Nanoscale Surface-Enhanced Raman Spectroscopy Investigation of a Polyphenol-Based Plasmonic Nanovector
Abstract
:1. Introduction
2. Materials and Methods
2.1. CA and CT Solutions for Raman Measurements
2.2. Polyphenol-Synthesized AgNPs
2.3. Hydroxylamine-Synthesized AgNPs
2.4. SEM Images
2.5. Dynamic Light Scattering, -Potential and Light Transmission Spectroscopy Measurements
2.6. Raman and SERS Spectroscopy
2.7. Density Functional Theory Calculations
3. Results and Discussion
3.1. pH-Response of Polyphenols by Raman Spectroscopy
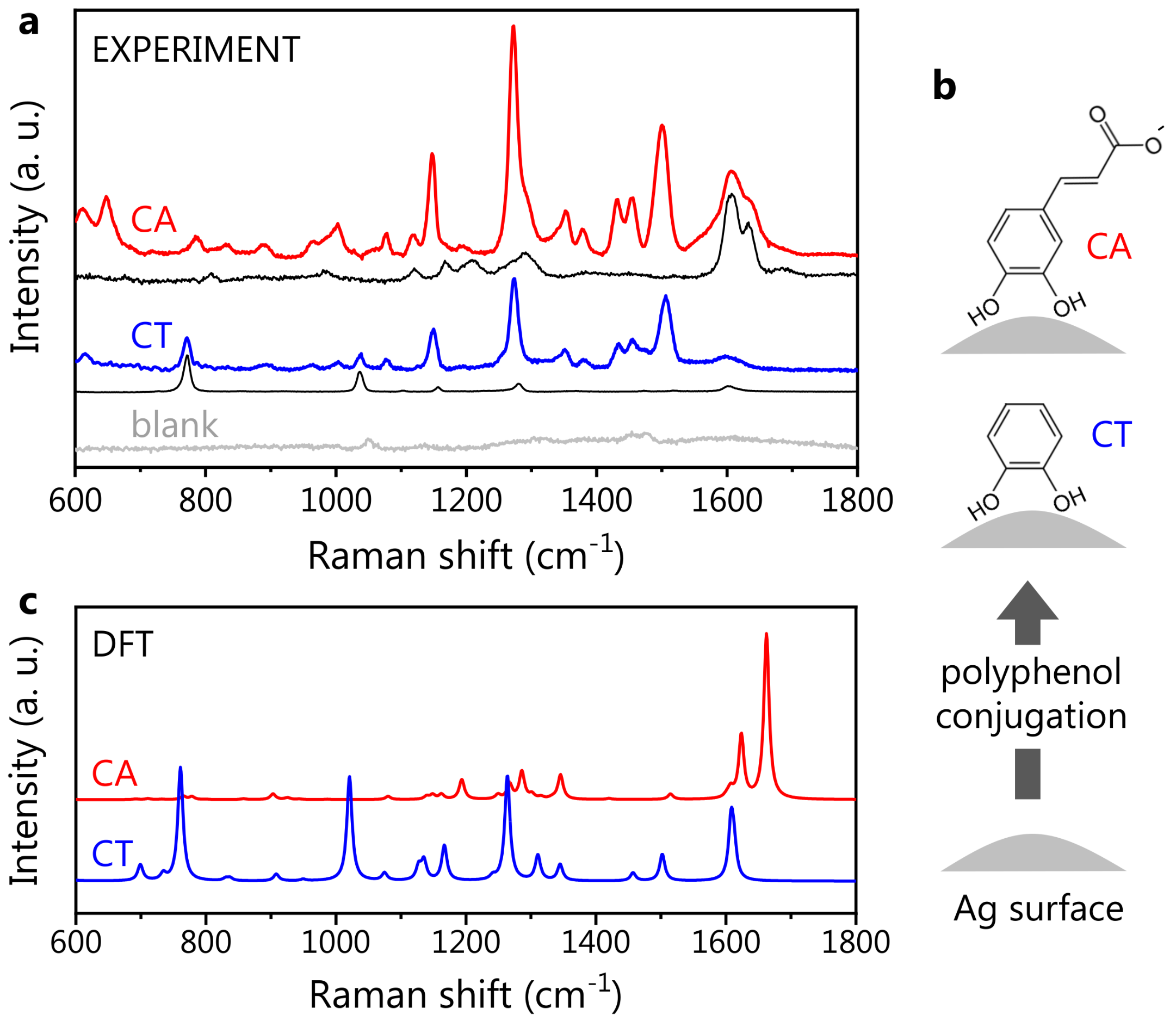
3.2. Reference Polyphenol SERS Spectra
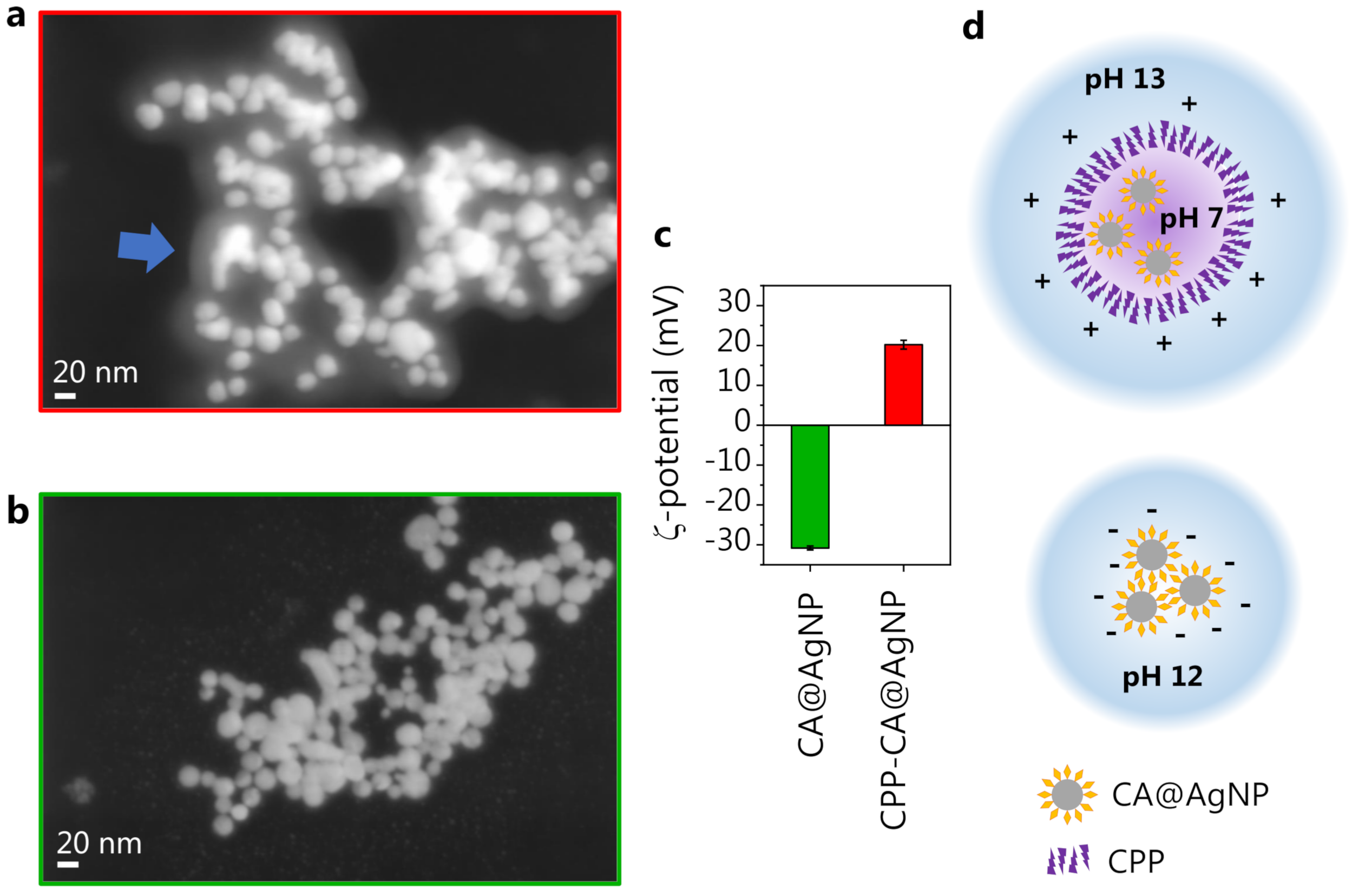
3.3. Caffeic Acid-Synthesized AgNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraga, C.G.; Galleano, M.; Verstraeten, S.V.; Oteiza, P.I. Basic biochemical mechanisms behind the health benefits of polyphenols. Mol. Asp. Med. 2010, 31, 43–445. [Google Scholar] [CrossRef] [PubMed]
- Santos-Sánchez, N.F.; Salas-Coronado, R.; Villanueva-Cañongo, C.; Hernández-Carlos, B. Antioxidant compounds and their antioxidant mechanism. In Antioxidants; IntehOpen: London, UK, 2019; Volume 10, pp. 23–396. [Google Scholar]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: Chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, B.; Lv, B. Antibacterial therapeutic agents composed of functional biological molecules. J. Chem. 2020, 2020, 6578579. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [Green Version]
- Seca, A.M.; Pinto, D.C. Plant secondary metabolites as anticancer agents: Successes in clinical trials and therapeutic application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef] [Green Version]
- Allawadhi, P.; Khurana, A.; Sayed, N.; Kumari, P.; Godugu, C. Isoproterenol-induced cardiac ischemia and fibrosis: Plant-based approaches for intervention. Phytother. Res. 2018, 32, 1908–1932. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Sang, W.; Xie, L.; Li, W.; Li, B.; Li, J.; Tian, H.; Yuan, Z.; Zhao, Q.; Dai, Y. Polyphenol-Based Nanomedicine Evokes Immune Activation for Combination Cancer Treatment. Angew. Chem. Int. Ed. 2021, 60, 1967–1975. [Google Scholar] [CrossRef]
- Kuzuhara, T.; Sei, Y.; Yamaguchi, K.; Suganuma, M.; Fujiki, H. DNA and RNA as new binding targets of green tea catechins. J. Biol. Chem. 2006, 281, 17446–17456. [Google Scholar] [CrossRef] [Green Version]
- Yi, J.; Li, S.; Wang, C.; Cao, N.; Qu, H.; Cheng, C.; Wang, Z.; Wang, L.; Zhou, L. Potential applications of polyphenols on main ncRNAs regulations as novel therapeutic strategy for cancer. Biomed. Pharmacother. 2019, 113, 108703. [Google Scholar] [CrossRef]
