Macrophage Polarization Status Impacts Nanoceria Cellular Distribution but Not Its Biotransformation or Ferritin Effects
Abstract
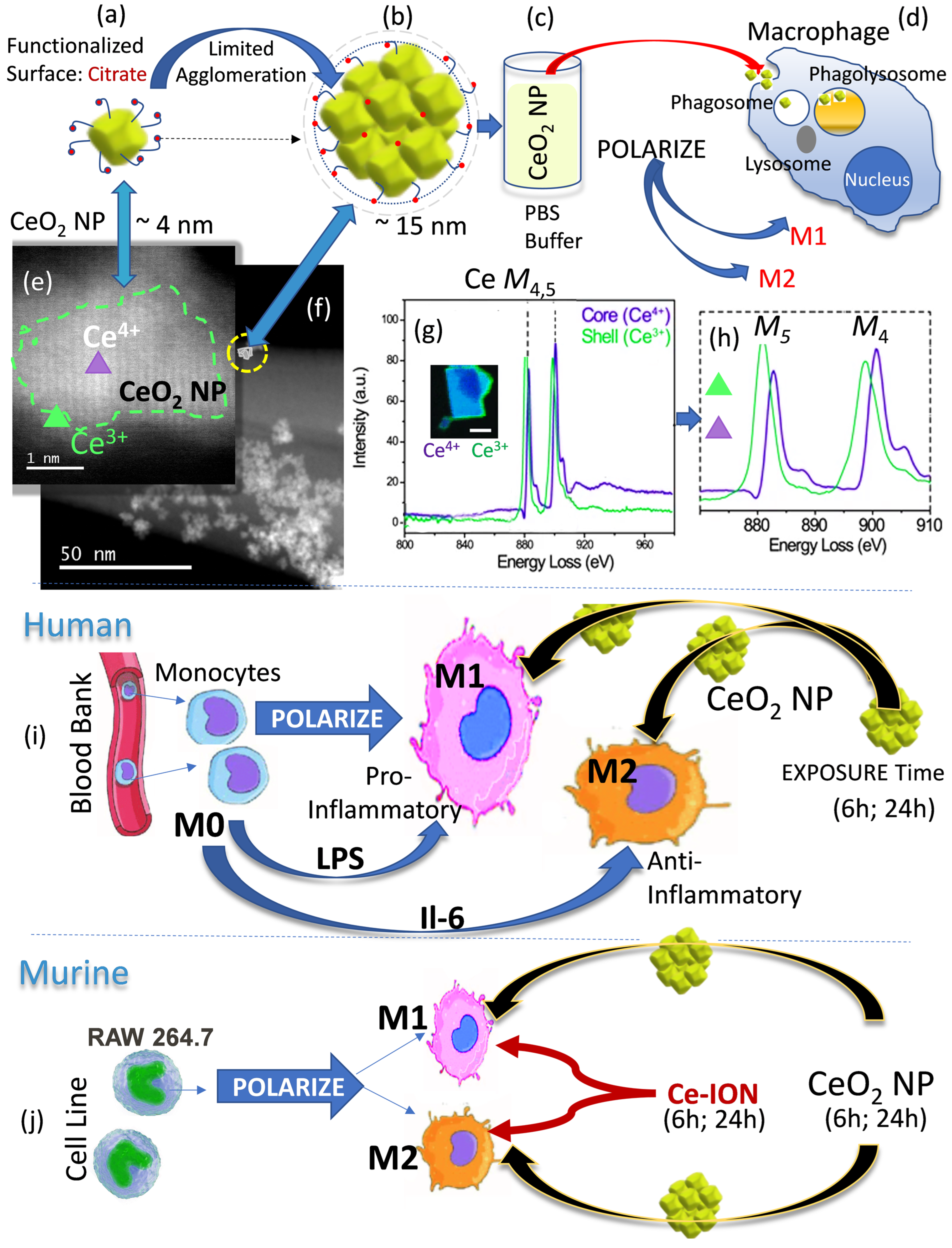
1. Introduction
2. Materials and Methods
2.1. Human Peripheral Blood Monocyte (M0) Source, Macrophage Differentiation, and Polarization to M1- and M2-like Cells
2.2. Murine Macrophage Polarization to M1 and M2 Cells and Polarization Verification
2.3. Nanoparticle Synthesis
2.4. Cell Treatment Procedures
2.5. Analytical Imaging Techniques for NPs in Tissues
3. Results and Discussion
3.1. Nanoceria Physicochemical Properties
3.2. Macrophage Polarization and Nanoceria Exposure
3.3. Analytical Electron Microscopy of CeO2 NP Uptake and Biotransformation
3.4. Nanoceria Cellular Distribution
3.5. Nanoceria and Cerium Ion Effects on Macrophage Polarization
3.6. Ferritin Biomineralization after CeO2 NP and Ce-Ion Exposure
4. Conclusions and Further Applications
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahapatro, A.; Singh, D.K. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J. Nanobiotechnol. 2011, 9, 55. [Google Scholar] [CrossRef]
- Kaittanis, C.; Shaffer, T.M.; Thorek, D.L.J.; Grimm, J. Dawn of advanced molecular medicine: Nanotechnological advancements in cancer imaging and therapy. Crit. Rev. Oncog. 2014, 19, 143–176. [Google Scholar] [CrossRef]
- Taki, A.; Smooker, P. Small wonders-the use of nanoparticles for delivering antigen. Vaccines 2015, 3, 638–661. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From monocytes to M1/M2 macrophages: Phenotypical vs. functional differentiation. Front. Immunol. 2014, 5, 1–22. [Google Scholar] [CrossRef]
- Nielsen, M.C.; Andersen, M.N.; Moller, H.J. Monocyte isolation techniques significantly impact the phenotype of both isolated monocytes and derived macrophages in vitro. Immunology 2020, 159, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Foroozandeh, P.; Aziz, A.A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 2018, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sohaebuddin, S.K.; Thevenot, P.T.; Baker, D.; Eaton, J.W.; Tang, L. Nanomaterial cytotoxicity is composition, size, and cell type dependent. Part. Fibre Toxicol. 2010, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Knoll, R.; Schultze, J.L.; Schulte-Schrepping, J. Monocytes and macrophages in COVID-19. Front. Immunol. 2021, 12, 720109. [Google Scholar] [CrossRef]
- Thakur, N.; Manna, P.; Das, J. Synthesis and biomedical applications of nanoceria, a redox active nanoparticle. J. Nanobiotechnol. 2019, 17, 1–27. [Google Scholar] [CrossRef]
- Nel, A.E.; Madler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Seaton, A.; Tran, L.; Aitken, R.; Donaldson, K. Nanoparticles, human health hazard and regulation. J. R. Soc. Interface 2010, 7, S119. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.R.; Greenamyre, J.T. The role of environmental exposures in neurodegeneration and neurodegenerative diseases. Toxicol. Sci. 2011, 124, 225–250. [Google Scholar] [CrossRef]
