Novel Mitochondria-Targeted Amphiphilic Aminophosphonium Salts and Lipids Nanoparticles: Synthesis, Antitumor Activity and Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Chemistry
General Procedure for the Synthesis of Alkyl Diethylaminophoshonium Derivatives 4a–e, 5a–d, 6a–e, 7
2.3. Preparation and Characterization of Lipid Nanoparticles
2.3.1. Chemicals
2.3.2. Preparation of Liposomes Modified by Aminophosphonium Salts
2.3.3. Preparation of SLN Modified by Aminophosphonium Salts
2.3.4. Characterization by DLS
2.3.5. In Vitro Rhodamine B Release Profile
2.4. Biological Study
2.4.1. Cells and Materials
2.4.2. Cell Toxicity Assay (MTT-Test)
2.4.3. Induction of Apoptotic Effects by Test Compounds and Flow Cytometry Assay
Cell Culture
Cell Apoptosis Assay
Mitochondrial Membrane Potential
Detection of Intracellular ROS
ELISA Assay
Cell Cycle Analysis
Hemolytic Assay
Cellular Uptake Study
Fluorescence Microscopy
Antimicrobial Activity
Statistical Analysis
In Vivo Study of Acute Toxicity of Aminophosphonic Salts
3. Results and Discussion
3.1. Chemistry
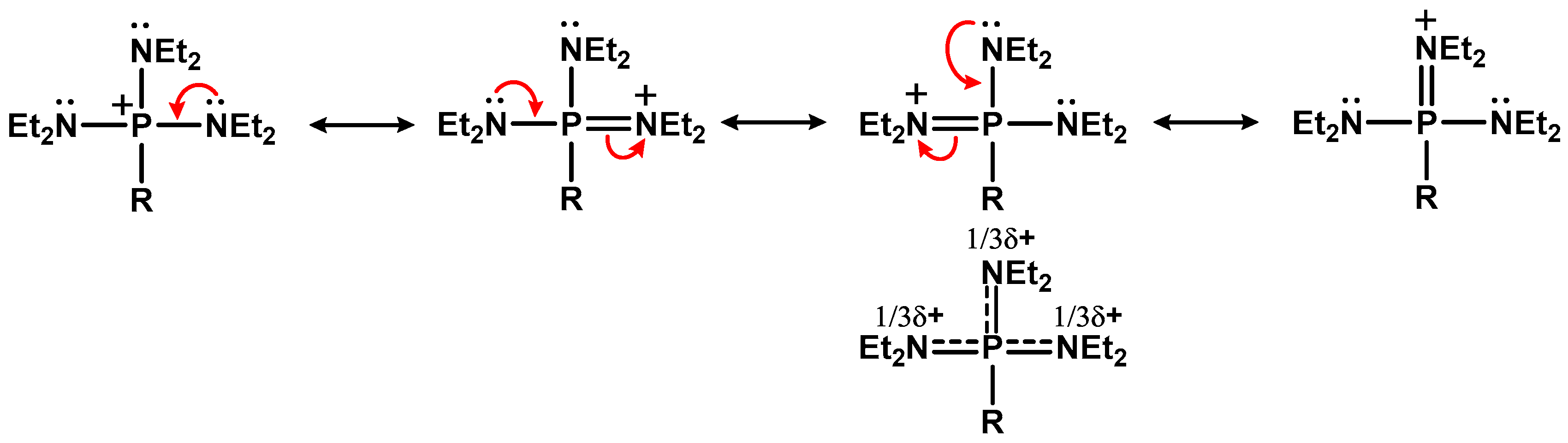
3.1.1. Synthesis of Alkyl Diethylaminophoshonium Salts
3.1.2. Aminophosphonium Salt Decorated Liposomes and Solid Lipid Nanoparticles
3.2. Biology
3.2.1. Cytotoxic Effects of Aminophosphonium Salts
3.2.2. Cellular Uptake of Liposomes and SLN Modified by Aminophosphonium Salts
3.2.3. Apoptosis Assay
3.2.4. Effect of Aminophosphonium Salts on Mitochondrial Potential
3.2.5. Effect of Aminophosphonium Salts on ROS Level in Cancer Cells
3.2.6. Measurements of Caspase-9 and Caspase-8 by ELISA
3.2.7. Effect of Aminophosphonium Salts on Cell Cycle in Cancer Cells
3.2.8. Antibacterial Activity of Aminophosphonium Salts
3.2.9. Hemolytic Action of Test Compounds
3.2.10. In Vivo Study of Acute Toxicity of Aminophosphonic Salts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pourahmad, J.; Salimi, A.; Seydi, E. Mitochondrial Targeting for Drug Development. In Toxicology Studies—Cells, Drugs and Environment; InTech: London, UK, 2015. [Google Scholar]
- Cojocaru, K.-A.; Luchian, I.; Goriuc, A.; Antoci, L.-M.; Ciobanu, C.-G.; Popescu, R.; Vlad, C.-E.; Blaj, M.; Foia, L.G. Mitochondrial Dysfunction, Oxidative Stress, and Therapeutic Strategies in Diabetes, Obesity, and Cardiovascular Disease. Antioxidants 2023, 12, 658. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, T.; Deore, S.L.; Kide, A.A.; Shende, B.A.; Sharma, R.; Dadarao Chakole, R.; Nemade, L.S.; Kishor Kale, N.; Borah, S.; Shrikant Deokar, S.; et al. Mitochondrial dysfunction and oxidative stress in Alzheimer’s disease, and Parkinson’s disease, Huntington’s disease and Amyotrophic Lateral Sclerosis—An updated review. Mitochondrion 2023, 71, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Danieli, M.G.; Antonelli, E.; Piga, M.A.; Cozzi, M.F.; Allegra, A.; Gangemi, S. Oxidative stress, mitochondrial dysfunction, and respiratory chain enzyme defects in inflammatory myopathies. Autoimmun. Rev. 2023, 22, 103308. [Google Scholar] [CrossRef]
- Behl, T.; Makkar, R.; Anwer, M.K.; Hassani, R.; Khuwaja, G.; Khalid, A.; Mohan, S.; Alhazmi, H.A.; Sachdeva, M.; Rachamalla, M. Mitochondrial Dysfunction: A Cellular and Molecular Hub in Pathology of Metabolic Diseases and Infection. J. Clin. Med. 2023, 12, 2882. [Google Scholar] [CrossRef] [PubMed]
- Amorim, J.A.; Coppotelli, G.; Rolo, A.P.; Palmeira, C.M.; Ross, J.M.; Sinclair, D.A. Mitochondrial and metabolic dysfunction in ageing and age-related diseases. Nat. Rev. Endocrinol. 2022, 18, 243–258. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Guo, Y.; Shi, X.; Chen, X.; Feng, W.; Wu, L.-L.; Zhang, J.; Yu, S.; Wang, Y.; et al. An Overview: The Diversified Role of Mitochondria in Cancer Metabolism. Int. J. Biol. Sci. 2023, 19, 897–915. [Google Scholar] [CrossRef]
- Martini, H.; Passos, J.F. Cellular senescence: All roads lead to mitochondria. FEBS J. 2023, 290, 1186–1202. [Google Scholar] [CrossRef]
- Grasso, D.; Zampieri, L.X.; Capelôa, T.; Van de Velde, J.A.; Sonveaux, P. Mitochondria in cancer. Cell Stress 2020, 4, 114–146. [Google Scholar] [CrossRef]
- Nath, S.; Villadsen, J. Oxidative phosphorylation revisited. Biotechnol. Bioeng. 2015, 112, 429–437. [Google Scholar] [CrossRef]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef]
- Obi, C.D.; Bhuiyan, T.; Dailey, H.A.; Medlock, A.E. Ferrochelatase: Mapping the Intersection of Iron and Porphyrin Metabolism in the Mitochondria. Front. Cell Dev. Biol. 2022, 10, 961. [Google Scholar] [CrossRef]
- Melchinger, P.; Garcia, B.M. Mitochondria are midfield players in steroid synthesis. Int. J. Biochem. Cell Biol. 2023, 160, 106431. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS signalling: Adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018, 52, 507–543. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Haskins, N.; Bhuvanendran, S.; Anselmi, C.; Gams, A.; Kanholm, T.; Kocher, K.M.; LoTempio, J.; Krohmaly, K.I.; Sohai, D.; Stearrett, N.; et al. Mitochondrial Enzymes of the Urea Cycle Cluster at the Inner Mitochondrial Membrane. Front. Physiol. 2021, 11, 542950. [Google Scholar] [CrossRef] [PubMed]
- Matuz-Mares, D.; González-Andrade, M.; Araiza-Villanueva, M.G.; Vilchis-Landeros, M.M.; Vázquez-Meza, H. Mitochondrial Calcium: Effects of Its Imbalance in Disease. Antioxidants 2022, 11, 801. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R. The Mitochondrial Pathway of Apoptosis. Cold Spring Harb. Perspect. Biol. 2022, 14, a041038. [Google Scholar] [CrossRef]
- Green, D.R. The Mitochondrial Pathway of Apoptosis Part II: The BCL-2 Protein Family. Cold Spring Harb. Perspect. Biol. 2022, 14, a041046. [Google Scholar] [CrossRef]
- Zhou, Q.; Tao, C.; Yuan, J.; Pan, F.; Wang, R. Ferroptosis, a subtle talk between immune system and cancer cells: To be or not to be? Biomed. Pharmacother. 2023, 165, 115251. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Wu, L.; Yang, L.; Yang, L.; Wang, J. The diversified role of mitochondria in ferroptosis in cancer. Cell Death Dis. 2023, 14, 519. [Google Scholar] [CrossRef]
- Wang, H.; Liu, C.; Zhao, Y.; Gao, G. Mitochondria regulation in ferroptosis. Eur. J. Cell Biol. 2020, 99, 151058. [Google Scholar] [CrossRef]
- Baines, C.P. Role of the Mitochondrion in Programmed Necrosis. Front. Physiol. 2010, 1, 156. [Google Scholar] [CrossRef] [PubMed]
- Faizan, M.I.; Ahmad, T. Altered mitochondrial calcium handling and cell death by necroptosis: An emerging paradigm. Mitochondrion 2021, 57, 47–62. [Google Scholar] [CrossRef]