- Fasolato, C.; Giantulli, S.; Capocefalo, A.; Toumia, Y.; Notariello, D.; Mazzarda, F.; Silvestri, I.; Postorino, P.; Domenici, F. Antifolate SERS-active nanovectors: Quantitative drug nanostructuring and selective cell targeting for effective theranostics. Nanoscale 2019, 11, 15224–15233. [Google Scholar] [CrossRef]
- Wu, Y.; Wan, S.; Chen, Y.; Fan, J.; Li, Y.; Wang, T.; Yuan, Z.; Yang, Q.; Qin, H.; Xu, J.; et al. Biomimetic lipidic nanovectors for effective asparaginase supramolecule delivery. Nanomed. Nanotechnol. Biol. Med. 2022, 41, 102518. [Google Scholar] [CrossRef]
- Yang, B.; Dong, Y.; Wang, F.; Zhang, Y. Nanoformulations to enhance the bioavailability and physiological functions of polyphenols. Molecules 2020, 25, 4613. [Google Scholar] [CrossRef]
- Ahamed, M.; AlSalhi, M.S.; Siddiqui, M. Silver nanoparticle applications and human health. Clin. Chim. Acta 2010, 411, 1841–1848. [Google Scholar] [CrossRef]
- Ravindran, A.; Chandran, P.; Khan, S.S. Biofunctionalized silver nanoparticles: Advances and prospects. Colloids Surf. Biointerfaces 2013, 105, 342–352. [Google Scholar] [CrossRef]
- Neri, F.; Scala, A.; Grimato, S.; Santoro, M.; Spadaro, S.; Barreca, F.; Cimino, F.; Speciale, A.; Saija, A.; Grassi, G.; et al. Biocompatible silver nanoparticles embedded in a PEG–PLA polymeric matrix for stimulated laser light drug release. J. Nanoparticle Res. 2016, 18, 153. [Google Scholar] [CrossRef]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver nanoparticles and their antibacterial applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef]
- Liu, B.; Sharma, M.; Yu, J.; Wang, L.; Shendre, S.; Sharma, A.; Izmir, M.; Delikanli, S.; Altintas, Y.; Dang, C.; et al. Management of electroluminescence from silver-doped colloidal quantum well light-emitting diodes. Cell Rep. Phys. Sci. 2022, 3, 100860. [Google Scholar] [CrossRef]
- Moulton, M.C.; Braydich-Stolle, L.K.; Nadagouda, M.N.; Kunzelman, S.; Hussain, S.M.; Varma, R.S. Synthesis, characterization and biocompatibility of “green” synthesized silver nanoparticles using tea polyphenols. Nanoscale 2010, 2, 763–770. [Google Scholar] [CrossRef]
- Rafique, M.; Sadaf, I.; Rafique, M.S.; Tahir, M.B. A review on green synthesis of silver nanoparticles and their applications. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1272–1291. [Google Scholar] [CrossRef]
- Della Pelle, F.; Scroccarello, A.; Sergi, M.; Mascini, M.; Del Carlo, M.; Compagnone, D. Simple and rapid silver nanoparticles based antioxidant capacity assays: Reactivity study for phenolic compounds. Food Chem. 2018, 256, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Nadaf, S.J.; Jadhav, N.R.; Naikwadi, H.S.; Savekar, P.L.; Sapkal, I.D.; Kambli, M.M.; Desai, I.A. Green Synthesis of Gold and Silver Nanoparticles: Updates on Research, Patents, and Future Prospects. OpenNano 2022, 8, 100076. [Google Scholar] [CrossRef]