- Teleanu, D.M.; Chircov, C.; Grumezescu, A.M.; Volceanov, A.; Teleanu, R.I. Impact of nanoparticles on brain health: An up to date overview. J. Clin. Med. Res. 2018, 7, 490. [Google Scholar] [CrossRef]
- Hu, G.; Guo, M.; Xu, J.; Wu, F.; Fan, J.; Huang, Q.; Yang, G.; Lv, Z.; Wang, X.; Jin, Y. Nanoparticles targeting macrophages as potential clinical therapeutic agents against cancer and inflammation. Front. Immunol. 2019, 10, 1998. [Google Scholar] [CrossRef]
- Laskin, D.L.; Sunil, V.R.; Gardner, C.R.; Laskin, J.D. Macrophages and tissue injury: Agents of defense or destruction? Annu. Rev. Pharmacol. Toxicol. 2011, 51, 267–288. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- MacParland, S.A.; Tsoi, K.M.; Ouyang, B.; Ma, X.Z.; Manuel, J.; Fawaz, A.; Ostrowski, M.A.; Alman, B.A.; Zilman, A.; Chan, W.C.; et al. Phenotype determines nanoparticle uptake by human macrophages from liver and blood. ACS Nano 2017, 11, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O. Regulators of macrophage activation. Eur. J. Immunol. 2011, 41, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Lichtnekert, J.; Kawakami, T.; Parks, W.C.; Duffield, J.S. Changes in macrophage phenotype as the immune response evolves. Curr. Opin. Pharmacol. 2013, 13, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Vannella, K.M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The metabolic signature of macrophage responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Fuchs, A.-K.; Syrovets, T.; Haas, K.A.; Loos, C.; Musyanovych, A.; Mailänder, V.; Landfester, K.; Simmet, T. Carboxyl- and amino-functionalized polystyrene nanoparticles differentially affect the polarization profile of M1 and M2 macrophage subsets. Biomaterials 2016, 85, 78–87. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Laskin, D.L. Macrophages and inflammatory mediators in chemical toxicity: A battle of forces. Chem. Res. Toxicol. 2009, 22, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. Macrophage polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Munder, M.; Eichmann, K.; Modolell, M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: Competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J. Immunol. 1998, 160, 5347–5354. [Google Scholar] [CrossRef]
- Miao, X.; Leng, X.; Zhang, Q. The current state of nanoparticle-induced macrophage polarization and reprogramming research. Int. J. Mol. Sci. 2017, 18, 336. [Google Scholar] [CrossRef]
- Mosser, D.M. The many faces of macrophage activation. J. Leukoc. Biol. 2003, 73, 209–212. [Google Scholar] [CrossRef]
- Zanganeh, S.; Hutter, G.; Spitler, R.; Lenkov, O.; Mahmoudi, M.; Shaw, A.; Pajarinen, J.S.; Nejadnik, H.; Goodman, S.; Moseley, M.; et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat. Nanotechnol. 2016, 11, 986–994. [Google Scholar] [CrossRef]
- Ye, Y.; Cota-Ruiz, K.; Cantu, J.M.; Valdes, C.; Gardea-Torresdey, J.L. Engineered nanomaterials’ fate assessment in biological matrices: Recent milestones in electron microscopy. ACS Sustain. Chem. Eng. 2021, 9, 4341–4356. [Google Scholar] [CrossRef]
