Engineering Materials and Devices for the Prevention, Diagnosis, and Treatment of COVID-19 and Infectious Diseases
Abstract
:1. Introduction
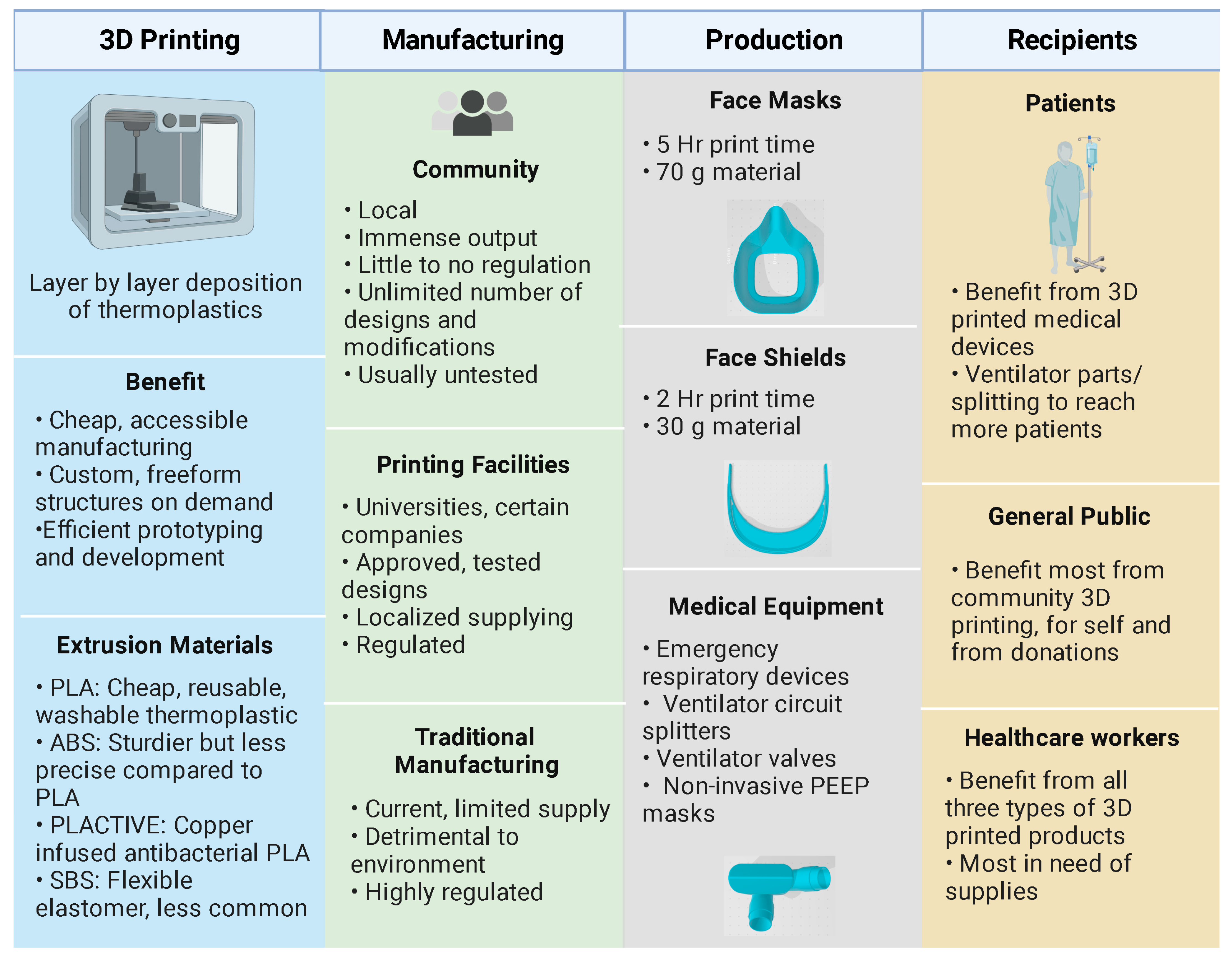
2. Three-Dimensional Printing of Protection Equipment
2.1. Materials for 3D-Printed PPE
2.2. Safety and Quality Considerations
2.3. Additive versus Conventional Manufacturing Technologies
2.4. Sterilization Considerations
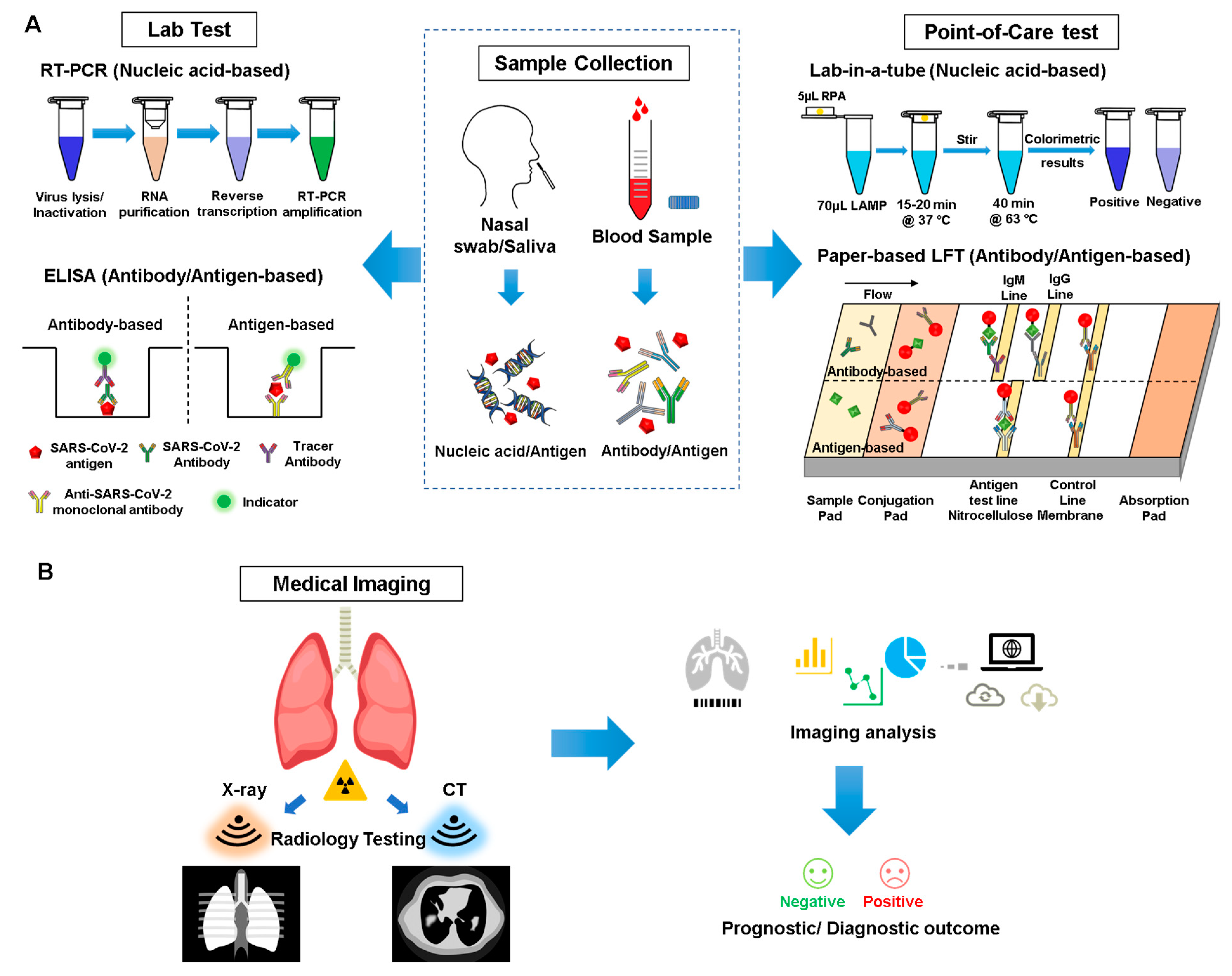
3. Detection and Diagnosis
3.1. Sampling/Serology Analysis
| Lab Test | |||||
|---|---|---|---|---|---|
| Technology | Sample | Target of Detection | Time Cost | Advantage | Disadvantage |
| RT-PCR | Nasal swab, tracheal aspirate, bronchoalveolar lavage specimens | Viral gene | 2–8 h |
|
|
| ELISA | Blood/serum | Antibody | 1–5 h |
|
|
| Serum, Nasopharyngeal swabs | Antigen |
| |||
| Point-of-Care | |||||
| Technology Platform | Sample | Target of Detection | Time Cost | Advantage | Disadvantage |
| LFA Paper-based strip [66] | Blood | Antibody | 5–20 min |
|
|
| Nasal or nasopharyngeal swabs | Antigen | ||||
| LAMP Lab-in-a-tube [67] | Nasal swab | Viral gene | 30 min |
|
|
| LAMP Silicon microfluidics chip [68] |
| ||||
| Electrochemistry Graphene field-effect transistor [69] | Nasopharyngeal swabs [70] | SARS-CoV-2 spike protein | 2 min |
|
|
3.2. Point-of-Care Devices for Diagnosis
3.3. CRISPR/Cas Technology
3.4. Medical Imaging and AI Assistance
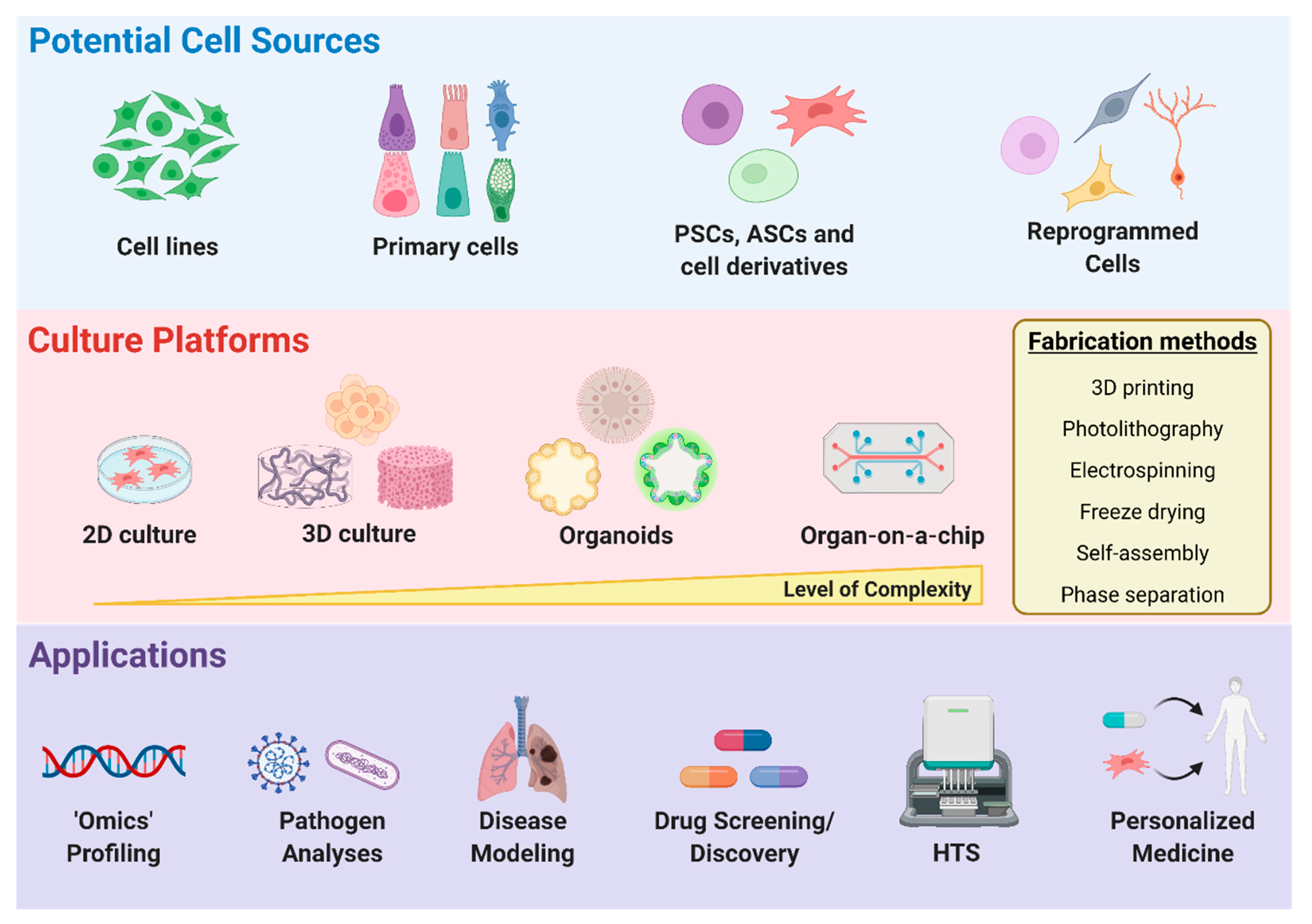
4. Disease Modeling and Drug Discovery
4.1. Cell Sources for Disease Modeling
4.2. Two-Dimensional vs. Three-Dimensional Microenvironments
Organoids
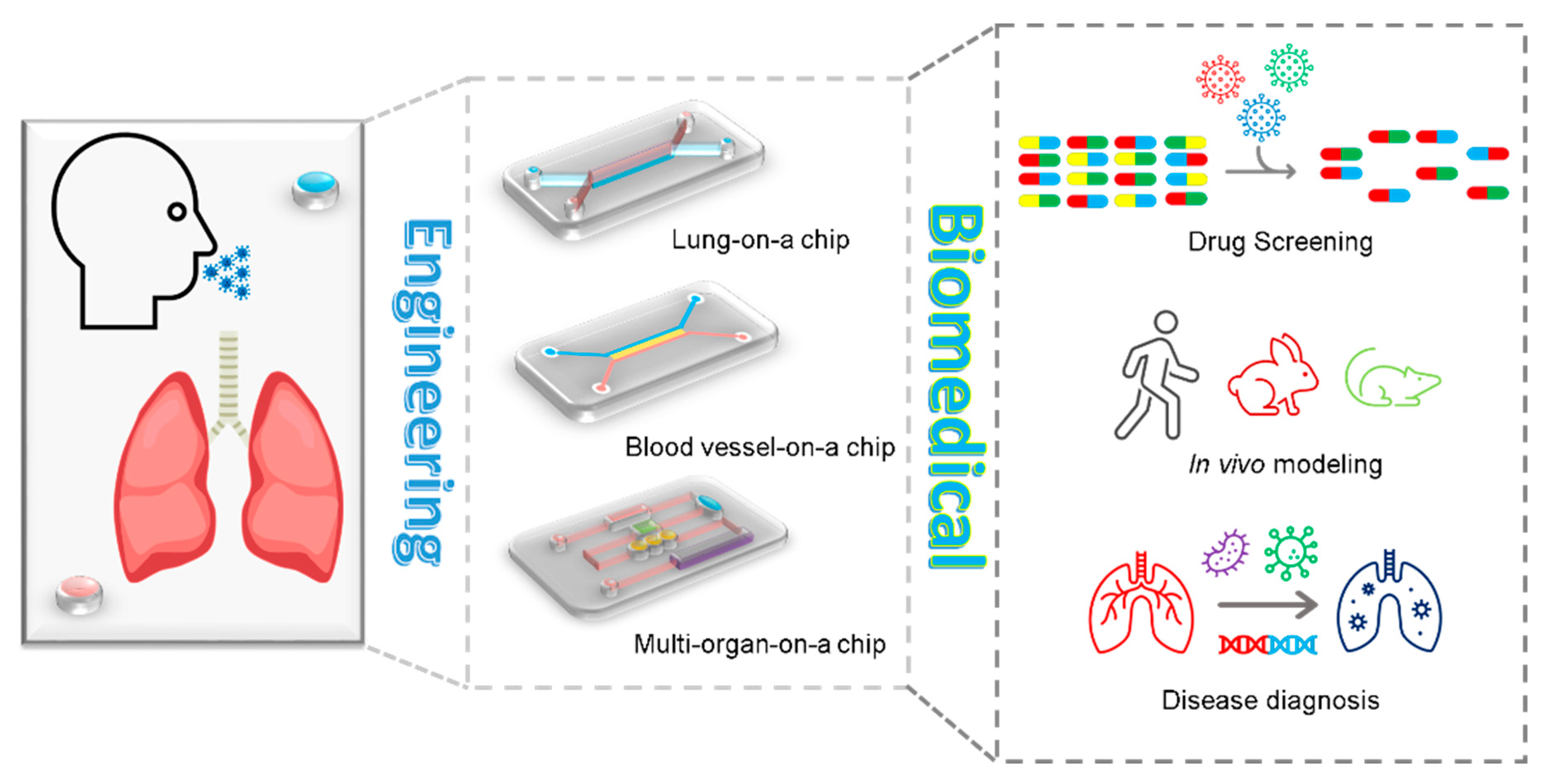
4.3. Organ-on-a-Chip
4.4. High-Throughput Screening
4.5. Machine Learning
5. Therapeutics Development
5.1. Antiviral, Anti-Inflammatory, and Anti-Thrombosis Therapy
5.2. Biomaterial-Based Therapies
5.3. Stem-Cell- and Tissue-Engineering-Based Therapies
6. Challenges and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus Disease (COVID-19) Dashboard|WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 3 January 2021).
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.D.; Wang, Z.Y.; Zhang, S.F.; Li, X.; Li, L.; Li, C.; Cui, Y.; Fu, R.B.; Dong, Y.Z.; Chi, X.Y.; et al. Aerosol and Surface Distribution of Severe Acute Respiratory Syndrome Coronavirus 2 in Hospital Wards, Wuhan, China, 2020. Emerg. Infect. Dis. 2020, 26, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Morawska, L.; Cao, J. Airborne Transmission of SARS-CoV-2: The World Should Face the Reality. Environ. Int. 2020, 139, 105730. [Google Scholar] [CrossRef]
