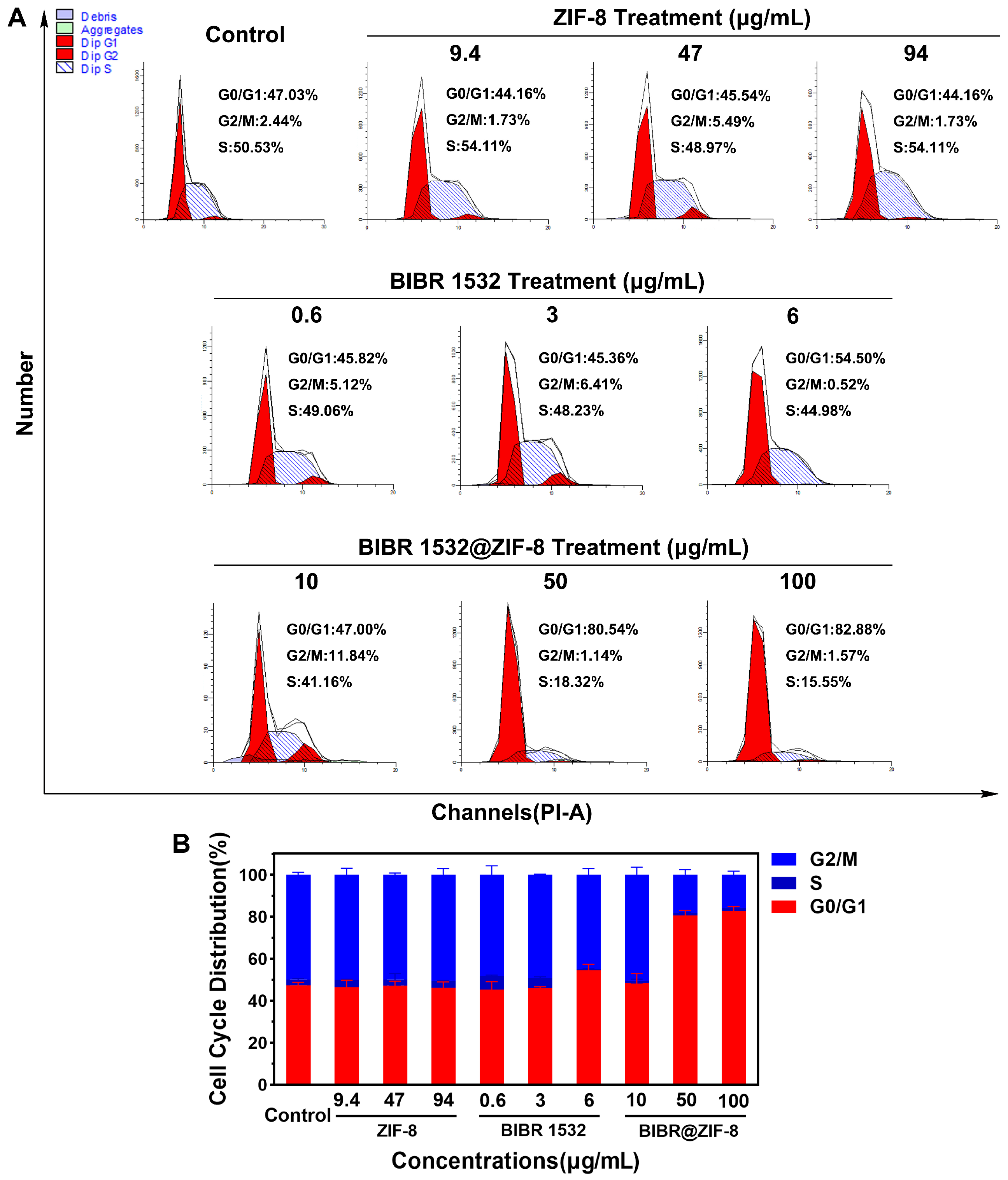
3.4. ZIF-8 Encapsulation Enhanced the Treatment Efficacy of BIBR 1532
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Hahn, W.C.; Meyerson, M. Telomerase activation, cellular immortalization and cancer. Ann. Med. 2001, 33, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.L.; Armando, R.G.; Cerrudo, C.S.; Ghiringhelli, P.D.; Gomez, D.E. Telomerase as a cancer target. Development of new molecules. Curr. Top. Med. Chem. 2016, 16, 2432–2440. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Folini, M.; Perego, P.; Supino, R.; Setti, E.; Daidone, M.G.; Zunino, F.; Zaffaroni, N. Telomerase activity and telomere length in human ovarian cancer and melanoma cell lines: Correlation with sensitivity to DNA damaging agents. Int. J. Oncol. 2000, 16, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Tauchi, T.; Sashida, G.; Sumi, M.; Abe, K.; Yamamoto, K.; Ohyashiki, J.H.; Ohyashiki, K. Telomerase inhibition enhances apoptosis in human acute leukemia cells: Possibility of antitelomerase therapy. Leukemia 2003, 17, 560–567. [Google Scholar] [CrossRef]
- Nowak, J.; Januszkiewicz, D.; Lewandowski, K.; Nowicka-Kujawska, K.; Pernak, M.; Rembowska, J.; Nowak, T.; Wysocki, J. Activity and expression of human telomerase in normal and malignant cells in gastric and colon cancer patients. Eur. J. Gastroenterol. Hepatol. 2003, 15, 75–80. [Google Scholar] [CrossRef]
- Ohmura, Y.; Aoe, M.; Andou, A.; Shimizu, N. Telomerase activity and Bcl-2 expression in non-small cell lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 2980–2987. [Google Scholar]
- Rosen, J.; Jakobs, P.; Ale-Agha, N.; Altschmied, J.; Haendeler, J. Non-canonical functions of telomerase reverse transcriptase—impact on redox homeostasis. Redox Biol. 2020, 34, 101543. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef]
- Guterres, A.N.; Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Krupp, G.; Bonatz, G.; Parwaresch, R. Telomerase, immortality and cancer. Biotechnol. Annu. Rev. 2000, 6, 103–140. [Google Scholar] [PubMed]
- Gurung, R.L.; Balakrishnan, L.; Bhattacharjee, R.N.; Manikandan, J.; Swaminathan, S.; Hande, M.P. Inhibition of poly (ADP-Ribose) polymerase-1 in telomerase deficient mouse embryonic fibroblasts increases arsenite-induced genome instability. Genome Integr. 2010, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Fakhoury, J.; Nimmo, G.A.; Autexier, C. Harnessing telomerase in cancer therapeutics. Anti-Cancer Agents Med. Chem. 2007, 7, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef]
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330. [Google Scholar] [CrossRef]
- Grach, A.A. Alternative telomere-lengthening mechanisms. Cytol. Genet. 2011, 45, 121–130. [Google Scholar] [CrossRef]
- Bryan, C.; Rice, C.; Hoffman, H.; Harkisheimer, M.; Sweeney, M.; Skordalakes, E. Structural basis of telomerase inhibition by the highly specific BIBR1532. Structure 2015, 23, 1934–1942. [Google Scholar] [CrossRef]