- Herd, H.L.; Bartlett, K.T.; Gustafson, J.A.; McGill, L.D.; Ghandehari, H. Macrophage silica nanoparticle response is phenotypically dependent. Biomaterials 2015, 53, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Qie, Y.; Yuan, H.; von Roemeling, C.A.; Chen, Y.; Liu, X.; Shih, K.D.; Knight, J.A.; Tun, H.W.; Wharen, R.E.; Jiang, W.; et al. Surface modification of nanoparticles enables selective evasion of phagocytic clearance by distinct macrophage phenotypes. Sci. Rep. 2016, 6, 26269. [Google Scholar] [CrossRef] [PubMed]
- Gallud, A.; Bondarenko, O.; Feliu, N.; Kupferschmidt, N.; Atluri, R.; Garcia-Bennett, A.; Fadeel, B. Macrophage activation status determines the internalization of mesoporous silica particles of different sizes: Exploring the role of different pattern recognition receptors. Biomaterials 2017, 121, 28–40. [Google Scholar] [CrossRef]
- Hoppstädter, J.; Seif, M.; Dembek, A.; Cavelius, C.; Huwer, H.; Kraegeloh, A.; Kiemer, A.K. M2 polarization enhances silica nanoparticle uptake by macrophages. Front. Pharmacol. 2015, 6, 1–12. [Google Scholar] [CrossRef]
- Medrano-Bosch, M.; Moreno-Lanceta, A.; Melgar-Lesmes, P. Nanoparticles to target and treat macrophages: The Ockham′s Concept? Pharmaceutics 2021, 13, 1340. [Google Scholar] [CrossRef]
- Lara, S.; Perez-Potti, A.; Herda, L.M.; Adumeau, L.; Dawson, K.A.; Yan, Y. Differential recognition of nanoparticle protein corona and modified low-density lipoprotein by macrophage receptor with collagenous structure. ACS Nano 2018, 12, 4930–4937. [Google Scholar] [CrossRef]
- Graham, U.M.; Dozier, A.K.; Oberdörster, G.; Yokel, R.A.; Molina, R.; Brain, J.D.; Pinto, J.M.; Weuve, J.; Bennett, D.A. Tissue specific fate of nanomaterials by advanced analytical imaging techniques-A review. Chem. Res. Toxicol. 2020, 33, 1145–1162. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.Y.; Song, S.Y.; Go, S.-h.; Sohn, H.S.; Baik, S.; Soh, M.; Kim, K.; Kim, D.; Kim, H.-C.; et al. Synergistic oxygen generation and reactive oxygen species scavenging by manganese ferrite/ceria co-decorated nanoparticles for rheumatoid arthritis treatment. ACS Nano 2019, 13, 3206–3217. [Google Scholar] [CrossRef]
- Wang, C.; Sun, J.; Chen, Q.; Chen, J.; Wan, H.; Ni, Q.; Yuan, H.; Wu, J.; Wang, Y.; Sun, B. Application of nanotechnology-based products in stroke. ACS Chem. Neurosci. 2023, 14, 2405–2415. [Google Scholar] [CrossRef]
- Graham, U.M.; Tseng, M.T.; Jasinski, J.B.; Yokel, R.A.; Unrine, J.M.; Davis, B.H.; Dozier, A.K.; Hardas, S.S.; Sultana, R.; Grulke, E.; et al. In vivo processing of ceria nanoparticles inside liver: Impact on free radical scavenging activity and oxidative stress. ChemPlusChem 2014, 79, 1083–1088. [Google Scholar] [CrossRef]
- Graham, U.M.; Yokel, R.A.; Dozier, A.K.; Drummy, L.; Mahalingam, K.; Tseng, M.T.; Birch, E.; Fernback, J. Analytical high-resolution electron microscopy reveals organ specific nanoceria bioprocessing. Toxicol. Pathol. 2018, 46, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Balfourier, A.; Luciani, N.; Wang, G.; Lelong, G.; Ersen, O.; Khelfa, A.; Alloyeau, D.; Gazeau, F.; Carn, F. Unexpected intracellular biodegradation and recrystallization of gold nanoparticles. Proc. Natl. Acad. Sci. USA 2020, 117, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Barth, K.A.; Waterfield, J.D.; Brunette, D.M. The effect of surface roughness on RAW 264.7 macrophage phenotype. J. Biomed. Mater. Res. A 2013, 101A, 2679–2688. [Google Scholar] [CrossRef] [PubMed]