- Zhang, W.; Du, R.-H.; Li, B.; Zheng, X.-S.; Yang, X.-L.; Hu, B.; Wang, Y.-Y.; Xiao, G.-F.; Yan, B.; Shi, Z.-L.; et al. Molecular and Serological Investigation of 2019-NCoV Infected Patients: Implication of Multiple Shedding Routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (Previously 2019-NCoV) Infection by a Highly Potent Pan-Coronavirus Fusion Inhibitor Targeting Its Spike Protein That Harbors a High Capacity to Mediate Membrane Fusion. Cell Res. 2020, 30, 343–355. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasllieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greeneugh, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA-J. Am. Med. Assoc. 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Gandhi, M.; Yokoe, D.S.; Havlir, D.V. Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control COVID-19. N. Engl. J. Med. 2020, 382, 2158–2160. [Google Scholar] [CrossRef]
- Rothan, H.A.; Byrareddy, S.N. The Epidemiology and Pathogenesis of Coronavirus Disease (COVID-19) Outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Polak, S.B.; Van Gool, I.C.; Cohen, D.; von der Thüsen, J.H.; van Paassen, J. A Systematic Review of Pathological Findings in COVID-19: A Pathophysiological Timeline and Possible Mechanisms of Disease Progression. Mod. Pathol. 2020, 33, 2128–2138. [Google Scholar] [CrossRef]
- Molinaro, R.; Pasto, A.; Taraballi, F.; Giordano, F.; Azzi, J.A.; Tasciotti, E.; Corbo, C. Biomimetic Nanoparticles Potentiate the Anti-Inflammatory Properties of Dexamethasone and Reduce the Cytokine Storm Syndrome: An Additional Weapon against COVID-19? Nanomaterials 2020, 10, 2301. [Google Scholar] [CrossRef] [PubMed]
- Salton, F.; Confalonieri, P.; Campisciano, G.; Cifaldi, R.; Rizzardi, C.; Generali, D.; Pozzan, R.; Tavano, S.; Bozzi, C.; Lapadula, G.; et al. Cytokine Profiles as Potential Prognostic and Therapeutic Markers in SARS-CoV-2-Induced ARDS. J. Clin. Med. 2022, 11, 2951. [Google Scholar] [CrossRef] [PubMed]
- Rolland-Debord, C.; Piéroni, L.; Bejar, F.; Milon, A.; Choinier, P.; Blin, E.; Bravais, J.; Halitim, P.; Letellier, A.; Camuset, J.; et al. Cell and Cytokine Analyses from Bronchoalveolar Lavage in Non-Critical COVID-19 Pneumonia. Intern. Emerg. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- High Proportion of Healthcare Workers with COVID-19 in Italy Is a Stark Warning to the World: Protecting Nurses and Their Colleagues Must Be the Number One Priority|ICN-International Council of Nurses. Available online: https://www.icn.ch/news/high-proportion-healthcare-workers-covid-19-italy-stark-warning-world-protecting-nurses-and (accessed on 17 September 2020).