- Scroccarello, A.; Junior, B.M.H.; Della Pelle, F.; Ciancetta, J.; Ferraro, G.; Fratini, E.; Valbonetti, L.; Copez, C.C.; Compagnone, D. Effect of phenolic compounds-capped AgNPs on growth inhibition of Aspergillus niger. Colloids Surf. B Biointerfaces 2021, 199, 111533. [Google Scholar] [CrossRef] [PubMed]
- Molina-Hernández, J.B.; Scroccarello, A.; Della Pelle, F.; De Flaviis, R.; Compagnone, D.; Del Carlo, M.; Paparella, A.; López, C.C. Synergistic antifungal activity of catechin and silver nanoparticles on Aspergillus niger isolated from coffee seeds. LWT 2022, 169, 113990. [Google Scholar] [CrossRef]
- Ivanova, N.; Gugleva, V.; Dobreva, M.; Pehlivanov, I.; Stefanov, S.; Andonova, V. Silver nanoparticles as multi-functional drug delivery systems. In Nanomedicines; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
- Zhang, X.F.; Liu, Z.G.; Shen, W.; Gurunathan, S. Silver nanoparticles: Synthesis, characterization, properties, applications, and therapeutic approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef] [PubMed]
- Galdopórpora, J.M.; Ibar, A.; Tuttolomondo, M.V.; Desimone, M.F. Dual-effect core–shell polyphenol coated silver nanoparticles for tissue engineering. Nano-Struct. Nano-Objects 2021, 26, 100716. [Google Scholar] [CrossRef]
- Hashemi, Z.; Shirzadi-Ahodashti, M.; Mortazavi-Derazkola, S.; Ebrahimzadeh, M.A. Sustainable biosynthesis of metallic silver nanoparticles using barberry phenolic extract: Optimization and evaluation of photocatalytic, in vitro cytotoxicity, and antibacterial activities against multidrug-resistant bacteria. Inorg. Chem. Commun. 2022, 139, 109320. [Google Scholar] [CrossRef]
- Li, D.; Liu, Z.; Yuan, Y.; Liu, Y.; Niu, F. Green synthesis of gallic acid-coated silver nanoparticles with high antimicrobial activity and low cytotoxicity to normal cells. Process. Biochem. 2015, 50, 357–366. [Google Scholar] [CrossRef]
- Singh, C.; Kumar, J.; Kumar, P.; Chauhan, B.S.; Tiwari, K.N.; Mishra, S.K.; Srikrishna, S.; Saini, R.; Nath, G.; Singh, J. Green synthesis of silver nanoparticles using aqueous leaf extract of Premna integrifolia (L.) rich in polyphenols and evaluation of their antioxidant, antibacterial and cytotoxic activity. Biotechnol. Biotechnol. Equip. 2019, 33, 359–371. [Google Scholar] [CrossRef] [Green Version]
- Cialla-May, D.; Zheng, X.S.; Weber, K.; Popp, J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: From cells to clinics. Chem. Soc. Rev. 2017, 46, 3945–3961. [Google Scholar] [CrossRef]