- Corraliza, I.M.; Campo, M.L.; Soler, G.; Modolell, M. Determination of arginase activity in macrophages: A micromethod. J. Immunol. Methods 1994, 174, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Yokel, R.A.; Ensor, M.L.; Vekaria, H.J.; Sullivan, P.G.; Feola, D.J.; Stromberg, A.; Tseng, M.T.; Harrison, D.A. Cerium dioxide, a Jekyll and Hyde nanomaterial, can increase basal and decrease elevated inflammation and oxidative stress. Nanomedicine 2022, 43, 102565. [Google Scholar] [CrossRef]
- Koo, S.-J.; Chowdhury, I.H.; Szczesny, B.; Wan, X.; Garg, N.J. Macrophages promote oxidative metabolism to drive nitric oxide generation in response to Trypanosoma Cruzi. Infect. Immun. 2016, 84, 3527–3541. [Google Scholar] [CrossRef]
- Scrima, R.; Menga, M.; Pacelli, C.; Agriesti, F.; Cela, O.; Piccoli, C.; Cotoia, A.; De Gregorio, A.; Gefter, J.V.; Cinnella, G.; et al. Para-hydroxyphenylpyruvate inhibits the pro-inflammatory stimulation of macrophage preventing LPS-mediated nitro-oxidative unbalance and immunometabolic shift. PLoS ONE 2017, 12, e0188683. [Google Scholar] [CrossRef]
- Biswas, S.; Adrian, M.; Evdokimov, K.; Schledzewski, K.; Weber, J.; Winkler, M.; Goerdt, S.; Géraud, C. Counter-regulation of the ligand-receptor pair Leda-1/Pianp and Pilralpha during the LPS-mediated immune response of murine macrophages. Biochem. Biophys. Res. Commun. 2015, 464, 1078–1083. [Google Scholar] [CrossRef]
- Saxena, R.K.; Vallyathan, V.; Lewis, D.M. Evidence for lipopolysaccharide-induced differentiation of RAW264.7 murine macrophage cell line into dendritic like cells. J. Biosci. 2003, 28, 129–134. [Google Scholar] [CrossRef]
- McWhorter, F.Y.; Wang, T.; Nguyen, P.; Chung, T.; Liu, W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. USA 2013, 110, 17253–17258. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, D.; Tredici, I.G.; Ghigna, P.; Castillio-Michel, H.; Falqui, A.; Di Benedetto, C.; Alberti, G.; Ricci, V.; Anselmi-Tamburini, U.; Sommi, P. Dependence of the Ce(III)/Ce(IV) ratio on intracellular localization in ceria nanoparticles internalized by human cells. Nanoscale 2017, 9, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Hancock, M.L.; Yokel, R.A.; Beck, M.J.; Calahan, J.L.; Jarrells, T.W.; Munson, E.J.; Olaniyan, G.A.; Grulke, E.A. The characterization of purified citrate-coated cerium oxide nanoparticles prepared via hydrothermal synthesis. Appl. Surf. Sci. 2021, 535, 147681. [Google Scholar] [CrossRef]
- Hui, Y.; Yi, X.; Wibowo, D.; Yang, G.; Middelberg, A.P.J.; Gao, H.; Zhao, C.-X. Nanoparticle elasticity regulates phagocytosis and cancer cell uptake. Sci. Adv. 2020, 6, eaaz4316. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Ong, S.P.; Hautier, G.; Chen, W.; Richards, W.D.; Dacek, S.; Cholia, S.; Gunter, D.; Skinner, D.; Ceder, G.; et al. Commentary: The Materials Project: A materials genome approach to accelerating materials innovation. APL Mater. 2013, 1, 011002. [Google Scholar] [CrossRef]