- Yousif, L.F.; Stewart, K.M.; Kelley, S.O. Targeting Mitochondria with Organelle-Specific Compounds: Strategies and Applications. ChemBioChem 2009, 10, 1939–1950. [Google Scholar] [CrossRef] [PubMed]
- Madak, J.; Neamati, N. Membrane Permeable Lipophilic Cations as Mitochondrial Directing Groups. Curr. Top. Med. Chem. 2015, 15, 745–766. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef] [PubMed]
- Pawar, A.; Korake, S.; Pawar, A.; Kamble, R. Delocalized Lipophilic Cation Triphenyl Phosphonium: Promising Molecule for Mitochondria Targeting. Curr. Drug Deliv. 2023, 20, 1217–1223. [Google Scholar] [CrossRef]
- Begum, H.M.; Shen, K. Intracellular and microenvironmental regulation of mitochondrial membrane potential in cancer cells. WIREs Mech. Dis. 2023, 15, e1595. [Google Scholar] [CrossRef]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef]
- Wang, Y.; Patti, G.J. The Warburg effect: A signature of mitochondrial overload. Trends Cell Biol. 2023. [Google Scholar] [CrossRef]
- Cheng, X.; Feng, D.; Lv, J.; Cui, X.; Wang, Y.; Wang, Q.; Zhang, L. Application Prospects of Triphenylphosphine-Based Mitochondria-Targeted Cancer Therapy. Cancers 2023, 15, 666. [Google Scholar] [CrossRef] [PubMed]
- Zaib, S.; Hayyat, A.; Ali, N.; Gul, A.; Naveed, M.; Khan, I. Role of Mitochondrial Membrane Potential and Lactate Dehydrogenase A in Apoptosis. Anticancer. Agents Med. Chem. 2022, 22, 2048–2062. [Google Scholar] [CrossRef] [PubMed]
- Khailova, L.S.; Nazarov, P.A.; Sumbatyan, N.V.; Korshunova, G.A.; Rokitskaya, T.I.; Dedukhova, V.I.; Antonenko, Y.N.; Skulachev, V.P. Uncoupling and toxic action of alkyltriphenylphosphonium cations on mitochondria and the bacterium Bacillus subtilis as a function of alkyl chain length. Biochemistry 2015, 80, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Tellería, F.; Mansilla, S.; Méndez, D.; Sepúlveda, M.; Araya-Maturana, R.; Castro, L.; Trostchansky, A.; Fuentes, E. The Use of Triphenyl Phosphonium Cation Enhances the Mitochondrial Antiplatelet Effect of the Compound Magnolol. Pharmaceuticals 2023, 16, 210. [Google Scholar] [CrossRef]
- Montecino-Garrido, H.; Sepúlveda, M.; Méndez, D.; Monroy-Cárdenas, M.; Alfaro, S.; González-Avendaño, M.; Caballero, J.; Urra, F.A.; Araya-Maturana, R.; Fuentes, E. Assessing mitochondria-targeted acyl hydroquinones on the mitochondrial platelet function and cytotoxic activity: Role of the linker length. Free Radic. Biol. Med. 2023, 208, 26–36. [Google Scholar] [CrossRef]
- Uno, S.; Harkiss, A.H.; Chowdhury, R.; Caldwell, S.T.; Prime, T.A.; James, A.M.; Gallagher, B.; Prudent, J.; Hartley, R.C.; Murphy, M.P. Incorporating a Polyethyleneglycol Linker to Enhance the Hydrophilicity of Mitochondria-Targeted Triphenylphosphonium Constructs. ChemBioChem 2023, 24, e202200774. [Google Scholar] [CrossRef]
- Yang, S.; Yoon, N.G.; Park, M.-A.; Yun, J.; Im, J.Y.; Kang, B.H.; Kang, S. Triphenylphosphonium conjugation to a TRAP1 inhibitor, 2-amino-6-chloro-7,9-dihydro-8H-purin-8-one increases antiproliferative activity. Bioorg. Chem. 2022, 126, 105856. [Google Scholar] [CrossRef]
- Tsepaeva, O.V.; Salikhova, T.I.; Grigor’eva, L.R.; Ponomaryov, D.V.; Dang, T.; Ishkaeva, R.A.; Abdullin, T.I.; Nemtarev, A.V.; Mironov, V.F. Synthesis and in vitro evaluation of triphenylphosphonium derivatives of acetylsalicylic and salicylic acids: Structure-dependent interactions with cancer cells, bacteria, and mitochondria. Med. Chem. Res. 2021, 30, 925–939. [Google Scholar] [CrossRef]
- Ong, H.C.; Coimbra, J.T.S.; Kwek, G.; Ramos, M.J.; Xing, B.; Fernandes, P.A.; García, F. Alkyl vs. aryl modifications: A comparative study on modular modifications of triphenylphosphonium mitochondrial vectors. RSC Chem. Biol. 2021, 2, 1643–1650. [Google Scholar] [CrossRef]
- Strobykina, I.Y.; Voloshina, A.D.; Andreeva, O.V.; Sapunova, A.S.; Lyubina, A.P.; Amerhanova, S.K.; Belenok, M.G.; Saifina, L.F.; Semenov, V.E.; Kataev, V.E. Synthesis, antimicrobial activity and cytotoxicity of triphenylphosphonium (TPP) conjugates of 1,2,3-triazolyl nucleoside analogues. Bioorg. Chem. 2021, 116, 105328. [Google Scholar] [CrossRef]
- Millard, M.; Pathania, D.; Shabaik, Y.; Taheri, L.; Deng, J.; Neamati, N. Preclinical Evaluation of Novel Triphenylphosphonium Salts with Broad-Spectrum Activity. PLoS ONE 2010, 5, e13131. [Google Scholar] [CrossRef] [PubMed]
- Trnka, J.; Elkalaf, M.; Anděl, M. Lipophilic Triphenylphosphonium Cations Inhibit Mitochondrial Electron Transport Chain and Induce Mitochondrial Proton Leak. PLoS ONE 2015, 10, e0121837. [Google Scholar] [CrossRef] [PubMed]
- Gardner, Z.S.; Schumacher, T.J.; Ronayne, C.T.; Kumpati, G.P.; Williams, M.J.; Yoshimura, A.; Palle, H.; Mani, C.; Rumbley, J.; Mereddy, V.R. Synthesis and biological evaluation of novel 2-alkoxycarbonylallylester phosphonium derivatives as potential anticancer agents. Bioorg. Med. Chem. Lett. 2021, 45, 128136. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, C.A.; Fink, B.D.; Gibbs, B.E.; Chheda, P.R.; Wu, M.; Sivitz, W.I.; Kerns, R.J. A Novel Triphenylphosphonium Carrier to Target Mitochondria without Uncoupling Oxidative Phosphorylation. J. Med. Chem. 2021, 64, 662–676. [Google Scholar] [CrossRef]
- AbuEid, M.; Keyes, R.F.; McAllister, D.; Peterson, F.; Kadamberi, I.P.; Sprague, D.J.; Chaluvally-Raghavan, P.; Smith, B.C.; Dwinell, M.B. Fluorinated triphenylphosphonium analogs improve cell selectivity and in vivo detection of mito-metformin. iScience 2022, 25, 105670. [Google Scholar] [CrossRef]
- Rokitskaya, T.I.; Kotova, E.A.; Luzhkov, V.B.; Kirsanov, R.S.; Aleksandrova, E.V.; Korshunova, G.A.; Tashlitsky, V.N.; Antonenko, Y.N. Lipophilic ion aromaticity is not important for permeability across lipid membranes. Biochim. Biophys. Acta -Biomembr. 2021, 1863, 183483. [Google Scholar] [CrossRef]
- Rokitskaya, T.I.; Aleksandrova, E.V.; Korshunova, G.A.; Khailova, L.S.; Tashlitsky, V.N.; Luzhkov, V.B.; Antonenko, Y.N. Membrane Permeability of Modified Butyltriphenylphosphonium Cations. J. Phys. Chem. B 2022, 126, 412–422. [Google Scholar] [CrossRef]
- Rokitskaya, T.I.; Luzhkov, V.B.; Korshunova, G.A.; Tashlitsky, V.N.; Antonenko, Y.N. Effect of methyl and halogen substituents on the transmembrane movement of lipophilic ions. Phys. Chem. Chem. Phys. 2019, 21, 23355–23363. [Google Scholar] [CrossRef]
- Milane, L.; Dolare, S.; Jahan, T.; Amiji, M. Mitochondrial nanomedicine: Subcellular organelle-specific delivery of molecular medicines. Nanomed. Nanotechnol. Biol. Med. 2021, 37, 102422. [Google Scholar] [CrossRef]