- Salmi, M.; Akmal, J.S.; Pei, E.; Wolff, J.; Jaribion, A.; Khajavi, S.H. 3D Printing in COVID-19: Productivity Estimation of the Most Promising Open Source Solutions in Emergency Situations. Appl. Sci. 2020, 10, 4004. [Google Scholar] [CrossRef]
- Novak, J.I.; Loy, J. A Quantitative Analysis of 3D Printed Face Shields and Masks during COVID-19. Emerald Open Res. 2020, 2, 42. [Google Scholar] [CrossRef]
- Zhuang, Z.; Bradtmiller, B. Head-and-Face Anthropometric Survey of U.S. Respirator Users. J. Occup. Environ. Hyg. 2005, 2, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Coffey, C.; Ann, R. The Effect of Subject Characteristics and Respirator Features on Respirator Fit. J. Occup. Environ. Hyg. 2005, 2, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Swennen, G.R.J.; Pottel, L.; Haers, P.E. Custom-Made 3D-Printed Face Masks in Case of Pandemic Crisis Situations with a Lack of Commercially Available FFP2/3 Masks. Int. J. Oral Maxillofac. Surg. 2020, 49, 673–677. [Google Scholar] [CrossRef]
- Nold, J.; Metzger, M.C.; Schwarz, S.; Wesemann, C.; Wemken, G.; Pieralli, S.; Kernen, F.; Weingart, J.; Schirmeister, C.G.; Schumann, S.; et al. Air Seal Performance of Personalized and Statistically Shaped 3D-Printed Face Masks Compared with Market-Available Surgical and FFP2 Masks. Sci. Rep. 2021, 11, 19347. [Google Scholar] [CrossRef]
- Banerjee, S.S.; Burbine, S.; Shivaprakash, N.K.; Mead, J. 3D-Printable PP/SEBS Thermoplastic Elastomeric Blends: Preparation and Properties. Polymers 2019, 11, 347. [Google Scholar] [CrossRef]
- Ishack, S.; Lipner, S.R. Applications of 3D Printing Technology to Address COVID-19–Related Supply Shortages. Am. J. Med. 2020, 133, 771. [Google Scholar] [CrossRef]
- He, H.; Gao, M.; Illés, B.; Molnar, K. 3D Printed and Electrospun, Transparent, Hierarchical Polylactic Acid Mask Nanoporous Filter. Int. J. Bioprinting 2020, 6, 278. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, D.M.; James Rao, Y.; Mitic, K.; Obaid, S.N.; Pierce, D.; Huckenpahler, J.; Berger, J.; Goyal, S.; Loew, M.H. Rapid Prototyping of Reusable 3D-Printed N95 Equivalent Respirators at the George Washington University. Preprints 2020, 2020030444. [Google Scholar] [CrossRef]
- Bachtiar, E.O.; Erol, O.; Millrod, M.; Tao, R.; Gracias, D.H.; Romer, L.H.; Kang, S.H. 3D Printing and Characterization of a Soft and Biostable Elastomer with High Flexibility and Strength for Biomedical Applications. J. Mech. Behav. Biomed. Mater. 2020, 104, 103649. [Google Scholar] [CrossRef] [PubMed]
- Clifton, W.; Damon, A.; Martin, A.K. Considerations and Cautions for Three-Dimensional-Printed Personal Protective Equipment in the COVID-19 Crisis. 3D Print Addit. Manuf. 2020, 7, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Mitik-Dineva, N.; Wang, J.; Truong, V.K.; Stoddart, P.; Malherbe, F.; Crawford, R.J.; Ivanova, E.P. Escherichia Coli, Pseudomonas Aeruginosa, and Staphylococcus Aureus Attachment Patterns on Glass Surfaces with Nanoscale Roughness. Curr. Microbiol. 2009, 58, 268–273. [Google Scholar] [CrossRef]
- Hall, D.C.; Palmer, P.; Ji, H.F.; Ehrlich, G.D.; Król, J.E. Bacterial Biofilm Growth on 3D-Printed Materials. Front. Microbiol. 2021, 12, 646303. [Google Scholar] [CrossRef] [PubMed]
- Teughels, W.; Van Assche, N.; Sliepen, I.; Quirynen, M. Effect of Material Characteristics and/or Surface Topography on Biofilm Development. Clin. Oral Implant. Res. 2006, 17 (Suppl. S2), 68–81. [Google Scholar] [CrossRef]
- Shah, J.; Snider, B.; Clarke, T.; Kozutsky, S.; Lacki, M.; Hosseini, A. Large-Scale 3D Printers for Additive Manufacturing: Design Considerations and Challenges. Int. J. Adv. Manuf. Technol. 2019, 104, 3679–3693. [Google Scholar] [CrossRef]
- Chohan, J.S.; Singh, R. Pre and Post Processing Techniques to Improve Surface Characteristics of FDM Parts: A State of Art Review and Future Applications. Rapid Prototyp. J. 2017, 23, 495–513. [Google Scholar] [CrossRef]
- Dizon, J.R.C.; Gache, C.C.L.; Cascolan, H.M.S.; Cancino, L.T.; Advincula, R.C. Post-Processing of 3D-Printed Polymers. Technologies 2021, 9, 61. [Google Scholar] [CrossRef]
- Franchetti, M.; Kress, C. An Economic Analysis Comparing the Cost Feasibility of Replacing Injection Molding Processes with Emerging Additive Manufacturing Techniques. Int. J. Adv. Manuf. Technol. 2017, 88, 2573–2579. [Google Scholar] [CrossRef]
- Fiedler, B.A. Managing Medical Devices within a Regulatory Framework; Elsevier Inc.: Amsterdam, The Netherlands, 2016; ISBN 9780128041925. [Google Scholar]
- Cuiffo, M.A.; Snyder, J.; Elliott, A.M.; Romero, N.; Kannan, S.; Halada, G.P. Impact of the Fused Deposition (FDM) Printing Process on Polylactic Acid (PLA) Chemistry and Structure. Appl. Sci. 2017, 7, 579. [Google Scholar] [CrossRef]
- Tambrallimath, V.; Keshavamurthy, R.; D, S.; Koppad, P.G.; Kumar, G.P. Thermal Behavior of PC-ABS Based Graphene Filled Polymer Nanocomposite Synthesized by FDM Process. Compos. Commun. 2019, 15, 129–134. [Google Scholar] [CrossRef]
- Wiseman, J.; Rawther, T.; Langbart, M.; Kernohan, M.; Ngo, Q. Sterilization of Bedside 3D-Printed Devices for Use in the Operating Room. Ann. 3D Print. Med. 2022, 5, 100045. [Google Scholar] [CrossRef]
- Luchini, K.; Sloan, S.N.B.; Mauro, R.; Sargsyan, A.; Newman, A.; Persaud, P.; Hawkins, D.; Wolff, D.; Staudinger, J.; Creamer, B.A. Sterilization and Sanitizing of 3D-Printed Personal Protective Equipment Using Polypropylene and a Single Wall Design. 3D Print. Med. 2021, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Xiao, W.; Zhao, M.; Yu, X.; Wang, H.; Wang, Q.; Chu, S.; Cui, Y. Can N95 Respirators Be Reused after Disinfection? How Many Times? ACS Nano 2020, 14, 6348–6356. [Google Scholar] [CrossRef]
- Pirker, L.; Krajnc, A.P.; Malec, J.; Radulović, V.; Gradišek, A.; Jelen, A.; Remškar, M.; Mekjavić, I.B.; Kovač, J.; Mozetič, M.; et al. Sterilization of Polypropylene Membranes of Facepiece Respirators by Ionizing Radiation. J. Memb. Sci. 2020, 619, 118756. [Google Scholar] [CrossRef]
- Al-Hadyan, K.; Alsbeih, G.; Al-Harbi, N.; Judia, S.B.; Al-Ghamdi, M.; Almousa, A.; Alsharif, I.; Bakheet, R.; Al-Romaih, K.; Al-Mozaini, M.; et al. Effect of Gamma Irradiation on Filtering Facepiece Respirators and SARS-CoV-2 Detection. Sci. Rep. 2021, 11, 19888. [Google Scholar] [CrossRef]
- Zhang, X.C.; Cameron, R.E. The Morphology of Irradiated Isotactic Polypropylene. J. Appl. Polym. Sci. 1999, 74, 2234–2242. [Google Scholar] [CrossRef]
- Rankouhi, B.; Javadpour, S.; Delfanian, F.; McTaggart, R.; Letcher, T. Experimental Investigation of Mechanical Performance and Printability of Gamma-Irradiated Additively Manufactured ABS. JMEP 2018, 27, 3643–3654. [Google Scholar] [CrossRef]
- West, C.; McTaggart, R.; Letcher, T.; Raynie, D.; Roy, R. Effects of Gamma Irradiation upon the Mechanical and Chemical Properties of 3D-Printed Samples of Polylactic Acid. J. Manuf. Sci. Eng. 2019, 141, 041002. [Google Scholar] [CrossRef]
- Valente, T.A.M.; Silva, D.M.; Gomes, P.S.; Fernandes, M.H.; Santos, J.D.; Sencadas, V. Effect of Sterilization Methods on Electrospun Poly(Lactic Acid) (PLA) Fiber Alignment for Biomedical Applications. ACS Appl. Mater. Interfaces 2016, 8, 3241–3249. [Google Scholar] [CrossRef]
- Toro, M.; Cardona, A.; Restrepo, D.; Buitrago, L. Does Vaporized Hydrogen Peroxide Sterilization Affect the Geometrical Properties of Anatomic Models and Guides 3D Printed from Computed Tomography Images? 3D Print. Med. 2021, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Shea, G.K.-H.; Wu, K.L.-K.; Li, I.W.-S.; Leung, M.-F.; Ko, A.L.-P.; Tse, L.; Pang, S.S.-Y.; Kwan, K.Y.-H.; Wong, T.-M.; Leung, F.K.-L.; et al. A Review of the Manufacturing Process and Infection Rate of 3D-Printed Models and Guides Sterilized by Hydrogen Peroxide Plasma and Utilized Intra-Operatively. 3D Print. Med. 2020, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Bosc, R.; Tortolano, L.; Hersant, B.; Oudjhani, M.; Leplay, C.; Woerther, P.L.; Aguilar, P.; Leguen, R.; Meningaud, J.P. Bacteriological and Mechanical Impact of the Sterrad Sterilization Method on Personalized 3D Printed Guides for Mandibular Reconstruction. Sci. Rep. 2021, 11, 581. [Google Scholar] [CrossRef] [PubMed]
- Peniston, S.J.; Choi, S.J. Effect of Sterilization on the Physicochemical Properties of Molded Poly(L-Lactic Acid). J. Biomed. Mater. Res. B. Appl. Biomater. 2007, 80, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, S.; Guignot, C.; Tabrizian, M.; Ferrier, D.; Yagoubi, N.; Yahia, L. Plasma-Based Sterilization: Effect on Surface and Bulk Properties and Hydrolitic Stability of Reprocessed Polyurethane Electrophysiology Catheters. J. Biomed. Mater. Res. 2000, 52, 774–782. [Google Scholar] [CrossRef]
- Lerouge, S.; Tabrizian, M.; Wertheimer, M.R.; Marchand, R.; Yahia, L. Safety of Plasma-Based Sterilization: Surface Modifications of Polymeric Medical Devices Induced by Sterrad and Plazlyte Processes. Biomed. Mater. Eng. 2002, 12, 3–13. [Google Scholar]
- Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases. Available online: https://apps.who.int/iris/bitstream/handle/10665/331329/WHO-COVID-19-laboratory-2020.4-eng.pdf (accessed on 4 January 2021).