- Ding, X.; Cheng, J.; Pang, Q.; Wei, X.; Zhang, X.; Wang, P.; Yuan, Z.; Qian, D. BIBR1532, a selective telomerase inhibitor, enhances radiosensitivity of non-small cell lung cancer through increasing telomere dysfunction and ATM/CHK1 inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, 861–874. [Google Scholar] [CrossRef]
- Pascolo, E.; Wenz, C.; Lingner, J.; Hauel, N.; Priepke, H.; Kauffmann, I.; Garin-Chesa, P.; Rettig, W.J.; Damm, K.; Schnapp, A. mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. J. Biol. Chem. 2002, 277, 15566–15572. [Google Scholar] [CrossRef]
- Damm, K.; Hemmann, U.; Garin-Chesa, P.; Hauel, N.; Kauffmann, I.; Priepke, H.; Niestroj, C.; Daiber, C.; Enenkel, B.; Guilliard, B.; et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001, 20, 6958–6968. [Google Scholar] [CrossRef]
- El-Daly, H.; Kull, M.; Zimmermann, S.; Pantic, M.; Waller, C.F.; Martens, U.M. Selective cytotoxicity and telomere damage in leukemia cells using the telomerase inhibitor BIBR1532. Blood 2005, 105, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, H.; Sheng, X.B.; Liu, X.H.; Chen, F.H. Design, synthesis and SARs of novel telomerase inhibitors based on BIBR1532. Bioorg. Chem. 2020, 102, 104077. [Google Scholar]
- Shi, Y.; Sun, L.; Chen, G.; Zheng, D.; Li, L.; Wei, W. A combination of the telomerase inhibitor, BIBR1532, and paclitaxel synergistically inhibit cell proliferation in breast cancer cell lines. Target. Oncol. 2015, 10, 565–573. [Google Scholar] [PubMed]
- Liu, R.; Liu, J.; Wang, S.; Wang, Y.; Zhang, T.; Liu, Y.; Geng, X.; Wang, F. Combined treatment with emodin and a telomerase inhibitor induces significant telomere damage/dysfunction and cell death. Cell Death Dis. 2019, 10, 527. [Google Scholar] [PubMed]
- Giliopoulos, D.; Zamboulis, A.; Giannakoudakis, D.; Bikiaris, D.; Triantafyllidis, K. Polymer/metal organic framework (MOF) nanocomposites for biomedical applications. Molecules 2020, 25, 185. [Google Scholar]
- Ma, Y.; Su, Z.; Zhou, L.; He, L.; Hou, Z.; Zou, J.; Cai, Y.; Chang, D.; Xie, J.; Zhu, C.; et al. Biodegradable metal-organic-framework-gated organosilica for tumor-microenvironment-unlocked glutathione-depletion-enhanced synergistic therapy. Adv. Mater. 2022, 34, e2107560. [Google Scholar] [CrossRef]
- Yang, J.; Ma, S.; Xu, R.; Wei, Y.; Zhang, J.; Zuo, T.; Wang, Z.; Deng, H.; Yang, N.; Shen, Q. Smart biomimetic metal organic frameworks based on ROS-ferroptosis-glycolysis regulation for enhanced tumor chemo-immunotherapy. J. Control. Release Off. J. Control. Release Soc. 2021, 334, 21–33. [Google Scholar] [CrossRef]
- Pu, Y.; Yin, H.; Dong, C.; Xiang, H.; Wu, W.; Zhou, B.; Du, D.; Chen, Y.; Xu, H. Sono-controllable and ROS-Sensitive CRISPR-Cas9 genome editing for augmented/synergistic ultrasound tumor nanotherapy. Adv. Mater. 2021, 33, e2104641. [Google Scholar]
- Huang, R.; Cai, G.Q.; Li, J.; Li, X.S.; Liu, H.T.; Shang, X.L.; Zhou, J.D.; Nie, X.M.; Gui, R. Platelet membrane-camouflaged silver metal-organic framework drug system against infections caused by methicillin-resistant Staphylococcus aureus. J. Nanobiotechnol. 2021, 19, 229. [Google Scholar]