- Liew, S.S.; Qin, X.; Zhou, J.; Li, L.; Huang, W.; Yao, S.Q. Smart Design of Nanomaterials for Mitochondria-Targeted Nanotherapeutics. Angew. Chemie Int. Ed. 2021, 60, 2232–2256. [Google Scholar] [CrossRef]
- Allemailem, K.S.; Almatroudi, A.; Alsahli, M.A.; Aljaghwani, A.; El-Kady, A.M.; Rahmani, A.H.; Khan, A.A. Novel Strategies for Disrupting Cancer-Cell Functions with Mitochondria-Targeted Antitumor Drug–Loaded Nanoformulations. Int. J. Nanomedicine 2021, 16, 3907–3936. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Satrialdi; Hibino, M.; Sasaki, D.; Abe, J.; Harashima, H. Power of mitochondrial drug delivery systems to produce innovative nanomedicines. Adv. Drug Deliv. Rev. 2020, 154–155, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, F.; Zhang, T.; Yu, M.; Sun, Y. Recent advances in anti-multidrug resistance for nano-drug delivery system. Drug Deliv. 2022, 29, 1684–1697. [Google Scholar] [CrossRef] [PubMed]
- Buchke, S.; Sharma, M.; Bora, A.; Relekar, M.; Bhanu, P.; Kumar, J. Mitochondria-Targeted, Nanoparticle-Based Drug-Delivery Systems: Therapeutics for Mitochondrial Disorders. Life 2022, 12, 657. [Google Scholar] [CrossRef]
- Mitragotri, S.; Anderson, D.G.; Chen, X.; Chow, E.K.; Ho, D.; Kabanov, A.V.; Karp, J.M.; Kataoka, K.; Mirkin, C.A.; Petrosko, S.H.; et al. Accelerating the Translation of Nanomaterials in Biomedicine. ACS Nano 2015, 9, 6644–6654. [Google Scholar] [CrossRef]
- Azarnezhad, A.; Samadian, H.; Jaymand, M.; Sobhani, M.; Ahmadi, A. Toxicological profile of lipid-based nanostructures: Are they considered as completely safe nanocarriers? Crit. Rev. Toxicol. 2020, 50, 148–176. [Google Scholar] [CrossRef]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef]
- Halwani, A.A. Development of Pharmaceutical Nanomedicines: From the Bench to the Market. Pharmaceutics 2022, 14, 106. [Google Scholar] [CrossRef]
- German-Cortés, J.; Vilar-Hernández, M.; Rafael, D.; Abasolo, I.; Andrade, F. Solid Lipid Nanoparticles: Multitasking Nano-Carriers for Cancer Treatment. Pharmaceutics 2023, 15, 831. [Google Scholar] [CrossRef]
- Moosavian, S.A.; Bianconi, V.; Pirro, M.; Sahebkar, A. Challenges and pitfalls in the development of liposomal delivery systems for cancer therapy. Semin. Cancer Biol. 2021, 69, 337–348. [Google Scholar] [CrossRef]
- Pashirova, T.N.; Nemtarev, A.V.; Souto, E.B.; Mironov, V.F. Triarylphosphonium compounds—Effective vectors for mitochondrial-directed delivery systems: Decoration strategies and prospects for clinical application. Russ. Chem. Rev. 2023, 92, RCR5095. [Google Scholar] [CrossRef]
- Mironov, V.F.; Nemtarev, A.V.; Tsepaeva, O.V.; Dimukhametov, M.N.; Litvinov, I.A.; Voloshina, A.D.; Pashirova, T.N.; Titov, E.A.; Lyubina, A.P.; Amerhanova, S.K.; et al. Rational Design 2-Hydroxypropylphosphonium Salts as Cancer Cell Mitochondria-Targeted Vectors: Synthesis, Structure, and Biological Properties. Molecules 2021, 26, 6350. [Google Scholar] [CrossRef] [PubMed]
- Tsepaeva, O.V.; Nemtarev, A.V.; Pashirova, T.N.; Khokhlachev, M.V.; Lyubina, A.P.; Amerkhanova, S.K.; Voloshina, A.D.; Mironov, V.F. Novel triphenylphosphonium amphiphilic conjugates of glycerolipid type: Synthesis, cytotoxic and antibacterial activity, and targeted cancer cell delivery. RSC Med. Chem. 2023, 14, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Grützmacher, H.; Marchand, C.M. Heteroatom stabilized carbenium ions. Coord. Chem. Rev. 1997, 163, 287–344. [Google Scholar] [CrossRef]
- Hudson, H.R. Quasi-phosphonium intermediates and compounds. In Topics in Phosphorus Chemistry; Intersci. Publication: New York, NY, USA; London, UK; Sydney, Australia; Toronto, ON, Canada, 1983; Volume 11, pp. 339–435. [Google Scholar]
- Nesterov, L.V.; Kessel, A.Y.; Mutalapova, R.I. Synthesis of Quasi-Phosphonium salts. J. Gen. Chem. USSR 1969, 39, 2394–2397. [Google Scholar]
- Marchenko, A.P.; Koidan, G.N.; Pinchuk, A.M. Reaction of tetrakis(dialkylamido)phosphonium bromides with bases. J. Gen. Chem. USSR 1984, 54, 2691–2696. [Google Scholar]
- Schwesinger, R.; Schlemper, H.; Hasenfratz, C.; Willaredt, J.; Dambacher, T.; Breuer, T.; Ottaway, C.; Fletschinger, M.; Boele, J.; Fritz, H.; et al. Extremely Strong, Uncharged Auxiliary Bases; Monomeric and Polymer-Supported Polyaminophosphazenes (P2–P5). Liebigs Ann. 1996, 1996, 1055–1081. [Google Scholar] [CrossRef]
- Kong, X.; Wadhwa, K.; Verkade, J.G.; Schmidt-Rohr, K. Determination of the Structure of a Novel Anion Exchange Fuel Cell Membrane by Solid-State Nuclear Magnetic Resonance Spectroscopy. Macromolecules 2009, 42, 1659–1664. [Google Scholar] [CrossRef]
- Stuebe, C.; Lankelma, H.P. Preparation of Some Hexaalkyl-phosphorous, Phosphoric and Phosphorothioic Triamides 1. J. Am. Chem. Soc. 1956, 78, 976–977. [Google Scholar] [CrossRef]
- Ewart, G.; Payne, D.S.; Porte, A.L.; Lane, A.P. 783. Some aminophosphines. J. Chem. Soc. 1962, 3984–3990. [Google Scholar] [CrossRef]
- Petrov, K.A.; Nifant’ev, E.E.; Lysenko, T.N.; Evdakov, V.P. Synthesis of esters of phosphorous and phosphonous acids by alcoholysis of their amides. Zh. Obshch. Khim. (J. Gen. Chem. USSR) 1961, 31, 2377–2380. [Google Scholar]
- Kostag, M.; Liebert, T.; El Seoud, O.A.; Heinze, T. Efficient Cellulose Solvent: Quaternary Ammonium Chlorides. Macromol. Rapid Commun. 2013, 34, 1580–1584. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhao, G.; Liu, F.; Zhang, S. Physicochemical Characterization of MFm−-Based Ammonium Ionic Liquids. J. Chem. Eng. Data 2013, 58, 1505–1515. [Google Scholar] [CrossRef]
- SAINTPlus. Data Reduction and Correction Program (Version 7.31A); APEX2 Version 2.1; Bruker AXS Inc.: Madison, WI, USA, 2006. [Google Scholar]
- Sheldrick, G.M. SADABS; Version 2.10; University of Göttingen: Göttingen, Germany, 2003. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef]
- Ballantyne, A.D.; Brisdon, A.K.; Dryfe, R.A.W. Immiscible electrolyte systems based on asymmetric hydrophobic room temperature ionic liquids. Chem. Commun. 2008, 40, 4980–4982. [Google Scholar] [CrossRef]
- Voloshina, A.D.; Gumerova, S.K.; Sapunova, A.S.; Kulik, N.V.; Mirgorodskaya, A.B.; Kotenko, A.A.; Prokopyeva, T.M.; Mikhailov, V.A.; Zakharova, L.Y.; Sinyashin, O.G. The structure—Activity correlation in the family of dicationic imidazolium surfactants: Antimicrobial properties and cytotoxic effect. Biochim. Biophys. Acta-Gen. Subj. 2020, 1864, 129728. [Google Scholar] [CrossRef]
- EC Committee Guidance on Management and Care of Animals Used for Experiments and Other Research Purposes; 2007/526/EC of 18 June 2007; European Commission: Brussels, Belgium, 2007.