- Gandhi, R.T.; Lynch, J.B.; del Rio, C. Mild or Moderate COVID-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef]
- Krammer, F.; Simon, V. Serology Assays to Manage COVID-19. Science 2020, 368, 1060–1061. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Song, J.; El-Tholoth, M.; Li, Y.; Graham-Wooten, J.; Liang, Y.; Li, J.; Li, W.; Weiss, S.R.; Collman, R.G.; Bau, H.H. Single- and Two-Stage, Closed-Tube, Point-of-Care, Molecular Detection of SARS-CoV-2. Anal. Chem. 2021, 93, 13063–13071. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Ganguli, A.; Nguyen, J.; Brisbin, R.; Shanmugam, K.; Hirschberg, D.L.; Wheeler, M.B.; Bashir, R.; Nash, D.M.; Cunningham, B.T. Smartphone-Based Multiplex 30-Minute Nucleic Acid Test of Live Virus from Nasal Swab Extract. Lab Chip 2020, 20, 1621–1627. [Google Scholar] [CrossRef]
- Zhang, X.; Qi, Q.; Jing, Q.; Ao, S.; Zhang, Z.; Ding, M.; Wu, M.; Liu, K.; Wang, W.; Ling, Y.; et al. Electrical Probing of COVID-19 Spike Protein Receptor Binding Domain via a Graphene Field-Effect Transistor. arXiv 2020, arXiv:2003.12529. [Google Scholar]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; Astudillo, M.G.; Yang, D.; Miller, T.E.; Feldman, J.; Hauser, B.M.; Caradonna, T.M.; Clayton, K.L.; Nitido, A.D.; et al. COVID-19-Neutralizing Antibodies Predict Disease Severity and Survival. Cell 2021, 184, 476–488.e11. [Google Scholar] [CrossRef]
- Lisboa Bastos, M.; Tavaziva, G.; Abidi, S.K.; Campbell, J.R.; Haraoui, L.P.; Johnston, J.C.; Lan, Z.; Law, S.; MacLean, E.; Trajman, A.; et al. Diagnostic Accuracy of Serological Tests for COVID-19: Systematic Review and Meta-Analysis. BMJ 2020, 370, 2516. [Google Scholar] [CrossRef]
- Theel, E.S.; Harring, J.; Hilgart, H.; Granger, D. Performance Characteristics of Four High-Throughput Immunoassays for Detection of IgG Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01243-20. [Google Scholar] [CrossRef]
- Bryan, A.; Pepper, G.; Wener, M.H.; Fink, S.L.; Morishima, C.; Chaudhary, A.; Jerome, K.R.; Mathias, P.C.; Greninger, A.L. Performance Characteristics of the Abbott Architect SARS-CoV-2 IgG Assay and Seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020, 58, 10–128. [Google Scholar] [CrossRef]
- EUA Authorized Serology Test Performance|FDA. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance (accessed on 6 January 2021).
- Babson Diagnostics AC19G1 EUA Summary. Available online: https://www.fda.gov/media/139446/download (accessed on 6 January 2021).
- VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack-Instructions for Use. Available online: https://www.fda.gov/media/137363/download (accessed on 30 October 2021).
- Serology Test Evaluation Report for “WANTAI SARS-CoV-2 Ab ELISA” from Beijing Wantai Biological Pharmacy Enterprise Co., Ltd. Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3319-a001.pdf (accessed on 6 January 2021).
- Platelia SARS-CoV-2 Total Ab. Available online: https://www.fda.gov/media/137493/download (accessed on 6 January 2021).
- Serology Test Evaluation Report for “CareStart COVID-19 IgM/IgG Rapid Diagnostic Test for the Detection of SARS-CoV-2 IgM/IgG Ab” from Access Bio Inc. Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3299-a001.pdf (accessed on 6 January 2021).
- Serology Test Evaluation Report for “COVID-19 IgG/IgM Rapid Test Cassette” from Hangzhou Biotest Biotech. Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3252-a001.pdf (accessed on 6 January 2021).
- Serology Test Evaluation Report for “COVID-19 IgG/IgM Rapid Test Cassette” from Healgen. Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3247-a001.pdf (accessed on 6 January 2021).
- Serology Test Evaluation Report for “Sienna COVID-19 IgG/IgM Rapid Test Cassette (Whole Blood/Serum/Plasma)” from Salofa Oy. Available online: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3293-a001.pdf (accessed on 6 January 2021).
- Kubina, R.; Dziedzic, A. Molecular and Serological Tests for COVID-19. A Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics 2020, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R. Development of Point-of-Care Biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Zhuoyue, W.; Xin, Z.; Xinge, Y. IFA in Testing Specific Antibody of SARS Coronavirus. S. China J. Prev. Med. 2003, 29, 36–37. [Google Scholar]
- Zarei, M. Advances in Point-of-Care Technologies for Molecular Diagnostics. Biosens. Bioelectron. 2017, 98, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, H.; Ni, S.; Korabečná, M.; Yobas, L.; Neuzil, P. The Vision of Point-of-Care PCR Tests for the COVID-19 Pandemic and Beyond. TrAC-Trends Anal. Chem. 2020, 130, 115984. [Google Scholar] [CrossRef]
- Darwish, N.T.; Sekaran, S.D.; Khor, S.M. Point-of-Care Tests: A Review of Advances in the Emerging Diagnostic Tools for Dengue Virus Infection. Sens. Actuators B Chem. 2018, 255, 3316–3331. [Google Scholar] [CrossRef]
- Rasmi, Y.; Li, X.; Khan, J.; Ozer, T.; Choi, J.R. Emerging Point-of-Care Biosensors for Rapid Diagnosis of COVID-19: Current Progress, Challenges, and Future Prospects. Anal. Bioanal. Chem. 2021, 413, 4137–4159. [Google Scholar] [CrossRef]
- Dincer, C.; Bruch, R.; Kling, A.; Dittrich, P.S.; Urban, G.A. Multiplexed Point-of-Care Testing—XPOCT. Trends Biotechnol. 2017, 35, 728–742. [Google Scholar] [CrossRef]
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; Stewart de Ramirez, S.A.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735. [Google Scholar] [CrossRef]
- Choi, J.; Yong, K.; Choi, J.; Cowie, A. Emerging Point-of-Care Technologies for Food Safety Analysis. Sensors 2019, 19, 817. [Google Scholar] [CrossRef]
- Tymm, C.; Zhou, J.; Tadimety, A.; Burklund, A.; Zhang, J.X.J. Scalable COVID-19 Detection Enabled by Lab-on-Chip Biosensors. Cell. Mol. Bioeng. 2020, 13, 313. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Reverse Transcription Loop-Mediated Isothermal Amplification Combined with Nanoparticles-Based Biosensor for Diagnosis of COVID-19. medRxiv 2020, 166, 112437. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, M.; Razavi Bazaz, S.; Zhand, S.; Sayyadi, N.; Jin, D.; Stewart, M.P.; Ebrahimi Warkiani, M. Point of Care Diagnostics in the Age of COVID-19. Diagnostics 2020, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, J. Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective. J. Clin. Microbiol. 2021, 59, 10–128. [Google Scholar] [CrossRef]
- Tan, E.K.W.; Au, Y.Z.; Moghaddam, G.K.; Occhipinti, L.G.; Lowe, C.R. Towards Closed-Loop Integration of Point-of-Care Technologies. Trends Biotechnol. 2019, 37, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Min, J.; Yang, Y.; Xu, C.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 1981. [Google Scholar] [CrossRef] [PubMed]
- CDC’s Diagnostic Test for COVID-19 Only and Supplies|CDC. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 5 September 2020).