- Fernandes, T.; Fateixa, S.; Nogueira, H.I.; Daniel-da Silva, A.L.; Trindade, T. Dendrimer-Based Gold Nanostructures for SERS Detection of Pesticides in Water. Eur. J. Inorg. Chem. 2020, 2020, 1153–1162. [Google Scholar] [CrossRef]
- Mota, F.L.; Queimada, A.J.; Pinho, S.P.; Macedo, E.A. Aqueous solubility of some natural phenolic compounds. Ind. Eng. Chem. Res. 2008, 47, 5182–5189. [Google Scholar] [CrossRef] [Green Version]
- Leopold, N.; Lendl, B. A new method for fast preparation of highly surface-enhanced Raman scattering (SERS) active silver colloids at room temperature by reduction of silver nitrate with hydroxylamine hydrochloride. J. Phys. Chem. B 2003, 107, 5723–5727. [Google Scholar] [CrossRef]
- Sarra, A.; Stanchieri, G.D.P.; De Marcellis, A.; Bordi, F.; Postorino, P.; Palange, E. Laser transmission spectroscopy based on tunable-gain dual-channel dual-phase LIA for biological nanoparticles characterization. IEEE Trans. Biomed. Circuits Syst. 2021, 15, 177–187. [Google Scholar] [CrossRef]
- Fasolato, C.; Domenici, F.; Sennato, S.; Mura, F.; De Angelis, L.; Luongo, F.; Costantini, F.; Bordi, F.; Postorino, P. Dimensional scale effects on surface enhanced Raman scattering efficiency of self-assembled silver nanoparticle clusters. Appl. Phys. Lett. 2014, 105, 073105. [Google Scholar] [CrossRef]
- Chen, T.; Wang, H.; Chen, G.; Wang, Y.; Feng, Y.; Teo, W.S.; Wu, T.; Chen, H. Hotspot-induced transformation of surface-enhanced Raman scattering fingerprints. ACS Nano 2010, 4, 3087–3094. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian˜16 Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Moskovits, M. Surface-enhanced Raman spectroscopy: A brief retrospective. J. Raman Spectrosc. 2005, 36, 485–496. [Google Scholar] [CrossRef]
- Dreuw, A.; Head-Gordon, M. Single-reference ab initio methods for the calculation of excited states of large molecules. Chem. Rev. 2005, 105, 4009–4037. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [Green Version]
- Henderson, T.M.; Izmaylov, A.F.; Scalmani, G.; Scuseria, G.E. Can short-range hybrids describe long-range-dependent properties? J. Chem. Phys. 2009, 131, 044108. [Google Scholar] [CrossRef]
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Dunning, T.H., Jr. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J. Chem. Phys. 1989, 90, 1007–1023. [Google Scholar] [CrossRef]
- Kendall, R.A.; Dunning, T.H., Jr.; Harrison, R.J. Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J. Chem. Phys. 1992, 96, 6796–6806. [Google Scholar] [CrossRef] [Green Version]