- National Research Council. Guide for the Care and Use of Laboratory Animals; National Academies Press: Washington, DC, USA, 2011; ISBN 978-0-309-15400-0.
- A Guide to Preclinical Drug Research, Part One; Mironov, A. (Ed.) Grif and K: Moscow, Russia, 2012. [Google Scholar]
- Dye, W.T. Herbicidal Alkyl-Amino-Phosphonium Halides. U.S. Patent 2774658, 18 December 1956. [Google Scholar]
- Phosphonium Halides and Process for the Production Thereof. G.B. Patent 716678, 1954.
- Divinskaya, L.P.; Limanov, V.E.; Skvortsova, E.K.; Putyatina, G.M.; Starkov, A.V.; Grinshtein, N.I.; Nifant’ev, E.E. Search for bactericidal preparations in the series of organophosphorus compounds. J. Gen. Chem. USSR 1966, 36, 1244–1246. [Google Scholar]
- Pashirova, T.N.; Zueva, I.V.; Petrov, K.A.; Lukashenko, S.S.; Nizameev, I.R.; Kulik, N.V.; Voloshina, A.D.; Almasy, L.; Kadirov, M.K.; Masson, P.; et al. Mixed cationic liposomes for brain delivery of drugs by the intranasal route: The acetylcholinesterase reactivator 2-PAM as encapsulated drug model. Colloids Surf. B Biointerfaces 2018, 171, 358–367. [Google Scholar] [CrossRef]
- Pashirova, T.N.; Burilova, E.A.; Tagasheva, R.G.; Zueva, I.V.; Gibadullina, E.M.; Nizameev, I.R.; Sudakov, I.A.; Vyshtakalyuk, A.B.; Voloshina, A.D.; Kadirov, M.K.; et al. Delivery nanosystems based on sterically hindered phenol derivatives containing a quaternary ammonium moiety: Synthesis, cholinesterase inhibition and antioxidant activity. Chem. Biol. Interact. 2019, 310, 108753. [Google Scholar] [CrossRef] [PubMed]
- Peña-Morán, O.; Villarreal, M.; Álvarez-Berber, L.; Meneses-Acosta, A.; Rodríguez-López, V. Cytotoxicity, Post-Treatment Recovery, and Selectivity Analysis of Naturally Occurring Podophyllotoxins from Bursera fagaroides var. fagaroides on Breast Cancer Cell Lines. Molecules 2016, 21, 1013. [Google Scholar] [CrossRef] [PubMed]
- Letai, A. Apoptosis and Cancer. Annu. Rev. Cancer Biol. 2017, 1, 275–294. [Google Scholar] [CrossRef]
- Morana, O.; Wood, W.; Gregory, C.D. The Apoptosis Paradox in Cancer. Int. J. Mol. Sci. 2022, 23, 1328. [Google Scholar] [CrossRef] [PubMed]
- Ngoi, N.Y.L.; Choong, C.; Lee, J.; Bellot, G.; Wong, A.L.A.; Goh, B.C.; Pervaiz, S. Targeting Mitochondrial Apoptosis to Overcome Treatment Resistance in Cancer. Cancers 2020, 12, 574. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Lim, B. Targeting Apoptosis in Cancer. Curr. Oncol. Rep. 2022, 24, 273–284. [Google Scholar] [CrossRef]
- Singh, V.; Khurana, A.; Navik, U.; Allawadhi, P.; Bharani, K.K.; Weiskirchen, R. Apoptosis and Pharmacological Therapies for Targeting Thereof for Cancer Therapeutics. Sci 2022, 4, 15. [Google Scholar] [CrossRef]
- Wang, R.-A.; Li, Q.-L.; Li, Z.-S.; Zheng, P.-J.; Zhang, H.-Z.; Huang, X.-F.; Chi, S.-M.; Yang, A.-G.; Cui, R. Apoptosis drives cancer cells proliferate and metastasize. J. Cell. Mol. Med. 2013, 17, 205–211. [Google Scholar] [CrossRef]
- Shoshan-Barmatz, V.; Arif, T.; Shteinfer-Kuzmine, A. Apoptotic proteins with non-apoptotic activity: Expression and function in cancer. Apoptosis 2023, 28, 730–753. [Google Scholar] [CrossRef] [PubMed]
- Cetraro, P.; Plaza-Diaz, J.; MacKenzie, A.; Abadía-Molina, F. A Review of the Current Impact of Inhibitors of Apoptosis Proteins and Their Repression in Cancer. Cancers 2022, 14, 1671. [Google Scholar] [CrossRef] [PubMed]
- Sarsour, E.H.; Kumar, M.G.; Chaudhuri, L.; Kalen, A.L.; Goswami, P.C. Redox Control of the Cell Cycle in Health and Disease. Antioxid. Redox Signal. 2009, 11, 2985–3011. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, I.Z. Free Radicals and Oxidative Stress: Signaling Mechanisms, Redox Basis for Human Diseases, and Cell Cycle Regulation. Curr. Mol. Med. 2023, 23, 13–35. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Koteva, K.; Wright, G.D. An unusual class of anthracyclines potentiate Gram-positive antibiotics in intrinsically resistant Gram-negative bacteria. J. Antimicrob. Chemother. 2014, 69, 1844–1855. [Google Scholar] [CrossRef]