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.; Ballesteros, E.; Sfeir, M.; Liu, C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV Virus. Biorxiv 2020. [Google Scholar] [CrossRef]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.S.F.; Kemball, M.E.; et al. Streamlined Inactivation, Amplification, and Cas13-Based Detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Segel, M.; Bruneau, R.; Huang, M.-L.W.; Kim, N.-G.; Yu, X.; Li, J.; Walker, B.D.; et al. Point-of-Care Testing for COVID-19 Using SHERLOCK Diagnostics. Medrxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical Validation of a Cas13-Based Assay for the Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 2, 13. [Google Scholar] [CrossRef]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020, 14, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.J.; Baden, L.R.; Morrissey, S.; Campion, E.W. Medical Journals and the 2019-NCoV Outbreak. N. Engl. J. Med. 2020, 382, 866. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus Disease (COVID-19): RRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False-Negative of RT-PCR and Prolonged Nucleic Acid Conversion in COVID-19: Rather than Recurrence. J. Med. Virol. 2020, 92, 1755–1756. [Google Scholar] [CrossRef]
- Ng, M.-Y.; Lee, E.Y.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Lui, M.M.; Lo, C.S.-Y.; Leung, B.; Khong, P.-L.; et al. Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic Detection of Coronavirus Disease (COVID-19) Using X-Ray Images and Deep Convolutional Neural Networks. Pattern Anal. Appl. 2020, 24, 1207–1220. [Google Scholar] [CrossRef]
- Kanne, J.P. Chest CT Findings in 2019 Novel Coronavirus (2019-NCoV) Infections from Wuhan, China: Key Points for the Radiologist. Radiology 2020, 295, 16–17. [Google Scholar] [CrossRef]
- Bhayana, R.; Som, A.; Li, M.D.; Carey, D.E.; Anderson, M.A.; Blake, M.A.; Catalano, O.; Gee, M.S.; Hahn, P.F.; Harisinghani, M.; et al. Abdominal Imaging Findings in COVID-19: Preliminary Observations. Radiology 2020, 297, E207–E215. [Google Scholar] [CrossRef]
- Jacobi, A.; Chung, M.; Bernheim, A.; Eber, C. Portable Chest X-Ray in Coronavirus Disease-19 (COVID-19): A Pictorial Review. Clin. Imaging 2020, 64, 35–42. [Google Scholar] [CrossRef]
- Naudé, W. Artificial Intelligence vs COVID-19: Limitations, Constraints and Pitfalls. AI Soc. 2020, 35, 761–765. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Foresti, R.; Rossi, S.; Pinelli, S.; Alinovi, R.; Barozzi, M.; Sciancalepore, C.; Galetti, M.; Caffarra, C.; Lagonegro, P.; Scavia, G.; et al. Highly-Defined Bioprinting of Long-Term Vascularized Scaffolds with Bio-Trap: Complex Geometry Functionalization and Process Parameters with Computer Aided Tissue Engineering. Materialia 2020, 9, 100560. [Google Scholar] [CrossRef]
- Rainer, A.; Moroni, L. Computer-Aided Tissue Engineering; Springer: Berlin/Heidelberg, Germany, 2021; Volume 2147. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Isolation and Characterization of SARS-CoV-2 from the First US COVID-19 Patient. bioRxiv 2020. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced Isolation of SARS-CoV-2 by TMPRSS2- Expressing Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Wang, W.; Liu, Z.; Liang, C.; Wang, W.; Ye, F.; Huang, B.; Zhao, L.; Wang, H.; Zhou, W.; et al. Morphogenesis and Cytopathic Effect of SARS-CoV-2 Infection in Human Airway Epithelial Cells. Nat. Commun. 2020, 11, 3910. [Google Scholar] [CrossRef]
- Avior, Y.; Sagi, I.; Benvenisty, N. Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat. Rev. Mol. Cell Biol. 2016, 17, 170–182. [Google Scholar] [CrossRef]
- Sayed, N.; Liu, C.; Wu, J.C. Translation of Human-Induced Pluripotent Stem Cells: From Clinical Trial in a Dish to Precision Medicine. J. Am. Coll. Cardiol. 2016, 67, 2161–2176. [Google Scholar] [CrossRef]
- Ilic, D.; Ogilvie, C. Concise Review: Human Embryonic Stem Cells-What Have We Done? What Are We Doing? Where Are We Going? Stem Cells 2017, 35, 17–25. [Google Scholar] [CrossRef]
- Sharma, A.; Garcia, G.; Wang, Y.; Plummer, J.T.; Morizono, K.; Arumugaswami, V.; Svendsen, C.N. Human IPSC-Derived Cardiomyocytes Are Susceptible to SARS-CoV-2 Infection. Cell Rep. Med. 2020, 1, 100052. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffré, F.; et al. A Human Pluripotent Stem Cell-Based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e7. [Google Scholar] [CrossRef] [PubMed]
- Doss, M.X.; Sachinidis, A. Current Challenges of IPSC-Based Disease Modeling and Therapeutic Implications. Cells 2019, 8, 403. [Google Scholar] [CrossRef]
- Morris, S.A.; Daley, G.Q. A Blueprint for Engineering Cell Fate: Current Technologies to Reprogram Cell Identity. Cell Res. 2013, 23, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Du, Y.; Deng, H. Direct Lineage Reprogramming: Strategies, Mechanisms, and Applications. Cell Stem Cell 2015, 16, 119–134. [Google Scholar] [CrossRef]
- Ma, X.; Kong, L.; Zhu, S. Reprogramming Cell Fates by Small Molecules. Protein Cell 2017, 8, 328–348. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Soto, J.; Li, S. Biophysical Regulation of Cell Reprogramming. Curr. Opin. Chem. Eng. 2017, 15, 95–101. [Google Scholar] [CrossRef]
- Fang, J.; Hsueh, Y.Y.; Soto, J.; Sun, W.; Wang, J.; Gu, Z.; Khademhosseini, A.; Li, S. Engineering Biomaterials with Micro/Nanotechnologies for Cell Reprogramming. ACS Nano 2020, 14, 1296–1318. [Google Scholar] [CrossRef]
- Herdy, J.R.; Mertens, J.; Yao, J.; Ku, M.; Ladjevardi, S.; Kim, Y.; Paquola, A.C.M.; Lee, H.; McGrath, S.; Hetzer, M.W.; et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015, 17, 705–718. [Google Scholar]
- Mertens, J.; Reid, D.; Lau, S.; Kim, Y.; Gage, F.H. Aging in a Dish: IPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu. Rev. Genet. 2018, 52, 271–293. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Rosellini, A.; Freer, G.; Quaranta, P.; Dovere, V.; Menichini, M.; Maggi, F.; Mazzetti, P.; Pistello, M. Enhanced in Vitro Virus Expression Using 3-Dimensional Cell Culture Spheroids for Infection. J. Virol. Methods 2019, 265, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D Cell Culture Systems: Advantages and Applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, T.J.; McCarthy, M.; Cohrs, R.J.; Kaufer, B.B. 3D Tissue-like Assemblies: A Novel Approach to Investigate Virus-Cell Interactions. Methods 2015, 90, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.E.; Nagle, I.; Wilhelm, C. Magnetic Molding of Tumor Spheroids: Emerging Model for Cancer Screening. Biofabrication 2021, 13, 015018. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic Alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Uto, K.; Arakawa, C.K.; DeForest, C.A. Next-Generation Biomaterials for Culture and Manipulation of Stem Cells. Cold Spring Harb. Perspect. Biol. 2019, 12, a035691. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Li, Y.; Chen, T. Techniques for Fabrication and Construction of Three-Dimensional Scaffolds for Tissue Engineering. Int. J. Nanomed. 2013, 8, 337–350. [Google Scholar] [CrossRef]
- Bajaj, P.; Schweller, R.M.; Khademhosseini, A.; West, J.L.; Bashir, R. 3D Biofabrication Strategies for Tissue Engineering and Regenerative Medicine. Annu. Rev. Biomed. Eng. 2014, 16, 247–276. [Google Scholar] [CrossRef]
- Moroni, L.; Burdick, J.A.; Highley, C.; Lee, S.J.; Morimoto, Y.; Takeuchi, S.; Yoo, J.J. Biofabrication Strategies for 3D in Vitro Models and Regenerative Medicine. Nat. Rev. Mater. 2018, 3, 21–37. [Google Scholar] [CrossRef]
- Matai, I.; Kaur, G.; Seyedsalehi, A.; McClinton, A.; Laurencin, C.T. Progress in 3D Bioprinting Technology for Tissue/Organ Regenerative Engineering. Biomaterials 2020, 226, 119536. [Google Scholar] [CrossRef] [PubMed]
- Katsura, H.; Sontake, V.; Tata, A.; Kobayashi, Y.; Edwards, C.E.; Heaton, B.E.; Konkimalla, A.; Asakura, T.; Mikami, Y.; Fritch, E.J.; et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 2020, 27, 890–904.e8. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and Potential in Organoid Research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Kratochvil, M.J.; Seymour, A.J.; Li, T.L.; Paşca, S.P.; Kuo, C.J.; Heilshorn, S.C. Engineered Materials for Organoid Systems. Nat. Rev. Mater. 2019, 4, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Brassard, J.A.; Nikolaev, M.; Hübscher, T.; Hofer, M.; Lutolf, M.P. Recapitulating Macro-Scale Tissue Self-Organization through Organoid Bioprinting. Nat. Mater. 2020, 20, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Ito, Y.; Sakai, Y.; Saito, A.; Okuzaki, D.; Motooka, D.; Minami, S.; Kobayashi, T.; Yamamoto, T.; Okamoto, T.; et al. Generation of Human Bronchial Organoids for SARS-CoV-2 Research. bioRxiv 2020, 4. [Google Scholar] [CrossRef]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 Inhibitors Using Lung and Colonic Organoids. Nature 2020, 589, 270–275. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef]
- Zhao, B.; Ni, C.; Gao, R.; Wang, Y.; Yang, L.; Wei, J.; Lv, T.; Liang, J.; Zhang, Q.; Xu, W.; et al. Recapitulation of SARS-CoV-2 Infection and Cholangiocyte Damage with Human Liver Ductal Organoids. Protein Cell 2020, 11, 771–775. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of Bat and Human Intestinal Organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B.; et al. TMPRSS2 and TMPRSS4 Promote SARS-CoV-2 Infection of Human Small Intestinal Enterocytes. Sci. Immunol. 2020, 5, abc3582. [Google Scholar] [CrossRef] [PubMed]