- Yang, H.; Yu, Z.; Ji, S.; Yan, J.; Han, L.; Liu, Y.; Wang, Y.; Niu, Y.; Huo, Q.; Xu, M. Construction and evaluation of detachable bone-targeting MOF carriers for the delivery of proteasome inhibitors. RSC Adv. 2022, 12, 14707–14715. [Google Scholar] [CrossRef]
- Liang, K.; Ricco, R.; Doherty, C.M.; Styles, M.J.; Bell, S.; Kirby, N.; Mudie, S.; Haylock, D.; Hill, A.J.; Doonan, C.J.; et al. Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun. 2015, 6, 7240. [Google Scholar] [CrossRef] [PubMed]
- Nasi, H.; di Gregorio, M.C.; Wen, Q.; Shimon, L.J.W.; Kaplan-Ashiri, I.; Bendikov, T.; Leitus, G.; Kazes, M.; Oron, D.; Lahav, M.; et al. Directing the morphology, packing, and properties of chiral metal-organic frameworks by cation exchange. Angew. Chem. 2022, 61, e202205238. [Google Scholar]
- Ma, M.; Chen, J.; Liu, H.; Huang, Z.; Huang, F.; Li, Q.; Xu, Y. A review on chiral metal-organic frameworks: Synthesis and asymmetric applications. Nanoscale 2022, 14, 13405–13427. [Google Scholar] [CrossRef]
- di Gregorio, M.C.; Singh, V.; Shimon, L.J.W.; Lahav, M.; van der Boom, M.E. Crystallographic-morphological connections in star shaped metal-organic Frameworks. J. Am. Chem. Soc. 2022, 144, 22838–22843. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Chen, Z.; Dong, J.; Liu, Y.; Cui, Y. Chiral metal-organic frameworks. Chem. Rev. 2022, 122, 9078–9144. [Google Scholar] [CrossRef]
- Yang, J.; Dai, D.; Zhang, X.; Teng, L.; Ma, L.; Yang, Y.W. Multifunctional metal-organic framework (MOF)-based nanoplatforms for cancer therapy: From single to combination therapy. Theranostics 2023, 13, 295–323. [Google Scholar] [CrossRef]
- Pan, W.; Li, Z.; Qiu, S.; Dai, C.; Wu, S.; Zheng, X.; Guan, M.; Gao, F. Octahedral Pt-MOF with Au deposition for plasmonic effect and Schottky junction enhanced hydrogenothermal therapy of rheumatoid arthritis. Mater. Today Bio. 2022, 13, 100214. [Google Scholar] [CrossRef]
- Tanaka, S.; Kida, K.; Okita, M.; Ito, Y.; Miyake, Y. Size-controlled synthesis of zeolitic imidazolate framework-8 (ZIF-8) crystals in an aqueous system at room temperature. Chem. Lett. 2012, 41, 1337–1339. [Google Scholar] [CrossRef]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef]
- Hou, C.; Wang, Y.; Ding, Q.; Jiang, L.; Li, M.; Zhu, W.; Pan, D.; Zhu, H.; Liu, M. Facile synthesis of enzyme-embedded magnetic metal-organic frameworks as a reusable mimic multi-enzyme system: Mimetic peroxidase properties and colorimetric sensor. Nanoscale 2015, 7, 18770–18779. [Google Scholar] [CrossRef]
- Ding, L.; Lin, X.; Lin, Z.; Wu, Y.; Liu, X.; Liu, J.; Wu, M.; Zhang, X.; Zeng, Y. Cancer cell-targeted photosensitizer and therapeutic protein co-delivery nanoplatform based on a metal-organic framework for enhanced synergistic photodynamic and protein therapy. ACS Appl. Mater. Interfaces 2020, 12, 36906–36916. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, K.; Liu, P.; Chen, M.; Zhong, Y.; Ye, Q.; Wei, M.Q.; Zhao, H.; Tang, Z. Encapsulation of plasmid DNA by nanoscale metal-organic frameworks for efficient gene transportation and expression. Adv. Mater. 2019, 31, e1901570. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.A.; Abuçafy, M.P.; Barbosa, T.W.L.; da Silva, B.L.; Fulindi, R.B.; Isquibola, G.; da Costa, P.I.; Chiavacci, L.A. ZnO@ZIF-8 nanoparticles as nanocarrier of ciprofloxacin for antimicrobial activity. Pharmaceutics 2023, 15, 259. [Google Scholar] [CrossRef]