- Berezovskaya, I.V. Classification of Substances with Respect to Acute Toxicity for Parenteral Administration. Pharm. Chem. J. 2003, 37, 139–141. [Google Scholar] [CrossRef]

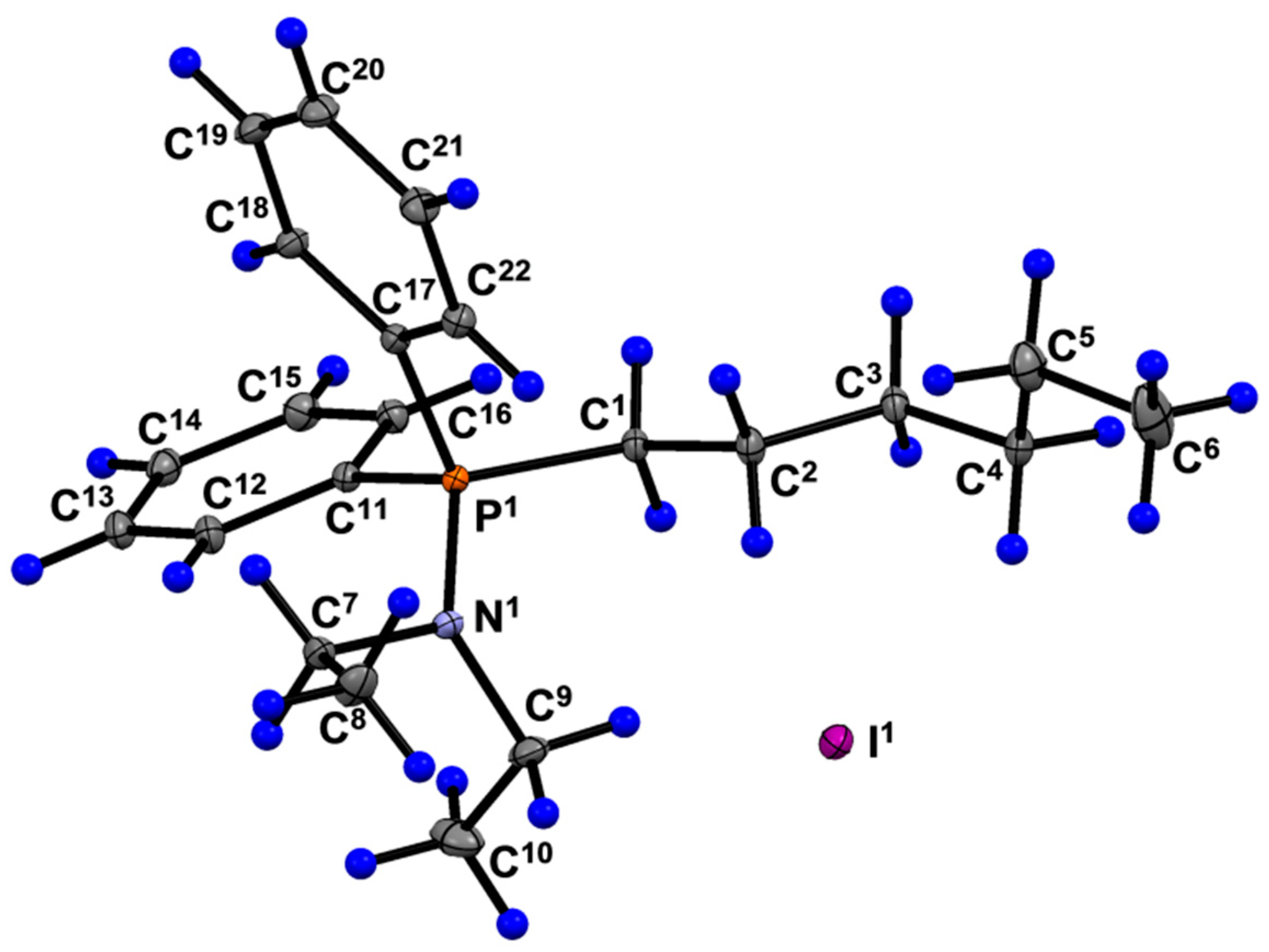












| Parameter | 6a | Parameter | 6a |
|---|---|---|---|
| Molecular formula | C22H33NP, I | Ranges of indices | |
| Sum Formula | C22H33INP | h | –13 ≤ h ≤ 13 |
| Molecular weight | 469.36 | k | –14 ≤ k ≤ 14 |
| Crystal system | triclinic | l | –20 ≤ l ≤ 20 |
| Space group | P-1 (No. 2) | Number of total reflections | 119,684 |
| Z | 2 | Independent reflections | 7633 |
| Unit cell parameters | Rint | 0.069 | |
| a/Å | 9.3261(5) | Completeness up to θ = 28.0° | 0.994 |
| b/Å | 9.9546(5) | Tmax/min | 0.7456/0.5131 |
| c/Å | 13.5060(7) | Number of observed reflections (I > 2σ(I)) | 7098 |
| α | 69.106(2) | Number of reflections/of contraints/number of parameters | 7633/0/229 |
| β/deg | 71.494(2) | GOOF | 1.068 |
| γ | 80.251(2) | R [I > 2σ(I)] | |
| V/Å3 | 1108.61(10) | R1 | 0.0213 |
| dcalc/g cm–3 | 1.406 | wR2 | 0.0553 |
| Absorption coefficient, μ/mm–1 | 1.521 | R (based on all reflections) | |
| F(000) | 480 | R1 and wR2 | 0.0237 and 0.0561 |
| Θ (min, max)/deg | 2.2, 32.0 | Residual electron density (ρmax/ρmin)/e Å–3 | 1.14/–0.50 |
| Composition | Ratio, (% w/w) | Zaverage, nm | PDI | ξ, mV |
|---|---|---|---|---|
| PC [75,76] | 100 | 119 ± 2 | 0.12 ± 0.02 | −7.0 ± 2 |
| PC/4a | 99.8/0.2 | 121 ± 0.5 | 0.09 ± 0.01 | −5.1 ± 0.2 |
| PC/4b | 99.8/0.2 | 124 ± 1 | 0.07 ± 0.01 | +4.3 ± 0.4 |
| PC/4c | 99.8/0.2 | 125 ± 1 | 0.14 ± 0.02 | +9.3 ± 0.3 |
| PC/4d | 99.8/0.2 | 134 ± 1 | 0.14 ± 0.01 | +13.9 ± 0.4 |
| PC/4e | 99.8/0.2 | 115 ± 1 | 0.12 ± 0.01 | +26.8 ± 3 |
| PC/5b | 99.8/0.2 | 129 ± 1 | 0.14 ± 0.01 | +3.2 ± 0.3 |
| PC/5c | 99.8/0.2 | 116 ± 2 | 0.11 ± 0.01 | +7.1 ± 0.2 |
| PC/5d | 99.8/0.2 | 126 ± 0.5 | 0.14 ± 0.02 | +13.3 ± 0.5 |
| PC/4d-Rhod | 99.8/0.2 | 122 ± 0.2 | 0.11 ± 0.01 | +19.0 ± 1.7 |
| PC/5b-Rhod | 99.8/0.2 | 116 ± 0.5 | 0.10 ± 0.02 | +7.9 ± 0.2 |
| PC/5c-Rhod | 99.8/0.2 | 118 ± 0.3 | 0.09 ± 0.03 | +2.6 ± 0.3 |
| PC/5d-Rhod | 99.8/0.2 | 117 ± 0.4 | 0.1 ± 0.01 | +14.2 ± 0.6 |
| PC/5d-Rhod * | 99.8/0.2 | 115 ± 0.2 | 0.1 ± 0.01 | −8.4 ± 0.6 |
| PC/8 | 99.8/0.2 | 117 ± 0.5 | 0.12 ± 0.02 | +16.3 ± 2.5 |
| PC/7 | 99.8/0.2 | 124 ± 0.1 | 0.15 ± 0.01 | −11.7 ± 1.6 |
| SLN [65] | 100 | 104 ± 1 | 0.24 ± 0.01 | −17.2 ± 1 |
| SLN/4b | 99.3/0.7 | 108 ± 0.3 | 0.31 ± 0.04 | −15.6 ± 0.5 |
| SLN/5b | 99.3/0.7 | 114 ± 1 | 0.24 ± 0.01 | −15.8 ± 0.7 |
| SLN/5c | 99.3/0.7 | 132 ± 2 | 0.32 ± 0.03 | −7.7 ± 0.5 |
| SLN/5d | 99.3/0.7 | 147 ± 2 | 0.47 ± 0.02 | −3.6 ± 0.1 |
| SLN-Cur | 100 | 125 ± 1 | 0.2 ± 0.01 | −28.0 ± 1 |
| SLN/5d-Cur | 99.8/0.2 | 123 ± 1 | 0.21 ± 0.01 | −18.5 ± 1 |
| Test Compounds | Cancer Cell Lines | Normal Cell Lines | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M-HeLa | MCF-7 | HuTu 80 | PANC-1 | A549 | PC3 | T98G | Hep G2 | SK-OV-3 | DU-145 | WI38 | Chang Liver | |
| 4a | 52.4 ± 3.7 (ns) | 26.0 ± 1.8 (ns) | 11.4 ± 0.9 (1.6 b) | 16.1 ± 1.3 (1.1 b) | 64.1 ± 4.5 (ns) | 44.0 ± 3.1 (ns) | 57.4 ± 4.0 (ns) | 46.6 ± 3.3 (ns) | 46.6 ± 3.5 (ns) | 26.2 ± 1.9 (ns) | 7.6 ± 0.6 | 17.7 ± 1.5 |
| PC/4a | 60 ± 12 (ns) | 28.3 ± 2.0 (ns) | 26.1 ± 1.8 (ns) | 24.6 ± 1.6 (ns) | 29.2 ± 1.9 (ns) | 15.5 ± 1.2 (ns) | 37.3 ± 2.8 (ns) | 29.1 ± 2.3 (ns) | 35.5 ± 2.5 (ns) | 68.5 ± 4.1 (ns) | 17.2 ± 2.4 | 6.5 ± 0.8 |