- Makovoz, B.; Moeller, R.; Zebitz Eriksen, A.; tenOever, B.R.; Blenkinsop, T.A. SARS-CoV-2 Infection of Ocular Cells from Human Adult Donor Eyes and HESC-Derived Eye Organoids. SSRN Electron. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bullen, C.K. Infectability of Human BrainSphere Neurons Suggests Neurotropism of SARS-CoV-2. ALTEX 2020, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Zhang, C.; Israelow, B.; Lu, P.; Weizman, O.-E.; Liu, F.; Dai, Y.; Szigeti-Buck, K.; Yasumoto, Y.; Wang, G.; et al. Neuroinvasive Potential of SARS-CoV-2 Revealed in a Human Brain Organoid Model. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mesci, P.; Macia, A.; Saleh, A.; Martin-Sancho, L.; Yin, X.; Snethlage, C.; Avansini, S.; Chanda, S.K.; Muotri, A. Sofosbuvir Protects Human Brain Organoids against SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 Targets Neurons of 3D Human Brain Organoids. EMBO J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef]
- Da, C.; Pedrosa, S.G.; Goto-Silva, L.; Gomes, I.C.; Souza, L.R.Q.; Vitória, G.; Ornelas, I.M.; Karmirian, K.; Mendes, M.A.; Salerno, J.A.; et al. Non-Permissive SARS-CoV-2 Infection of Neural Cells in the Developing Human Brain and Neurospheres Running Title: SARS-CoV-2 Infection in the Developing Brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Miller, A.J.; Spence, J.R. In Vitro Models to Study Human Lung Development, Disease and Homeostasis. Physiology 2017, 32, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Perrin, S. Make Mouse Studies Work. Nature 2014, 507, 423–425. [Google Scholar] [CrossRef] [PubMed]
- El-Ali, J.; Sorger, P.K.; Jensen, K.F. Cells on Chips. Nature 2006, 442, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in Organ-on-a-Chip Engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Nurani, A.; Zhu, D.; Goyal, G.; Gilpin, S.; et al. Human Organs-on-Chips as Tools for Repurposing Approved Drugs as Potential Influenza and COVID19 Therapeutics in Viral Pandemics. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lamprou, D.A. Emerging Technologies for Diagnostics and Drug Delivery in the Fight against COVID-19 and Other Pandemics. Expert Rev. Med. Devices 2020, 17, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Samanipour, R.; Kim, K. Organ-on-a-Chip Platforms for Drug Screening and Tissue Engineering. In Biomedical Engineering: Frontier Research and Converging Technologies; Springer International Publishing: Cham, Switzerland, 2015; Volume 9, pp. 209–233. ISBN 9783319218137. [Google Scholar]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Yuan Hsin, H.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-Time Monitoring of Metabolic Function in Liver-Onchip Microdevices Tracks the Dynamics of Mitochondrial Dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef]
- Choe, A.; Ha, S.K.; Choi, I.; Choi, N.; Sung, J.H. Microfluidic Gut-Liver Chip for Reproducing the First Pass Metabolism. Biomed. Microdevices 2017, 19, 4. [Google Scholar] [CrossRef]
- Ho, C.M.B.; Ng, S.H.; Li, K.H.H.; Yoon, Y.J. 3D Printed Microfluidics for Biological Applications. Lab Chip 2015, 15, 3627–3637. [Google Scholar] [CrossRef]
- Pasman, T.; Grijpma, D.; Stamatialis, D.; Poot, A. Flat and Microstructured Polymeric Membranes in Organs-on-Chips. J. R. Soc. Interface 2018, 15, 20180351. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Lian, Q.; Xu, F. Perspective: Fabrication of Integrated Organ-on-a-Chip via Bioprinting. Biomicrofluidics 2017, 11, 031301. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Yue, K.; Aleman, J.; Mollazadeh-Moghaddam, K.; Bakht, S.M.; Yang, J.; Jia, W.; Dell’Erba, V.; Assawes, P.; Shin, S.R.; et al. 3D Bioprinting for Tissue and Organ Fabrication. Ann. Biomed. Eng. 2017, 45, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Achberger, K.; Probst, C.; Haderspeck, J.C.; Bolz, S.; Rogal, J.; Chuchuy, J.; Nikolova, M.; Cora, V.; Antkowiak, L.; Haq, W.; et al. Merging Organoid and Organ-on-a-Chip Technology to Generate Complex Multi-Layer Tissue Models in a Human Retina-on-a-Chip Platform. Elife 2019, 8, e46188. [Google Scholar] [CrossRef] [PubMed]
- Waheed, S.; Cabot, J.M.; Macdonald, N.P.; Lewis, T.; Guijt, R.M.; Paull, B.; Breadmore, M.C. 3D Printed Microfluidic Devices: Enablers and Barriers. Lab Chip 2016, 16, 1993–2013. [Google Scholar] [CrossRef] [PubMed]
- Foresti, R.; Rossi, S.; Selleri, S. Bio Composite Materials: Nano Functionalization of 4D Bio Engineered Scaffold. In Proceedings of the 2019 IEEE International Conference on BioPhotonics (BioPhotonics), Taipei, Taiwan, 15–18 September 2019. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Segal, E. Lab-on-a-Chip Devices for Point-of-Care Medical Diagnostics. Adv. Biochem. Eng. Biotechnol. 2020, 179, 247–265. [Google Scholar]
- Murphy, B.; Morgan, S.; Luy, E.; Creelman, J.; Sieben, V. Lab-on-a-Chip Sensor for in Situ Nutrient Monitoring. In Proceedings of the OCEANS 2019 MTS/IEEE Seattle, OCEANS 2019, Seattle, WA, USA, 27–31 October 2019; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2019. [Google Scholar]
- Valenta, A.C.; D’Amico, C.I.; Dugan, C.E.; Grinias, J.P.; Kennedy, R.T. A Microfluidic Chip for On-Line Derivatization and Application to in Vivo Neurochemical Monitoring. Analyst 2021, 146, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-Integrated Organs-on-Chips Platform for Automated and Continual in Situ Monitoring of Organoid Behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Shen, L.; Niu, J.; Wang, C.; Huang, B.; Wang, W.; Zhu, N.; Deng, Y.; Wang, H.; Ye, F.; Cen, S.; et al. High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 2019, 93, 10–128. [Google Scholar] [CrossRef]
- Desbordes, S.C.; Studer, L. Adapting Human Pluripotent Stem Cells to High-Throughput and High-Content Screening. Nat. Protoc. 2013, 8, 111–130. [Google Scholar] [CrossRef]
- Paull, D.; Sevilla, A.; Zhou, H.; Hahn, A.K.; Kim, H.; Napolitano, C.; Tsankov, A.; Shang, L.; Krumholz, K.; Jagadeesan, P.; et al. Automated, High-Throughput Derivation, Characterization and Differentiation of Induced Pluripotent Stem Cells. Nat. Methods 2015, 12, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, X.; Niu, Q.; Mo, X.; Qui, M.; Ma, T.; Kuo, C.J.; Fu, H. Development of a Miniaturized 3D Organoid Culture Platform for Ultra-High Throughput Screening. J. Mol. Cell Biol. 2020, 12, 630–643. [Google Scholar] [CrossRef] [PubMed]
- Boehnke, K.; Iversen, P.W.; Schumacher, D.; Lallena, M.J.; Haro, R.; Amat, J.; Haybaeck, J.; Liebs, S.; Lange, M.; Schäfer, R.; et al. Assay Establishment and Validation of a High-Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures. J. Biomol. Screen. 2016, 21, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Czerniecki, S.M.; Cruz, N.M.; Harder, J.L.; Menon, R.; Annis, J.; Otto, E.A.; Gulieva, R.E.; Islas, L.V.; Kim, Y.K.; Tran, L.M.; et al. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 2018, 22, 929–940.e4. [Google Scholar] [CrossRef] [PubMed]
- Durens, M.; Nestor, J.; Williams, M.; Herold, K.; Niescier, R.F.; Lunden, J.W.; Phillips, A.W.; Lin, Y.C.; Dykxhoorn, D.M.; Nestor, M.W. High-Throughput Screening of Human Induced Pluripotent Stem Cell-Derived Brain Organoids. J. Neurosci. Methods 2020, 335, 108627. [Google Scholar] [CrossRef] [PubMed]
- Worthington, P.; Drake, K.M.; Li, Z.; Napper, A.D.; Pochan, D.J.; Langhans, S.A. Beta-Hairpin Hydrogels as Scaffolds for High-Throughput Drug Discovery in Three-Dimensional Cell Culture. Anal. Biochem. 2017, 535, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Mazzocchi, A.; Soker, S.; Skardal, A. 3D Bioprinting for High-Throughput Screening: Drug Screening, Disease Modeling, and Precision Medicine Applications. Appl. Phys. Rev. 2019, 6, 11302. [Google Scholar] [CrossRef] [PubMed]
- Shoichet, B.K. Virtual Screening of Chemical Libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Gosu, V.; Hong, S.; Choi, S. Role of Computer-Aided Drug Design in Modern Drug Discovery. Arch. Pharm. Res. 2015, 38, 1686–1701. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Ton, A.T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, minf.202000028. [Google Scholar] [CrossRef]
- Choudhary, S.; Malik, Y.S.; Tomar, S. Identification of SARS-CoV-2 Cell Entry Inhibitors by Drug Repurposing Using in Silico Structure-Based Virtual Screening Approach. Front. Immunol. 2020, 11, 1664. [Google Scholar] [CrossRef]
- Rismanbaf, A. Potential Treatments for COVID-19; a Narrative Literature Review. Arch. Acad. Emerg. Med. 2020, 8, 2–4. [Google Scholar] [CrossRef]
- Alimadadi, A.; Aryal, S.; Manandhar, I.; Munroe, P.B.; Joe, B.; Cheng, X. Artificial Intelligence and Machine Learning to Fight COVID-19. Physiol. Genom. 2020, 52, 200–202. [Google Scholar] [CrossRef] [PubMed]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of Machine Learning and Artificial Intelligence for COVID-19 (SARS-CoV-2) Pandemic: A Review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, G.S.; Soltysiak, M.P.M.; El Roz, H.; de Souza, C.P.E.; Hill, K.A.; Kari, L. Machine Learning Using Intrinsic Genomic Signatures for Rapid Classification of Novel Pathogens: COVID-19 Case Study. PLoS ONE 2020, 15, e0232391. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Liu, F.; Ma, X.; He, P.; Tang, W.; Tong, X.; Zuo, J. Overview of Therapeutic Drug Research for COVID-19 in China. Acta Pharmacol. Sin. 2020, 41, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Rizk, J.G.; Kalantar-Zadeh, K.; Mehra, M.R.; Lavie, C.J.; Rizk, Y.; Forthal, D.N. Pharmaco-Immunomodulatory Therapy in COVID-19. Drugs 2020, 80, 1267–1292. [Google Scholar] [CrossRef]
- SEARCH of: COVID-19-List Results-ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/results?cond=COVID-19 (accessed on 18 October 2020).