- Woon, D.E.; Dunning, T.H., Jr. Gaussian basis sets for use in correlated molecular calculations. III. The atoms aluminum through argon. J. Chem. Phys. 1993, 98, 1358–1371. [Google Scholar] [CrossRef] [Green Version]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Wadt, W.R.; Hay, P.J. Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi. J. Chem. Phys. 1985, 82, 284–298. [Google Scholar] [CrossRef]
- Johnson, R. D, III. NIST 101. Computational Chemistry Comparison and Benchmark Database; NIST: Gaithersburg, MD, USA, 1999. [Google Scholar]
- López-Tocón, I.; Valdivia, S.; Soto, J.; Otero, J.C.; Muniz-Miranda, F.; Menziani, M.C.; Muniz-Miranda, M. A DFT approach to the surface-enhanced Raman scattering of 4-cyanopyridine adsorbed on silver nanoparticles. Nanomaterials 2019, 9, 1211. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.H.; Lee, S. SERS and DFT investigation of the adsorption behavior of 4-mercaptobenzoic acid on silver colloids. Colloids Surf. Physicochem. Eng. Asp. 2015, 474, 29–35. [Google Scholar] [CrossRef]
- Mircescu, N.E.; Oltean, M.; Chiş, V.; Leopold, N. FTIR, FT-Raman, SERS and DFT study on melamine. Vib. Spectrosc. 2012, 62, 165–171. [Google Scholar] [CrossRef]
- Otto, A.; Billmann, J.; Eickmans, J.; Ertürk, U.; Pettenkofer, C. The “adatom model” of SERS (surface enhanced Raman scattering): The present status. Surf. Sci. 1984, 138, 319–338. [Google Scholar] [CrossRef]
- Yurenko, Y.P.; Zhurakivsky, R.O.; Ghomi, M.; Samijlenko, S.P.; Hovorun, D.M. Comprehensive conformational analysis of the nucleoside analogue 2′-β-deoxy-6-azacytidine by DFT and MP2 calculations. J. Phys. Chem. B 2007, 111, 6263–6271. [Google Scholar] [CrossRef]
- Goerigk, L.; Grimme, S. Efficient and accurate double-hybrid-meta-GGA Density Functionals - Evaluation with the extended GMTKN30 database for general main group thermochemistry, kinetics, and non-covalent interactions. J. Chem. Theory Comput. 2011, 7, 291–309. [Google Scholar] [CrossRef]
- Polavarapu, P.L. Ab initio vibrational Raman and Raman optical activity spectra. J. Phys. Chem. 1990, 94, 8106–8112. [Google Scholar] [CrossRef]
- Krishnakumar, V.; Keresztury, G.; Sundius, T.; Ramasamy, R. Simulation of IR and Raman spectra based on scaled DFT force fields: A case study of 2-(methylthio) benzonitrile, with emphasis on band assignment. J. Mol. Struct. 2004, 702, 9–21. [Google Scholar] [CrossRef]
- Williams, A.O.; Isaacs, R.J.; Stowell, K.M. Down-regulation of human topoisomerase IIα expression correlates with relative amounts of specificity factors Sp1 and Sp3 bound at proximal and distal promoter regions. BMC Mol. Biol. 2007, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, F.; Cordeiro, M.N.D. Improving vibrational mode interpretation using bayesian regression. J. Chem. Theory Comput. 2018, 15, 456–470. [Google Scholar] [CrossRef]