- Ju, G.; Liu, B.; Ji, M.; Jin, R.; Xu, X.; Xiao, Y.; Li, J.; Xu, D.; Huang, Y.; Hou, J. Folic acid-modified miR-491-5p-loaded ZIF-8 nanoparticles inhibit castration-resistant prostate cancer by regulating the expression of EPHX1. Front. Bioeng. Biotechnol. 2021, 9, 706536. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, Y.; Zeng, G.; Zhao, L.; Lai, Z. Rapid synthesis of zeolitic imidazolate framework-8 (ZIF-8) nanocrystals in an aqueous system. Chem. Commun. 2011, 47, 2071–2073. [Google Scholar] [CrossRef]
- Zhuang, J.; Kuo, C.H.; Chou, L.Y.; Liu, D.Y.; Weerapana, E.; Tsung, C.K. Optimized metal-organic-framework nanospheres for drug delivery: Evaluation of small-molecule encapsulation. ACS Nano 2014, 8, 2812–2819. [Google Scholar] [CrossRef] [PubMed]
- Novio, F.; Lorenzo, J.; Nador, F.; Wnuk, K.; Ruiz-Molina, D. Carboxyl group functionalized coordination polymer nanoparticles as efficient platforms for drug delivery. Chemistry 2014, 20, 15443–15450. [Google Scholar] [CrossRef]
- Sommer, S.; Buraczewska, I.; Kruszewski, M. Micronucleus assay: The state of art, and future directions. Int. J. Mol. Sci. 2020, 21, 1534. [Google Scholar] [CrossRef]
- Yoon, M.C.; Hook, V.; O’Donoghue, A.J. Cathepsin B dipeptidyl carboxypeptidase and endopeptidase activities demonstrated across a broad pH range. Biochemistry 2022, 61, 1904–1914. [Google Scholar] [CrossRef]
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006, 27, 495–502. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Li, X.; Qiu, K.; Jiao, F.; Liu, Y.; Kong, Q.; Liu, Y.; Wu, Y. Proteasome inhibition activates autophagy-lysosome pathway associated with TFEB dephosphorylation and nuclear translocation. Front. Cell Dev. Biol. 2019, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, M.A.; Jamshidi, M. Overexpression of lysosome-associated membrane protein 1 in oral squamous cell carcinoma and its correlation with tumor differentiation and metastasis. Iran. J. Otorhinolaryngol. 2022, 34, 3–8. [Google Scholar] [PubMed]
- Surrallés, J.; Catalán, J.; Creus, A.; Norppa, H.; Xamena, N.; Marcos, R. Micronuclei induced by alachlor, mitomycin C and vinblastine in human lymphocytes: Presence of centromeres and kinetochores and influence of staining technique. Mutagenesis 1995, 10, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Garaj-Vrhovac, V.; Kopjar, N.; Besendorfer, V.; Papes, D. Induction of micronuclei in human lymphocytes after occupational exposure to ultrasound. Chemosphere 1999, 38, 3541–3553. [Google Scholar] [CrossRef] [PubMed]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev, V.V., Jr.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-loop-based telomeric DNA damage. Nat. Commun. 2021, 12, 3542. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Arora, R.; Wischnewski, H.; Azzalin, C.M. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018, 25, 147–153. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, S.; Choi, J.H.; Na, K. ι-Carrageenan nanocomposites for enhanced stability and oral bioavailability of curcumin. Biomater. Res. 2021, 25, 32. [Google Scholar] [CrossRef]