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the MRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II Study of COVID-19 RNA Vaccine BNT162b1 in Adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.N.; Li, X.F.; Deng, Y.Q.; Zhao, H.; Huang, Y.J.; Yang, G.; Huang, W.J.; Gao, P.; Zhou, C.; Zhang, R.R.; et al. A Thermostable MRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283.e16. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. Concurrent Human Antibody and TH1 Type T-Cell Responses Elicited by a COVID-19 RNA Vaccine. medRxiv 2020. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for Translation: Non-Viral Materials for Therapeutic MRNA Delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines-a New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Petrovsky, N. Molecular Mechanisms for Enhanced DNA Vaccine Immunogenicity. Expert Rev. Vaccines 2016, 15, 313. [Google Scholar] [CrossRef] [PubMed]
- Kutzler, M.A.; Weiner, D.B. DNA Vaccines: Ready for Prime Time? Nat. Rev. Genet. 2008, 9, 776–788. [Google Scholar] [CrossRef]
- Cai, Y.; Rodriguez, S.; Hebel, H. DNA Vaccine Manufacture: Scale and Quality. Expert Rev. Vaccines 2009, 8, 1277–1291. [Google Scholar] [CrossRef]
- Middaugh, C.R.; Evans, R.K.; Montgomery, D.L.; Casimiro, D.R. Analysis of Plasmid DNA from a Pharmaceutical Perspective. J. Pharm. Sci. 1998, 87, 130–146. [Google Scholar] [CrossRef]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 Vaccine Development and a Potential Nanomaterial Path Forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Tian, J.-H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 Spike Glycoprotein Vaccine Candidate NVX-CoV2373 Immunogenicity in Baboons and Protection in Mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Bangaru, S.; Ozorowski, G.; Turner, H.L.; Antanasijevic, A.; Huang, D.; Wang, X.; Torres, J.L.; Diedrich, J.K.; Tian, J.H.; Portnoff, A.D.; et al. Structural Analysis of Full-Length SARS-CoV-2 Spike Protein from an Advanced Vaccine Candidate. Science 2020, 370, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Noad, R.; Roy, P. Virus-like Particles as Immunogens. Trends Microbiol. 2003, 11, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Hodgins, B.; Pillet, S.; Landry, N.; Ward, B.J. Prime-Pull Vaccination with a Plant-Derived Virus-like Particle Influenza Vaccine Elicits a Broad Immune Response and Protects Aged Mice from Death and Frailty after Challenge. Immun. Ageing 2019, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, B.J.; Bonar, M.M.; Costas-Cancelas, I.N.; Hunt, K.; Makarkov, A.I.; Chierzi, S.; Krawczyk, C.M.; Landry, N.; Ward, B.J.; Rouiller, I. Morphological Characterization of a Plant-Made Virus-like Particle Vaccine Bearing Influenza Virus Hemagglutinins by Electron Microscopy. Vaccine 2018, 36, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stevens, V.; Berry, J.D.; Crameri, G.; McEachern, J.; Tu, C.; Shi, Z.; Liang, G.; Weingartl, H.; Cardosa, J.; et al. Determination and Application of Immunodominant Regions of SARS Coronavirus Spike and Nucleocapsid Proteins Recognized by Sera from Different Animal Species. J. Immunol. Methods 2008, 331, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Hu, Y.; Chen, L.; Tong, Y.; Hu, J.; Cai, J.; Chan, K.-H.; Dou, Y.; Deng, J.; Gong, H.; et al. Mapping the Immunodominance Landscape of SARS-CoV-2 Spike Protein for the Design of Vaccines against COVID-19. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kalita, P.; Padhi, A.K.; Zhang, K.Y.J.; Tripathi, T. Design of a Peptide-Based Subunit Vaccine against Novel Coronavirus SARS-CoV-2. Microb. Pathog. 2020, 145, 104236. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in Adults with Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Preliminary Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Therapeutics and COVID-19: Living Guideline. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-therapeutics-2021.3 (accessed on 27 October 2021).
- Frediansyah, A.; Tiwari, R.; Sharun, K.; Dhama, K.; Harapan, H. Antivirals for COVID-19: A Critical Review. Clin. Epidemiol. Glob. Health 2021, 9, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wang, B.; Mao, J. The Pathogenesis and Treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C. Focus Shifts to Antibody Cocktails for COVID-19 Cytokine Storm. Nat. Biotechnol. 2020, 38, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Martin, J.C. Pathological Inflammation in Patients with COVID-19: A Key Role for Monocytes and Macrophages. Nat. Rev. Immunol. 2020, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Lang, F.M.; Lee, K.M.C.; Teijaro, J.R.; Becher, B.; Hamilton, J.A. GM-CSF-Based Treatments in COVID-19: Reconciling Opposing Therapeutic Approaches. Nat. Rev. Immunol. 2020, 20, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Qin, C.; Jin, R.; Sunney, X.; Correspondence, X. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020, 182, 73–84.e16. [Google Scholar] [CrossRef]
- Barnes, C.O.; West, A.P.; Huey-, K.E.; Robbiani, D.F.; Nussenzweig, M.C.; Correspondence, P.J.B.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842. [Google Scholar] [CrossRef]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A Human Neutralizing Antibody Targets the Receptor-Binding Site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A Neutralizing Human Antibody Binds to the N-Terminal Domain of the Spike Protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent Neutralizing Antibodies against Multiple Epitopes on SARS-CoV-2 Spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Klasse, P.J.; Burton, D.R. Antibodies to West Nile Virus: A Double-Edged Sword. Cell Host Microbe 2007, 1, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Yang, Y. The Potential Danger of Suboptimal Antibody Responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Chakhalian, D.; Shultz, R.B.; Miles, C.E.; Kohn, J. Opportunities for Biomaterials to Address the Challenges of COVID-19. J. Biomed. Mater. Res.-Part A 2020, 108, 1974–1990. [Google Scholar] [CrossRef]
- Zarubova, J.; Zhang, X.; Hoffman, T.; Hasani-Sadrabadi, M.M.; Li, S. Biomaterial-Based Immunoengineering to Fight COVID-19 and Infectious Diseases. Matter 2021, 4, 1528–1554. [Google Scholar] [CrossRef]
- Florindo, H.F.; Kleiner, R.; Vaskovich-Koubi, D.; Acúrcio, R.C.; Carreira, B.; Yeini, E.; Tiram, G.; Liubomirski, Y.; Satchi-Fainaro, R. Immune-Mediated Approaches against COVID-19. Nat. Nanotechnol. 2020, 15, 630–645. [Google Scholar] [CrossRef]
- Cagno, V.; Andreozzi, P.; D’Alicarnasso, M.; Silva, P.J.; Mueller, M.; Galloux, M.; Goffic, R.L.; Jones, S.T.; Vallino, M.; Hodek, J.; et al. Broad-Spectrum Non-Toxic Antiviral Nanoparticles with a Virucidal Inhibition Mechanism. Nat. Mater. 2018, 17, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.A.; Yoon, B.K.; Ouyang, L.; Wang, N.; Ferhan, A.R.; Kim, J.; Majima, T.; Cho, N.-J. Biomimetic Nanomaterial Strategies for Virus Targeting: Antiviral Therapies and Vaccines. Adv. Funct. Mater. 2020, 31, 2008352. [Google Scholar] [CrossRef]
- Nie, C.; Stadtmüller, M.; Yang, H.; Xia, Y.; Wolff, T.; Cheng, C.; Haag, R. Spiky Nanostructures with Geometry-Matching Topography for Virus Inhibition. Nano Lett. 2020, 20, 5367–5375. [Google Scholar] [CrossRef] [PubMed]
- Nanotechnology versus Coronavirus. Nat. Nanotechnol. 2020, 15, 617. [CrossRef] [PubMed]
- Talebian, S.; Wallace, G.G.; Schroeder, A.; Stellacci, F.; Conde, J. Nanotechnology-Based Disinfectants and Sensors for SARS-CoV-2. Nat. Nanotechnol. 2020, 15, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 MRNA Vaccine Design Enabled by Prototype Pathogen Preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Huang, X.; Zhang, Y.; Shen, S.; Feng, Z.; Dong, H.; Zhang, C.; Mo, R. Self-Regulated Hirudin Delivery for Anticoagulant Therapy. Sci. Adv. 2020, 6, eabc0382. [Google Scholar] [CrossRef]
- Kang, C.; Gwon, S.; Song, C.; Kang, P.M.; Park, S.C.; Jeon, J.; Hwang, D.W.; Lee, D. Fibrin-Targeted and H2O2-Responsive Nanoparticles as a Theranostics for Thrombosed Vessels. ACS Nano 2017, 11, 6194–6203. [Google Scholar] [CrossRef]
- Su, M.; Dai, Q.; Chen, C.; Zeng, Y.; Chu, C.; Liu, G. Nano-Medicine for Thrombosis: A Precise Diagnosis and Treatment Strategy. Nano-Micro Lett. 2020, 12, 96. [Google Scholar] [CrossRef]
- Gaharwar, A.K.; Singh, I.; Khademhosseini, A. Engineered Biomaterials for in Situ Tissue Regeneration. Nat. Rev. Mater. 2020, 5, 686–705. [Google Scholar] [CrossRef]
- Pober, J.S.; Sessa, W.C. Evolving Functions of Endothelial Cells in Inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and Cardiovascular Disease: From Basic Mechanisms to Clinical Perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M.; et al. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [PubMed]
- De Virgiliis, F.; Di Giovanni, S. Lung Innervation in the Eye of a Cytokine Storm: Neuroimmune Interactions and COVID-19. Nat. Rev. Neurol. 2020, 16, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Kronbichler, A.; Gauckler, P.; Windpessl, M.; Il Shin, J.; Jha, V.; Rovin, B.H.; Oberbauer, R. COVID-19: Implications for Immunosuppression in Kidney Disease and Transplantation. Nat. Rev. Nephrol. 2020, 16, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.M. COVID-19 and the Liver-Related Deaths to Come. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 523–525. [Google Scholar] [CrossRef]
- Mao, R.; Qiu, Y.; He, J.S.; Tan, J.Y.; Li, X.H.; Liang, J.; Shen, J.; Zhu, L.R.; Chen, Y.; Iacucci, M.; et al. Manifestations and Prognosis of Gastrointestinal and Liver Involvement in Patients with COVID-19: A Systematic Review and Meta-Analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial Cell Infection and Endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Choudhery, M.S.; Harris, D.T. Stem Cell Therapy for COVID-19: Possibilities and Challenges. Cell Biol. Int. 2020, 44, 2182–2191. [Google Scholar] [CrossRef]