| 4b | 1.8 ± 0.1 (2.2 b) | 2.9 ± 0.2 (1.4 b) | 3.2 ± 0.2 (1.3 b) | 2.3 ± 0.1 (1.7 b) | 15.4 ± 1.2 (ns) | 11.6 ± 0.9 (ns) | 5.9 ± 0.4 (ns) | 2.9 ± 0.2 (1.4 b) | 26.3 ± 1.8 (ns) | 4.2 ± 0.3 (ns) | 0.8 ± 0.06 | 4.0 ± 0.3 |
| PC/4b | 4.5 ± 0.3 (2.8 b) | 4.1 ± 0.3 (3.1 b) | 0.7 ± 0.06 (18 b) | 0.9 ± 0.08 (14 b) | 9.5 ± 0.8 (1.3 b) | 3.5 ± 0.4 (3.6 b) | 2.2 ± 0.1 (5.7 b) | 5.0 ± 0.4 (2.5 b) | 83.0 ± 5.8 (ns) | 1.6 ± 0.1 (8.0 b) | 2.6 ± 0.1 | 12.6 ± 1.2 |
| SLN/4b | 0.42 ± 0.04 (26 b) | 0.21 ± 0.02 (53 b) | 0.42 ± 0.04 (26 b) | 1.1 ± 0.1 (10 b) | 1.5 ± 0.1 (7.4 b) | 0.3 ± 0.03 (37 b) | 0.6 ± 0.06 (19 b) | 0.3 ± 0.03 (37 b) | 75 ± 6 (ns) | 0.2 ± 0.01 (56 b) | 2.1 ± 0.2 | 11.1 ± 1.2 |
| 4c | 1.7 ± 0.1 (2.8 b) | 1.9 ± 0.1 (2.5 b) | 2.6 ± 0.2 (1.8 b) | 1.7 ± 0.1 (2.8 b) | 21.1 ± 1.5 (ns) | 1.8 ± 0.1 (2.7 b) | 3.2 ± 0.2 (1.5 b) | 2.4 ± 0.1 (2.0 b) | 12.1 ± 1.1 (ns) | 1.3 ± 0.1 (ns) | 0.8 ± 0.07 | 4.8 ± 0.4 |
| PC/4c | 1.1 ± 0.09 (ns) | 0.3 ± 0.02 (30 b) | 0.6 ± 0.05 (15.6 b) | 0.7 ± 0.06 (13 b) | 4.2 ± 0.3 (2.2 b) | 1.1 ± 0.08 (8.3 b) | 1.9 ± 0.1 (4.8 b) | 1.1 ± 0.08 (8.3 b) | 99 ± 7 (ns) | 0.4 ± 0.03 (22.8 b) | 2.6 ± 0.2 | 9.1 ± 0.8 |
| 4d | 1.7 ± 0.1 (7.1 b) | 2.1 ± 0.2 (5.7 b) | 2.8 ± 0.2 (4.3 b) | 1.6 ± 0.1 (7.5 b) | 18.4 ± 1.3 (ns) | 7.3 ± 0.6 (1.6 b) | 1.8 ± 0.2 (6.7 b) | 2.3 ± 0.1 (5.2 b) | 10.0 ± 0.9 (1.2 b) | 1.2 ± 0.1 (10 b) | 0.9 ± 0.08 | 12.0 ± 0.9 |
| PC/4d | 0.7 ± 0.06 (2.6 a) | 0.4 ± 0.03 (4.5 a) | 0.24 ± 0.02 (7.5 a) | 0.26 ± 0.02 (7.0 a) | 3.7 ± 0.3 (ns) | 0.9 ± 0.08 (2.0 a) | 0.9 ± 0.07 (2.0 a) | 0.6 ± 0.05 (3.0 a) | 69.4 ± 4.9 (ns) | 0.15 ± 0.01 (12 a) | 1.8 ± 0.1 | 0.7 ± 0.06 |
| 4e | 0.4 ± 0.03 (4.3 b) | 0.9 ± 0.07 (1.9 b) | 0.06 ± 0.005 (28 b) | 0.4 ± 0.03 (4.3 b) | 1.7 ± 0.1 (1.0 b) | 1.7 ± 0.1 (1.0 b) | 1.0 ± 0.09 (1.7 b) | 1.5 ± 0.1 (1.1 b) | 1.7 ± 0.1 (1.0 b) | 0.4 ± 0.03 (4.3 b) | 1.2 ± 0.1 | 1.7 ± 0.1 |
| PC/4e | 62 ± 5.3 (ns) | 1.5 ± 0.8 (24 b) | 29.7 ± 2.4 (1.2 b) | 2.5 ± 0.3 (14 b) | 2.3 ± 0.1 (16 b) | 3.8 ± 0.3 (9.5 b) | 3.2 ± 0.2 (11 b) | 7.9 ± 0.6 (4.5 b) | 35.5 ± 2.8 (1 b) | 38.5 ± 3.2 (ns) | 7.1 ± 0.7 | 36.0 ± 2.9 |
| 5a | 23.0 ± 1.6 (2.7 a) | 17.1 ± 1.3 (3.7 a) | 0.3 ± 0.02 (210 a) | 17.8 ± 1.4 (3.5 a) | 17.0 ± 1.3 (3.7 a) | 4.9 ± 0.4 (13 a) | 30.3 ± 2.1 (3.7 a) | 16.5 ± 1.3 (3.8 a) | 20.0 ± 1.6 (3.2 a) | 6.3 ± 0.5 (10 a) | 63.0 ± 4.4 | 22.6 ± 1.6 |
| 5b | 1.8 ± 0.1 (2.2 b) | 2.6 ± 0.2 (1.5 b) | 4.0 ± 0.3 (1.0 b) | 1.2 ± 0.1 (3.3 b) | 20.0 ± 1.6 (ns) | 11.4 ± 0.9 (ns) | 4.3 ± 0.3 (1.0 b) | 2.9 ± 0.3 (1.4 b) | 25.6 ± 1.8 (ns) | 2.0 ± 0.1 (2.0 b) | 0.5 ± 0.4 | 4.0 ± 0.3 |
| PC/5b | 1.1 ± 0.08 (5.5 a) | 0.7 ± 0.06 (8.6 a) | 0.22 ± 0.01 (5.5 a) | 0.7 ± 0.06 (8.6 a) | 3.0 ± 0.2 (2.0 a) | 1.4 ± 0.1 (4.3 a) | 3.4 ± 0.2 (1.8 a) | 1.0 ± 0.09 (6.0 a) | 1.8 ± 0.1 (3.3 a) | 0.4 ± 0.03 (15 a) | 6.0 ± 0.5 | 4.6 ± 0.4 |
| SLP/5b | 2.0 ± 0.1 (1.8 a) | 1.3 ± 0.1 (2.8 a) | 0.9 ± 0.07 (4.0 a) | 1.2 ± 0.1 (3.0 a) | 2.5 ± 0.2 (1.4 a) | 1.6 ± 0.1 (2.3 a) | 5.0 ± 0.4 (ns) | 10.0 ± 0.9 (ns) | 1.7 ± 0.1 (2.1 a) | 1.1 ± 0.09 (3.3 a) | 3.6 ± 0.2 | 1.3 ± 0.1 |
| 5c | 1.7 ± 0.1 (2.4 b) | 3.1 ± 0.2 (1.3 b) | 2.8 ± 0.3 (1.5 b) | 2.8 ± 0.2 (1.5 b) | 8.7 ± 0.7 (ns) | 10.0 ± 0.9 (ns) | 2.7 ± 0.3 (1.5 b) | 2.3 ± 0.1 (1.8 b) | 14.8 ± 1.2 (ns) | 0.9 ± 0.07 (4.6 b) | 0.5 ± 0.4 | 4.1 ± 0.3 |
| PC/5c | 0.7 ± 0.06 (3.3 b) | 0.5 ± 0.04 (4.6 b) | 0.4 ± 0.03 (5.8 b) | 0.8 ± 0.06 (2.9 b) | 1.6 ± 0.1 (1.4 b) | 0.6 ± 0.05 (3.8 b) | 2.7 ± 0.2 (ns) | 0.6 ± 0.05 (3.8 b) | 83.1 ± 5.8 (ns) | 0.2 ± 0.01 (12 b) | 1.9 ± 0.1 | 2.3 ± 0.1 |
| 5d | 1.5 ± 0.1 (ns) | 0.7 ± 0.06 (1.9 b) | 2.5 ± 0.1 (ns) | 1.4 ± 0.1 (ns) | 8.9 ± 0.7 (ns) | 0.8 ± 0.06 (1.6 b) | 0.7 ± 0.05 (1.9 b) | 1.1 ± 0.09 (1.2 b) | 10.0 ± 0.8 (ns) | 0.45 ± 0.03 (2.9 b) | 0.3 ± 0.02 | 1.3 ± 0.1 |