- Que, H.; Hong, W.; Lan, T.; Zeng, H.; Chen, L.; Wan, D.; Bi, Z.; Ren, W.; Luo, M.; Yang, J.; et al. Tripterin liposome relieves severe acute respiratory syndrome as a potent COVID-19 treatment. Signal Transduct. Target. Ther. 2022, 7, 399. [Google Scholar] [CrossRef]
- Tian, C.; Guo, J.; Wang, G.; Sun, B.; Na, K.; Zhang, X.; Xu, Z.; Cheng, M.; He, Z.; Sun, J. Efficient intestinal digestion and on site tumor-bioactivation are the two important determinants for chylomicron-mediated lymph-targeting triglyceride-mimetic docetaxel oral prodrugs. Adv. Sci. 2019, 6, 1901810. [Google Scholar] [CrossRef]
- Mundaca-Uribe, R.; Karshalev, E.; de Ávila, B.E.-F.; Wei, X.; Nguyen, B.; Litvan, I.; Fang, R.H.; Zhang, L.; Wang, J. A Microstirring pill enhances bioavailability of orally administered drugs. Adv. Sci. 2021, 8, 2100389. [Google Scholar] [CrossRef]
- Yan, L.; Chen, X.; Wang, Z.; Zhang, X.; Zhu, X.; Zhou, M.; Chen, W.; Huang, L.; Roy, V.A.L.; Yu, P.K.N.; et al. Size controllable and surface tunable zeolitic imidazolate framework-8-poly(acrylic acid sodium salt) nanocomposites for pH responsive drug release and enhanced in vivo cancer treatment. ACS Appl. Mater. Interfaces 2017, 9, 32990–33000. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Zhao, X.; Su, Z.; Wang, C.; Lin, Y. Layer-by-layer decorated nanoscale ZIF-8 with high curcumin loading effectively inactivates gram-negative and gram-positive bacteria. ACS Appl. Bio Mater. 2020, 3, 3673–3680. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, Y.; Ding, W.; Li, F.; Lin, J.; Wu, M.; Wu, J.; Wen, L.P.; Qiu, B.; Wei, P.F.; et al. Rationally designed rapamycin-encapsulated ZIF-8 nanosystem for overcoming chemotherapy resistance. Biomaterials 2020, 258, 120308. [Google Scholar] [CrossRef]
- Moreira, C.; Oliveira, H.; Pires, L.R.; Simões, S.; Barbosa, M.A.; Pêgo, A.P. Improving chitosan-mediated gene transfer by the introduction of intracellular buffering moieties into the chitosan backbone. Acta Biomater. 2009, 5, 2995–3006. [Google Scholar] [CrossRef] [PubMed]
- Pack, D.W.; Putnam, D.; Langer, R. Design of imidazole-containing endosomolytic biopolymers for gene delivery. Biotechnol. Bioeng. 2000, 67, 217–223. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef]
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef]
- Berg, K.; Selbo, P.K.; Prasmickaite, L.; Tjelle, T.E.; Sandvig, K.; Moan, J.; Gaudernack, G.; Fodstad, O.; Kjølsrud, S.; Anholt, H.; et al. Photochemical internalization: A novel technology for delivery of macromolecules into cytosol. Cancer Res. 1999, 59, 1180–1183. [Google Scholar]
- Chen, J.; Li, J.; Zhou, J.; Lin, Z.; Cavalieri, F.; Czuba-Wojnilowicz, E.; Hu, Y.; Glab, A.; Ju, Y.; Richardson, J.J.; et al. Metal-phenolic coatings as a platform to trigger endosomal escape of nanoparticles. ACS Nano 2019, 13, 11653–11664. [Google Scholar] [CrossRef]