- Basiri, A.; Pazhouhnia, Z.; Beheshtizadeh, N.; Hoseinpour, M.; Saghazadeh, A.; Rezaei, N. Regenerative Medicine in COVID-19 Treatment: Real Opportunities and Range of Promises. Stem Cell Rev. Rep. 2020, 17, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Tatara, A.M. Role of Tissue Engineering in COVID-19 and Future Viral Outbreaks. Tissue Eng.-Part A 2020, 26, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Xu, R.; Wang, S.; Xu, Z.; Zhang, C.; Li, Y.; Yang, T.; Shi, L.; Fu, J.; Jiang, T.; et al. Human Umbilical Cord-Derived Mesenchymal Stem Cell Therapy in Patients with COVID-19: A Phase 1 Clinical Trial. Signal Transduct. Target. Ther. 2020, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Sterodimas, A. Adipose Stem Cells (ASCs) and Stromal Vascular Fraction (SVF) as a Potential Therapy in Combating (COVID-19)-Disease. Aging Dis. 2020, 11, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Jiang, Y.; Zhu, M.; Chen, L.; Zhou, X.; Zhou, C.; Ye, P.; Chen, X.; Wang, B.; Xu, Z.; et al. Clinical Study Using Mesenchymal Stem Cells for the Treatment of Patients with Severe COVID-19. Front. Med. 2020, 14, 664–673. [Google Scholar] [CrossRef]
- Esquivel, D.; Mishra, R.; Soni, P.; Seetharaman, R.; Mahmood, A.; Srivastava, A. Stem Cells Therapy as a Possible Therapeutic Option in Treating COVID-19 Patients. Stem Cell Rev. Rep. 2020, 17, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Yen, B.L.; Yen, M.L.; Wang, L.T.; Liu, K.J.; Sytwu, H.K. Current Status of Mesenchymal Stem Cell Therapy for Immune/Inflammatory Lung Disorders: Gleaning Insights for Possible Use in COVID-19. Stem Cells Transl. Med. 2020, 9, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Jayaramayya, K.; Mahalaxmi, I.; Subramaniam, M.D.; Raj, N.; Dayem, A.A.; Lim, K.M.; Kim, S.J.; An, J.Y.; Lee, Y.; Choi, Y.; et al. Immunomodulatory Effect of Mesenchymal Stem Cells and Mesenchymal Stem-Cell-Derived Exosomes for COVID-19 Treatment. BMB Rep. 2020, 53, 400–412. [Google Scholar]
- Leng, Z.; Zhu, R.; Hou, W.; Feng, Y.; Yang, Y.; Han, Q.; Shan, G.; Meng, F.; Du, D.; Wang, S.; et al. Transplantation of ACE2- Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020, 11, 216–228. [Google Scholar] [CrossRef]
- Lukomska, B.; Stanaszek, L.; Zuba-Surma, E.; Legosz, P.; Sarzynska, S.; Drela, K. Challenges and Controversies in Human Mesenchymal Stem Cell Therapy. Stem Cells Int. 2019, 2019, 9628536. [Google Scholar] [CrossRef]
- Kuate, S.; Cinatl, J.; Doerr, H.W.; Überla, K. Exosomal Vaccines Containing the S Protein of the SARS Coronavirus Induce High Levels of Neutralizing Antibodies. Virology 2007, 362, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Pinky; Gupta, S.; Krishnakumar, V.; Sharma, Y.; Dinda, A.K.; Mohanty, S. Mesenchymal Stem Cell Derived Exosomes: A Nano Platform for Therapeutics and Drug Delivery in Combating COVID-19. Stem Cell Rev. Rep. 2021, 17, 33–43. [Google Scholar] [CrossRef]
- Akbari, A.; Jabbari, N.; Sharifi, R.; Ahmadi, M.; Vahhabi, A.; Seyedzadeh, S.J.; Nawaz, M.; Szafert, S.; Mahmoodi, M.; Jabbari, E.; et al. Free and Hydrogel Encapsulated Exosome-Based Therapies in Regenerative Medicine. Life Sci. 2020, 249, 117447. [Google Scholar] [CrossRef] [PubMed]
- Khatri, M.; Richardson, L.A.; Meulia, T. Mesenchymal Stem Cell-Derived Extracellular Vesicles Attenuate Influenza Virus-Induced Acute Lung Injury in a Pig Model. Stem Cell Res. Ther. 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Ghilan, A.; Chiriac, A.P.; Nita, L.E.; Rusu, A.G.; Neamtu, I.; Chiriac, V.M. Trends in 3D Printing Processes for Biomedical Field: Opportunities and Challenges. J. Polym. Environ. 2020, 28, 1345–1367. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Zhang, Y.S.; Akpek, A.; Shin, S.R.; Khademhosseini, A. 4D Bioprinting: The next-Generation Technology for Biofabrication Enabled by Stimuli-Responsive Materials. Biofabrication 2017, 9, 012001. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Hu, C.; Jahan, S.; Yuan, B.; Saleh, M.S.; Ju, E.; Gao, S.; Panat, R. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2020, 33, 2006647. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-Chips: Into the next Decade. Nat. Rev. Drug Discov. 2020, 20, 345–361. [Google Scholar] [CrossRef]
- Tang, H.; Abouleila, Y.; Si, L.; Ortega-Prieto, A.M.; Mummery, C.L.; Ingber, D.E.; Mashaghi, A. Human Organs-on-Chips for Virology. Trends Microbiol. 2020, 28, 934–946. [Google Scholar] [CrossRef]
- Yang, J.W.; Shen, Y.C.; Lin, K.C.; Cheng, S.J.; Chen, S.L.; Chen, C.Y.; Kumar, P.V.; Lin, S.F.; Lu, H.E.; Chen, G.Y. Organ-on-a-Chip: Opportunities for Assessing the Toxicity of Particulate Matter. Front. Bioeng. Biotechnol. 2020, 8, 519. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling Cancer in Microfluidic Human Organs-on-Chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.M.; Gonagle, D.M.; Ward, S.E.; Preston, R.J.S.; O’Donnell, J.S. Endothelial Cells Orchestrate COVID-19 Coagulopathy. Lancet Haematol. 2020, 7, e553–e555. [Google Scholar] [CrossRef] [PubMed]
- Santonja, C.; Heras, F.; Núñez, L.; Requena, L. COVID-19 Chilblain-like Lesion: Immunohistochemical Demonstration of SARS-CoV-2 Spike Protein in Blood Vessel Endothelium and Sweat Gland Epithelium in a Polymerase Chain Reaction-Negative Patient. Br. J. Dermatol. 2020, 183, 778–780. [Google Scholar] [CrossRef] [PubMed]
- Battinelli, E.M. COVID-19 Concerns Aggregate around Platelets. Blood 2020, 136, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Ball, P. The Lightning-Fast Quest for COVID Vaccines—And What It Means for Other Diseases. Nature 2021, 589, 16–18. [Google Scholar] [CrossRef]
- Castells, M.C.; Phillips, E.J. Maintaining Safety with SARS-CoV-2 Vaccines. N. Engl. J. Med. 2020, 384, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; To, K.K.W.; Sze, K.H.; Yung, T.T.M.; Bian, M.; Lam, H.; Yeung, M.L.; Li, C.; Chu, H.; Yuen, K.Y. A Broad-Spectrum Virus- and Host-Targeting Peptide against Respiratory Viruses Including Influenza Virus and SARS-CoV-2. Nat. Commun. 2020, 11, 4252. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H.; Stevens, L.J.; et al. An Orally Bioavailable Broad-Spectrum Antiviral Inhibits SARS-CoV-2 in Human Airway Epithelial Cell Cultures and Multiple Coronaviruses in Mice. Sci. Transl. Med. 2020, 12, 5883. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.A.; et al. Feline Coronavirus Drug Inhibits the Main Protease of SARS-CoV-2 and Blocks Virus Replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, B.; Jiang, X.M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-Based Design of Antiviral Drug Candidates Targeting the SARS-CoV-2 Main Protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef]
| Name of Serology Test | Type of Test | High Throughput | Sensitivity | Specificity | Type of Antibodies | PPV/NPV with 5% Prevalence Assumption of Infection in Sample Population | Limit of Detection or Smallest Volume Needed for Assay | Refs. |
|---|---|---|---|---|---|---|---|---|
| Abbott—Architect SARS-CoV-IgG | Chemiluminescent | Yes | 100.0% | 99.6% | IgG | 93.4%/100% | Needs 100 μL of serum or plasma to be diluted | [73,74] |
| Babson Diagnostics aC19G1 | Chemiluminescent | Yes | 100.0% | 100.0% | IgG | 100%/100% | N/A | [75,76] |
| VITROS Anti-SARS-CoV-2 IgG test | Chemiluminescent | Yes | 90.0% | 100.0% | IgG | 100%/99.5% | 110 ng/well in assay with 20uL sample volume | [73,77] |
| EUROIMMUN | ELISA | Yes | 90.0% | 100.0% | IgG | 100%/99.5% | 10 μL of serum | [73] |
| WANTAI SARS-CoV-2 Ab ELISA | ELISA | No | 96.7% | 97.5% | Total antibody | 67.1%/99.8% | 100 μL needed of sample per test | [75,78] |
| Platelia SARS-CoV-2 Total Ab | ELISA | No | 98.0% | 99.3% | Total antibody | 88.6%/99.9% | 15 μL of specimen needed | [75,79] |
| Access BioCare Start COVID-19 | Lateral Flow | No | 98.4% | 98.9% | IgG/IgM | 82.5%/99.9% | N/A | [75,80] |
| RightSign COVID-19 IgG/IgM Rapid Test Cassette | Lateral Flow | No | 100% | 100% | IgG/IgM | 100%/100% | ~10 μL needed for assay | [75,81] |
| COVID-19 IgG/IgM Rapid Test Cassette | Lateral Flow | No | 100.0% | 97.5% | IgG/IgM | 67.8%/100% | ~10 μL for assay | [75,82] |
| Sienna-Clarity COVIBLOCK COVID-19 IgG/IgM Rapid Test Cassette | Lateral Flow | No | 93.3% | 98.8% | IgG/IgM | 79.7%/99.6% | ~10 μL | [75,83] |
| Platform | Benefits | Drawbacks | Citations |
|---|---|---|---|
| mRNA |
|
| [208,209,210,211,212,213,214] |
| DNA |
|
| [215,216,217,218] |
| Recombinant Protein |
|
| [219,220,221] |
| Virus-like Particles |
|
| [222,223,224] |
| Peptide |
|
| [225,226,227] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soto, J.; Linsley, C.; Song, Y.; Chen, B.; Fang, J.; Neyyan, J.; Davila, R.; Lee, B.; Wu, B.; Li, S. Engineering Materials and Devices for the Prevention, Diagnosis, and Treatment of COVID-19 and Infectious Diseases. Nanomaterials 2023, 13, 2455. https://doi.org/10.3390/nano13172455
Soto J, Linsley C, Song Y, Chen B, Fang J, Neyyan J, Davila R, Lee B, Wu B, Li S. Engineering Materials and Devices for the Prevention, Diagnosis, and Treatment of COVID-19 and Infectious Diseases. Nanomaterials. 2023; 13(17):2455. https://doi.org/10.3390/nano13172455
Chicago/Turabian StyleSoto, Jennifer, Chase Linsley, Yang Song, Binru Chen, Jun Fang, Josephine Neyyan, Raul Davila, Brandon Lee, Benjamin Wu, and Song Li. 2023. "Engineering Materials and Devices for the Prevention, Diagnosis, and Treatment of COVID-19 and Infectious Diseases" Nanomaterials 13, no. 17: 2455. https://doi.org/10.3390/nano13172455
APA StyleSoto, J., Linsley, C., Song, Y., Chen, B., Fang, J., Neyyan, J., Davila, R., Lee, B., Wu, B., & Li, S. (2023). Engineering Materials and Devices for the Prevention, Diagnosis, and Treatment of COVID-19 and Infectious Diseases. Nanomaterials, 13(17), 2455. https://doi.org/10.3390/nano13172455