| PC/5d | 0.8 ± 0.07 (1.9 a) | 0.7 ± 0.06 (2.1 a) | 0.4 ± 0.02 (3.8 a) | 0.5 ± 0.04 (3.0 a) | 2.6 ± 0.2 (ns) | 0.6 ± 0.05 (2.5 a) | 2.5 ± 0.2 (ns) | 2.7 ± 0.2 (ns) | 92.4 ± 6.5 (ns) | 0.3 ± 0.02 (5.0 a) | 1.5 ± 0.1 | 0.23 ± 0.01 |
| 6a | 5.8 ± 0.4 (4.8 a) | 17.0 ± 1.3 (1.6 a) | 0.1 ± 0.007 (277 a) | 5.5 ± 0.4 (5.0 a) | 48.2 ± 3.4 (ns) | 4.0 ± 0.3 (6.9 a) | 18.4 ± 1.5 (1.5 a) | 34.4 ± 2.4 (ns) | 36.8 ± 2.6 (ns) | 2.6 ± 0.3 (11 a) | 27.7 ± 1.9 | 15.2 ± 1.3 |
| 6b | 0.95 ± 0.08 (17 a) | 9.5 ± 0.8 (1.7 a) | 0.2 ± 0.01 (81 a) | 1.3 ± 0.1 (12 a) | 13.0 ± 1.1 (1.2 a) | 0.85 ± 0.07 (19 a) | 3.4 ± 0.2 (5.0 a) | 11.6 ± 0.9 (1.4 a) | 17.6 ± 1.4 (ns) | 0.8 ± 0.07 (20 a) | 16.1 ± 1.2 | 0.42 ± 0.03 |
| 6c | 0.9 ± 0.07 (1.6 a) | 4.2 ± 0.3 (2.0 a) | 0.2 ± 0.01 (42 a) | 1.3 ± 0.1 (6.4 a) | 11.4 ± 0.9 (ns) | 1.0 ± 0.08 (8.3 a) | 2.4 ± 0.1 (3.5 a) | 6.2 ± 0.5 (1.3 a) | 12.7 ± 1.1 (ns) | 1.3 ± 0.1 (6.4 a) | 8.3 ± 0.7 | 0.5 ± 0.04 |
| 6d | 0.24 ± 0.02 (1.6 a) | 1.6 ± 0.1 (ns) | 0.35 ± 0.02 (1.1 a) | 0.4 ± 0.03 (1.0 a) | 8.2 ± 0.7 (ns) | 1.4 ± 0.1 (ns) | 0.6 ± 0.05 (ns) | 1.3 ± 0.1 (ns) | 3.9 ± 0.4 (ns) | 0.43 ± 0.03 (1.0 a) | 0.4 ± 0.03 | 0.3 ± 0.02 |
| 6e | 0.4 ± 0.03 (7.0 a) | 1.2 ± 0.1 (2.3 a) | 0.07 ± 0.005 (40 a) | 0.3 ± 0.02 (9.3 a) | 2.1 ± 0.1 (1.3 a) | 1.5 ± 0.1 (1.9 a) | 1.1 ± 0.09 (2.5 a) | 1.4 ± 0.1 (2.0 a) | 1.6 ± 0.1 (1.8 a) | 0.4 ± 0.02 (7.0 a) | 2.8 ± 0.2 | 2.5 ± 0.2 |
| 7 | 16.1 ± 1.3 (3.0 a) | 29.1 ± 2.0 (1.7 a) | 1.9 ± 0.2 (26 a) | 44.6 ± 3.1 (1.1 a) | >100 (ns) | 32.7 ± 2.3 (1.5 a) | >100 (ns) | >100 (ns) | 69.0 ± 4.8 (ns) | 58.0 ± 4.1 (ns) | 49.0 ± 3.4 | 34.0 ± 2.4 |
| PC/7 | 148 ± 12 (ns) | 67 ± 5.4 (ns) | 54.3 ± 3.8 (ns) | 134.2 ± 10 (ns) | >100 (ns) | 41.7 ± (ns) | >100 (ns) | >100 (ns) | >100 (ns) | 99.2 ± (ns) | 12.6 ± 1.3 | 0.3 ± 0.02 |
| 8 | 0.45 ± 0.04 (20 a) | 4.2 ± 0.3 (2.2 a) | 0.09 ± 0.006 (102 a) | 0.3 ± 0.02 (31 a) | 8.7 ± 0.7 (1.1 a) | 0.4 ± 0.03 (23 a) | 0.6 ± 0.05 (15 a) | 1.6 ± 0.1 (5.8 a) | 9.8 ± 0.7 (ns) | 0.4 ± 0.03 (23 a) | 9.2 ± 0.8 | 3.0 ± 0.2 |
| PC/8 | 17.0 ± 1.5 (1.7 b) | 11.2 ± 0.9 (2.6 b) | 6.2 ± 0.5 (4.7 b) | 18.2 ± 1.4 (1.6 b) | 0.8 ± 0.06 (36 b) | 35.6 ± 2.2 (ns) | 1.2 ± 0.1 (24 b) | 37.2 ± 2.5 (ns) | 67.0 ± 4.7 (ns) | 2.5 ± 0.1 (12 b) | 28.0 ± 1.8 | 29.0 ± 2.0 |
| 9 | 31.1 ± 3.9 (1.5 a) | 37.8 ± 4.8 (1.2 a) | 13.1 ± 1.7 (3.5 a) | 39.6 ± 5.1 (1.2 a) | >100 | 34.9 ± 4.5 (1.3 a) | >100 | 53.9 ± 6.9 (ns) | 36.4 ± 4.7 (1.3 a) | 50.5 ± 6.4 (ns) | 45.8 ± 5.9 | 37.2 ± 4.8 |
| doxorubicin | 2.1 ± 0.1 (ns) | 0.4 ± 0.03 (1.3 b) | 0.2 ± 0.01 (2.5 b) | 2.2 ± 0.1 (ns) | 0.7 ± 0.06 (ns) | 1.4 ± 0.1 (ns) | 1.0 ± 0.09 (ns) | 0.2 ± 0.01 (2.5 b) | 6.7 ± 0.5 (ns) | 0.3 ± 0.02 (1.7 b) | 0.4 ± 0.03 | 0.5 ± 0.03 |
| Compounds | Minimum Inhibitory Concentration (MIC), μM | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | Yeast | ||||||
| Sa | Bc | Ef | MRSA-1 | MRSA-2 | Ec | Pa | Ca | |
| 4a | 62.5 ± 5.2 | 62.5 ± 5.3 | 250 ± 21 | 125 ± 10 | 125 ± 11 | – | – | – |
| 4b | 15.6 ± 1.3 | 7.8 ± 0.7 | 250 ± 19 | 15.6 ± 0.3 | 15.6 ± 1.2 | – | – | – |
| 4c | 3.9 ± 0.3 | 3.9 ± 0.3 | 62.5 ± 5.2 | 3.9 ± 0.4 | 7.8 ± 0.6 | 500 ± 48 | – | – |
| 4d | 1.9 ± 0.1 | 0.9 ± 0.07 | 15.6 ± 1.3 | 0.9 ± 0.07 | 1.9 ± 0.1 | 125 ± 9 | 500 ± 47 | 125 ± 11 |
| 4e | 0.25 ± 0.01 | 0.5 ± 0.04 | 0.5 ± 0.04 | 0.5 ± 0.04 | 0.25 ± 0.02 | 15.6 ± 1.3 | 7.8 ± 0.6 | 1.9 ± 0.1 |
| 9 | 34.3 ± 5.4 | 274 ± 48 | – | 137 ± 19 | 34.3 ± 5.3 | – | – | – |
| 5b | 7.8 ± 0.6 | 7.8 ± 0.6 | 125 ± 11 | 15.6 ± 1.2 | 15.6 ± 1.2 | – | 500 ± 46 | – |
| 5c | 3.9 ± 0.3 | 3.9 ± 0.3 | 31.3 ± 2.2 | 3.9 ± 0.3 | 3.9 ± 0.4 | 250 ± 19 | – | – |
| 5d | 0.9 ± 0.07 | 0.9 ± 0.08 | 15.6 ± 1.2 | 0.9 ± 0.08 | 0.9 ± 0.08 | 62.5 ± 5.4 | 500 ± 48 | 250 ± 21 |
| 6a | 62.5 ± 5.5 | 31.3 ± 2.3 | 125 ± 11 | 62.5 ± 5.3 | 62.5 ± 5.6 | – | – | – |