- Xi, Z.; Ahmad, E.; Zhang, W.; Li, J.; Wang, A.; Wang, N.; Zhu, C.; Huang, W.; Xu, L.; Yu, M.; et al. Dual-modified nanoparticles overcome sequential absorption barriers for oral insulin delivery. J. Control. Release Off. J. Control. Release Soc. 2022, 342, 1–13. [Google Scholar] [CrossRef]
- Wojnilowicz, M.; Glab, A.; Bertucci, A.; Caruso, F.; Cavalieri, F. Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering RNA. ACS Nano 2019, 13, 187–202. [Google Scholar] [CrossRef]
- Donahue, N.D.; Acar, H.; Wilhelm, S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 68–96. [Google Scholar] [CrossRef] [PubMed]
- Linnane, E.; Haddad, S.; Melle, F.; Mei, Z.; Fairen-Jimenez, D. The uptake of metal-organic frameworks: A journey into the cell. Chem. Soc. Rev. 2022, 51, 6065–6086. [Google Scholar] [CrossRef] [PubMed]
- Sousa de Almeida, M.; Susnik, E.; Drasler, B.; Taladriz-Blanco, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem. Soc. Rev. 2021, 50, 5397–5434. [Google Scholar] [CrossRef]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Espín, D.; Serrano, M. Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Lavanya, C.; Venkataswamy, M.M.; Sibin, M.K.; Srinivas Bharath, M.M.; Chetan, G.K. Down regulation of human telomerase reverse transcriptase (hTERT) expression by BIBR1532 in human glioblastoma LN18 cells. Cytotechnology 2018, 70, 1143–1154. [Google Scholar] [CrossRef]
- Wu, Y.; Zhong, D.; Li, Y.; Wu, H.; Xu, X.; Yang, J.; Gu, Z. Tumor-oriented telomerase-terminated nanoplatform as versatile strategy for multidrug resistance reversal in cancer treatment. Adv. Healthc. Mater. 2020, 9, e1901739. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Li, J.; Yan, L.; You, Y.; Zhao, F.; Cheng, J.; Yang, L.; Sun, Y.; Chang, Q.; Liu, R.; et al. Zeolitic Imidazolate Framework-8 (ZIF-8) as a Drug Delivery Vehicle for the Transport and Release of Telomerase Inhibitor BIBR 1532. Nanomaterials 2023, 13, 1779. https://doi.org/10.3390/nano13111779
Zhang S, Li J, Yan L, You Y, Zhao F, Cheng J, Yang L, Sun Y, Chang Q, Liu R, et al. Zeolitic Imidazolate Framework-8 (ZIF-8) as a Drug Delivery Vehicle for the Transport and Release of Telomerase Inhibitor BIBR 1532. Nanomaterials. 2023; 13(11):1779. https://doi.org/10.3390/nano13111779
Chicago/Turabian StyleZhang, Shunyu, Jinxia Li, Liang Yan, Yue You, Feng Zhao, Jixing Cheng, Limin Yang, Yanqi Sun, Qingchao Chang, Ru Liu, and et al. 2023. "Zeolitic Imidazolate Framework-8 (ZIF-8) as a Drug Delivery Vehicle for the Transport and Release of Telomerase Inhibitor BIBR 1532" Nanomaterials 13, no. 11: 1779. https://doi.org/10.3390/nano13111779
APA StyleZhang, S., Li, J., Yan, L., You, Y., Zhao, F., Cheng, J., Yang, L., Sun, Y., Chang, Q., Liu, R., & Li, Y. (2023). Zeolitic Imidazolate Framework-8 (ZIF-8) as a Drug Delivery Vehicle for the Transport and Release of Telomerase Inhibitor BIBR 1532. Nanomaterials, 13(11), 1779. https://doi.org/10.3390/nano13111779