- Szymanski, C.J.; Munusamy, P.; Mihai, C.; Xie, Y.; Hu, D.; Gilles, M.K.; Tyliszczak, T.; Thevuthasan, S.; Baer, D.R.; Orr, G. Shifts in oxidation states of cerium oxide nanoparticles detected inside intact hydrated cells and organelles. Biomaterials 2015, 62, 147–154. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, Y.; Zhang, Z.; He, X.; Zhang, J.; Guo, Z.; Tai, R.; Zhao, Y.; Chai, Z. Biotransformation of ceria nanoparticles in cucumber plants. ACS Nano 2012, 6, 9943–9950. [Google Scholar] [CrossRef]
- Brun, E.; Barreau, F.; Veronesi, G.; Fayard, B.; Sorieul, S.; Chaneac, C.; Carapito, C.; Rabilloud, T.; Mabondzo, A.; Herlin-Boime, N.; et al. Titanium dioxide nanoparticle impact and translocation through ex vivo, in vivo and in vitro gut epithelia. Part. Fibre Toxicol. 2014, 11, 13. [Google Scholar] [CrossRef]
- Yameen, B.; Choi, W.I.; Vilos, C.; Swami, A.; Shi, J.; Farokhzad, O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 2014, 190, 485–499. [Google Scholar] [CrossRef]
- Tsuda, A.; Venkata, N.K. The role of natural processes and surface energy of inhaled engineered nanoparticles on aggregation and corona formation. NanoImpact 2016, 2, 38–44. [Google Scholar] [CrossRef]
- Wachtel, E.; Lubomirsky, I. The elastic modulus of pure and doped ceria. Scr. Mater. 2011, 65, 112–117. [Google Scholar] [CrossRef]
- Gunawan, C.; Lord, M.S.; Lovell, E.; Wong, R.J.; Jung, M.S.; Oscar, D.; Mann, R.; Amal, R. Oxygen-vacancy engineering of cerium-oxide nanoparticles for antioxidant activity. ACS Omega 2019, 4, 9473–9479. [Google Scholar] [CrossRef] [PubMed]
- Schanen, B.C.; Das, S.; Reilly, C.M.; Warren, W.L.; Self, W.T.; Seal, S.; Drake, D.R., III. Immunomodulation and T helper TH1/TH2 response polarization by CeO2 and TiO2 nanoparticles. PLoS ONE 2013, 8, e62816. [Google Scholar] [CrossRef]
- Zeng, H.-L.; Shi, J.-M. The role of microglia in the progression of glaucomatous neurodegeneration- a review. Int. J Ophthalmol. 2018, 11, 143–149. [Google Scholar] [CrossRef]
- Yokel, R.A.; Tseng, M.T.; Butterfield, D.A.; Hancock, M.L.; Grulke, E.A.; Unrine, J.M.; Stromberg, A.J.; Dozier, A.K.; Graham, U.M. Nanoceria distribution and effects are mouse-strain dependent. Nanotoxicology 2020, 14, 827–846. [Google Scholar] [CrossRef]
- Rosario, C.; Zandman-Goddard, G.; Meyron-Holtz, E.G.; D’Cruz, D.P.; Shoenfeld, Y. The Hyperferritinemic Syndrome: Macrophage activation syndrome, Still’s disease, septic shock and catastrophic antiphospholipid syndrome. BMC Med. 2013, 11, 185. [Google Scholar] [CrossRef]
- Bradley, J.M.; Moore, G.R.; Le Brun, N.E. Mechanisms of iron mineralization in ferritins: One size does not fit all. JBIC J. Biol. Inorg. Chem. 2014, 19, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Honarmand Ebrahimi, K.; Bill, E.; Hagedoorn, P.-L.; Hagen, W.R. The catalytic center of ferritin regulates iron storage via Fe(II)-Fe(III) displacement. Nat. Chem. Biol. 2012, 8, 941–948. [Google Scholar] [CrossRef]