| 6b | 7.8 ± 0.6 | 3.9 ± 0.4 | 62.5 ± 5.5 | 7.8 ± 0.6 | 7.8 ± 0.6 | – | – | – |
| 6c | 3.9 ± 0.3 | 1.9 ± 0.1 | 31.3 ± 5.4 | 3.9 ± 0.3 | 3.9 ± 0.4 | 500 ± 46 | 250 ± 21 | 500 ± 47 |
| 6d | 0.9 ± 0.08 | 0.9 ± 0.08 | 3.9 ± 0.3 | 0.5 ± 0.03 | 0.5 ± 0.03 | 62.5 ± 5.3 | 125 ± 10 | 62.5 ± 5.4 |
| 6e | 0.5 ± 0.04 | 0.9 ± 0.08 | 0.5 ± 0.03 | 0.5 ± 0.04 | 0.5 ± 0.03 | 31.3 ± 2.4 | 3.9 ± 0.3 | 1.9 ± 0.1 |
| 8 | 0.9 ± 0.07 | 0.9 ± 0.07 | 15.6 ± 1.2 | 1.9 ± 0.1 | 3.9 ± 0.3 | 500 ± 47 | – | – |
| 7 | 250 ± 20 | 500 ± 47 | – | 250 ± 19 | 250 ± 20 | – | – | – |
| Norfloxacin | 7.5 ± 0.5 | 24.4 ± 2.1 | 24.4 ± 2.2 | – | 7.5 ± 0.5 | 4.7 ± 0.02 | 12.1 ± 1.1 | – |
| Ketoconazole | – | – | – | – | – | – | – | 7.3 ± 0.5 |
| Minimum bactericidal and fungicidal concentrations (MBC, MFC), μM | ||||||||
| 4a | 250 ± 20 | – | 500 ± 46 | – | 125 ± 10 | – | – | – |
| 4b | 31.3 ± 2.3 | – | – | 500 ± 47 | 125 ± 9 | – | – | – |
| 4c | 250 ± 18 | >500 | 500 ± 48 | 125 ± 10 | 250 ± 20 | 500 ± 48 | – | – |
| 4d | 31.3 ± 2.5 | 250 ± 22 | >500 | 62.5 ± 5.4 | 250 ± 18 | 250 ± 19 | 500 ± 48 | 250 ± 18 |
| 4e | 0.9 ± 0.07 | 7.8 ± 0.6 | 1.9 ± 0.1 | 0.5 ± 0.04 | 0.25 ± 0.02 | 62.5 ± 5.2 | 7.8 ± 0.6 | 7.8 ± 0.6 |
| 9 | 137 ± 21 | – | – | 137 ± 18 | 137 ± 21 | – | – | – |
| 5b | 250 ± 20 | – | – | 500 ± 48 | 250 ± 20 | 500 ± 49 | – | – |
| 5c | 125 ± 10 | 500 ± 48 | 500 ± 48 | 62.5 ± 5.5 | 62.5 ± 5.4 | 500 ± 48 | >500 | – |
| 5d | 125 ± 9 | 500 ± 47 | 125 ± 9 | 125 ± 11 | 62.5 ± 5.2 | 62.5 ± 5.3 | 500 ± 47 | – |
| 6a | 125 ± 10 | 500 ± 46 | – | 500 ± 47 | 62.5 ± 5.6 | – | – | – |
| 6b | 125 ± 11 | – | – | 31.3 ± 2.6 | 31.3 ± 2.5 | – | – | – |
| 6c | 125 ± 11 | >500 | – | 62.5 ± 5.4 | 7.8 ± 0.7 | 500 ± 47 | 250 ± 21 | – |
| 6d | 62.5 ± 5.4 | 250 ± 21 | 250 ± 19 | 0.9 ± 0.07 | 0.9 ± 0.08 | 62.5 ± 5.5 | 125 ± 9 | 250 ± 19 |
| 6e | 0.5 ± 0.04 | 31.3 ± 2.5 | 0.9 ± 0.1 | 1.9 ± 0.1 | 0.5 ± 0.04 | 125 ± 11 | 3.9 ± 0.4 | 15.6 ± 1.2 |
| 8 | 62.5 ± 5.3 | 500 ± 47 | – | 15.6 ± 1.2 | 15.6 ± 1.3 | – | – | – |
| 7 | – | – | – | – | 500 ± 47 | – | – | – |
| Norfloxacin | 7.5 ± 0.6 | 24.4 ± 2.1 | 24.4 ± 1.9 | – | 7.5 ± 0.6 | 24.4 ± 2.3 | 49.0 ± 4.2 | |
| Ketoconazole | 7.3 ± 0.5 | |||||||
| Test Compound | HC50 (μM) |
|---|---|
| 4b | >100 |
| 5b | >100 |
| 6b | >100 |
| Gramicidin S | 9.4 ± 0.8 |
| Compound | LD50, mg/kg |
|---|---|
| 4a | 31.7 (28.3÷35.8) |
| 4b | 43.4 (41.8÷44.8) |
| 4d | 5.9 (3.7÷7.6) |
| 5a | 44.7 (43.0÷46.3) |
| 5b | 27.9 (26.9÷29.9) |
| 6b | 102.5 (98.7÷105.8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mironov, V.F.; Dimukhametov, M.N.; Nemtarev, A.V.; Pashirova, T.N.; Tsepaeva, O.V.; Voloshina, A.D.; Vyshtakalyuk, A.B.; Litvinov, I.A.; Lyubina, A.P.; Sapunova, A.S.; et al. Novel Mitochondria-Targeted Amphiphilic Aminophosphonium Salts and Lipids Nanoparticles: Synthesis, Antitumor Activity and Toxicity. Nanomaterials 2023, 13, 2840. https://doi.org/10.3390/nano13212840
Mironov VF, Dimukhametov MN, Nemtarev AV, Pashirova TN, Tsepaeva OV, Voloshina AD, Vyshtakalyuk AB, Litvinov IA, Lyubina AP, Sapunova AS, et al. Novel Mitochondria-Targeted Amphiphilic Aminophosphonium Salts and Lipids Nanoparticles: Synthesis, Antitumor Activity and Toxicity. Nanomaterials. 2023; 13(21):2840. https://doi.org/10.3390/nano13212840
Chicago/Turabian StyleMironov, Vladimir F., Mudaris N. Dimukhametov, Andrey V. Nemtarev, Tatiana N. Pashirova, Olga V. Tsepaeva, Alexandra D. Voloshina, Alexandra B. Vyshtakalyuk, Igor A. Litvinov, Anna P. Lyubina, Anastasiia S. Sapunova, and et al. 2023. "Novel Mitochondria-Targeted Amphiphilic Aminophosphonium Salts and Lipids Nanoparticles: Synthesis, Antitumor Activity and Toxicity" Nanomaterials 13, no. 21: 2840. https://doi.org/10.3390/nano13212840