- Williams, T.; Kelley, C.; Bröker, H.B.; Campbell, J.; Cunningham, R.; Denholm, D.; Elber, G.; Fearick, R.; Grammes, C.; Hart, L.; et al. Gnuplot 4.6. An Interactive Plotting Program. 2012. Available online: http://www.gnuplot.info/ (accessed on 1 July 2022).
- Genaro-Mattos, T.C.; Maurício, Â.Q.; Rettori, D.; Alonso, A.; Hermes-Lima, M. Antioxidant activity of caffeic acid against iron-induced free radical generation—A chemical approach. PLoS ONE 2015, 10, e0129963. [Google Scholar]
- Beltran, J.; Sanli, N.; Fonrodona, G.; Barron, D.; Özkan, G.; Barbosa, J. Spectrophotometric, potentiometric and chromatographic pKa values of polyphenolic acids in water and acetonitrile–water media. Anal. Chim. Acta 2003, 484, 253–264. [Google Scholar] [CrossRef]
- Miertuš, S.; Scrocco, E.; Tomasi, J. Electrostatic interaction of a solute with a continuum. A direct utilization of ab initio molecular potentials for the prevision of solvent effects. Chem. Phys. 1981, 55, 117–129. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef] [PubMed]
- Nurchi, V.M.; Pivetta, T.; Lachowicz, J.I.; Crisponi, G. Effect of substituents on complex stability aimed at designing new iron (III) and aluminum (III) chelators. J. Inorg. Biochem. 2009, 103, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Charkoudian, L.K.; Franz, K.J. Fe (III)-coordination properties of neuromelanin components: 5, 6-dihydroxyindole and 5, 6-dihydroxyindole-2-carboxylic acid. Inorg. Chem. 2006, 45, 3657–3664. [Google Scholar] [CrossRef] [PubMed]
- Panicker, C.Y.; Varghese, H.T.; Philip, D.; Nogueira, H.I. FT-IR, FT-Raman and SERS spectra of pyridine-3-sulfonic acid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2006, 64, 744–747. [Google Scholar] [CrossRef]
- Kang, J.S.; Hwang, S.Y.; Lee, C.J.; Lee, M.S. SERS of dithiocarbamate pesticides adsorbed on silver surface; Thiram. Bull. Korean Chem. Soc. 2002, 23, 1604–1610. [Google Scholar]
- Muniz-Miranda, M. On the occurrence of the central line (∼1025 cm−1) in the SERS spectra of pyridine adsorbed on silver hydrosols. Chem. Phys. Lett. 2001, 340, 437–443. [Google Scholar] [CrossRef]
- Capocefalo, A.; Mammucari, D.; Brasili, F.; Fasolato, C.; Bordi, F.; Postorino, P.; Domenici, F. Exploring the potentiality of a SERS-active pH nano-biosensor. Front. Chem. 2019, 7, 413. [Google Scholar] [CrossRef] [Green Version]