- Maher, B.A.; Ahmed, I.A.M.; Karloukovski, V.; MacLaren, D.A.; Foulds, P.G.; Allsop, D.; Mann, D.M.A.; Torres-Jardon, R.; Calderon-Garciduenas, L. Magnetite pollution nanoparticles in the human brain. Proc. Natl. Acad. Sci. USA 2016, 113, 10797–10801. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Merad, M.; Martin, J.C. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Pastora, J.; Weigand, M.; Kim, J.; Wu, X.; Strayer, J.; Palmer, A.F.; Zborowski, M.; Yazer, M.; Chalmers, J.J. Hyperferritinemia in critically ill COVID-19 patients-Is ferritin the product of inflammation or a pathogenic mediator? Clin. Chim. Acta 2020, 509, 249–251. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Diao, B.; Wang, R.; Wang, G.; Wang, C.; Tan, Y.; Liu, L.; Wang, C.; Liu, Y.; Liu, Y.; et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv 2020, 1–18. [Google Scholar] [CrossRef]






| 6 h | M0 Control (19.44) | M0 CeO2 NPs (22.42) | M0 Ce-Ion (19.81) | M1 Control (32.22) | M1 CeO2 NPs (30.03) | M1 Ce-Ion (35.71) | M2 Control (18.40) | M2 CeO2 NPs (17.58) | M2 Ce-Ion 24.04) |
|---|---|---|---|---|---|---|---|---|---|
| M0 control | - | NS | NS | **** | **** | **** | NS | NS | NS |
| M0 CeO2 NPs | NS | - | NS | **** | * | **** | NS | NS | NS |
| M0 Ce-ion | NS | NS | - | **** | *** | **** | NS | NS | NS |
| M1 control | **** | **** | **** | - | NS | NS | **** | **** | ** |
| M1 CeO2 NPs | NS | NS | NS | **** | - | NS | **** | **** | NS |
| M1 Ce-ion | **** | **** | **** | NS | **** | - | **** | **** | **** |
| M2 control | NS | NS | NS | **** | NS | **** | - | NS | NS |
| M2 CeO2 NPs | **** | NS | NS | **** | **** | **** | NS | - | * |
| M2 Ce-ion | * | NS | NS | **** | * | **** | NS | NS | - |
| 24 h | M0 control (20.88) | M0 CeO2 NPs (18.35) | M0 Ce-ion (18.37) | M1 control (28.89) | M1 CeO2 NPs (21.61) | M1 Ce-ion (30.19) | M2 control (18.60) | M2 CeO2 NPs (15.76) | M2 Ce-ion (17.39) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, U.M.; Dozier, A.K.; Feola, D.J.; Tseng, M.T.; Yokel, R.A. Macrophage Polarization Status Impacts Nanoceria Cellular Distribution but Not Its Biotransformation or Ferritin Effects. Nanomaterials 2023, 13, 2298. https://doi.org/10.3390/nano13162298
Graham UM, Dozier AK, Feola DJ, Tseng MT, Yokel RA. Macrophage Polarization Status Impacts Nanoceria Cellular Distribution but Not Its Biotransformation or Ferritin Effects. Nanomaterials. 2023; 13(16):2298. https://doi.org/10.3390/nano13162298
Chicago/Turabian StyleGraham, Uschi M., Alan K. Dozier, David J. Feola, Michael T. Tseng, and Robert A. Yokel. 2023. "Macrophage Polarization Status Impacts Nanoceria Cellular Distribution but Not Its Biotransformation or Ferritin Effects" Nanomaterials 13, no. 16: 2298. https://doi.org/10.3390/nano13162298
APA StyleGraham, U. M., Dozier, A. K., Feola, D. J., Tseng, M. T., & Yokel, R. A. (2023). Macrophage Polarization Status Impacts Nanoceria Cellular Distribution but Not Its Biotransformation or Ferritin Effects. Nanomaterials, 13(16), 2298. https://doi.org/10.3390/nano13162298