- Birke, R.L.; Lombardi, J.R. Simulation of SERS by a DFT study: A comparison of static and near-resonance Raman for 4-mercaptopyridine on small Ag clusters. J. Opt. 2015, 17, 114004. [Google Scholar] [CrossRef]
- Della Pelle, F.; Scroccarello, A.; Scarano, S.; Compagnone, D. Silver nanoparticles-based plasmonic assay for the determination of sugar content in food matrices. Anal. Chim. Acta 2019, 1051, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Sharaf, O.Z.; Taylor, R.A.; Abu-Nada, E. On the colloidal and chemical stability of solar nanofluids: From nanoscale interactions to recent advances. Phys. Rep. 2020, 867, 1–84. [Google Scholar] [CrossRef]
- Liu, Y.; Yue, S.; Wang, Y.N.; Wang, Y.; Xu, Z.R. A multicolor-SERS dual-mode pH sensor based on smart nano-in-micro particles. Sens. Actuators B Chem. 2020, 310, 127889. [Google Scholar] [CrossRef]
- Cai, Y.; Gui, C.; Samedov, K.; Su, H.; Gu, X.; Li, S.; Luo, W.; Sung, H.H.; Lam, J.W.; Kwok, R.T.; et al. An acidic pH independent piperazine–TPE AIEgen as a unique bioprobe for lysosome tracing. Chem. Sci. 2017, 8, 7593–7603. [Google Scholar] [CrossRef] [Green Version]
- Baltazar, G.C.; Guha, S.; Lu, W.; Lim, J.; Boesze-Battaglia, K.; Laties, A.M.; Tyagi, P.; Kompella, U.B.; Mitchell, C.H. Acidic nanoparticles are trafficked to lysosomes and restore an acidic lysosomal pH and degradative function to compromised ARPE-19 cells. PLoS ONE 2012, 7, e49635. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.; Morgan, L.R. Tumor physiology and charge dynamics of anticancer drugs: Implications for camptothecin-based drug development. Curr. Med. Chem. 2011, 18, 1367–1372. [Google Scholar] [CrossRef]
- Sánchez-Cortés, S.; Garcıa-Ramos, L.V. Photoinduced coupling and adsorption of caffeic acid on silver surface studied by surface-enhanced Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 1999, 55, 2935–2941. [Google Scholar] [CrossRef]
- Sánchez-Cortés, S.; Garcıa-Ramos, L.V. FT surface-enhanced Raman evidence of the oxidative condensation reactions of caffeic acid in solution and on silver surface. Appl. Spectrosc. 2000, 54, 230–238. [Google Scholar] [CrossRef]
- Sánchez-Cortés, S.; Francioso, O.; Garcıa-Ramos, J.V.; Ciavatta, C.; Gessa, C. Catechol polymerization in the presence of silver surface. Colloids Surf. A Physicochem. Eng. Asp. 2001, 176, 177–184. [Google Scholar] [CrossRef]
- Zhang, Z.; Kneipp, J. Surface Molecular Patterning by Plasmon-Catalyzed Reactions. ACS Appl. Mater. Interfaces 2021, 13, 43708–43714. [Google Scholar] [CrossRef]
- Stewart, A.; Murray, S.; Bell, S.E.J. Simple preparation of positively charged silver nanoparticles for detection of anions by surface-enhanced Raman spectroscopy. Analyst 2015, 140, 2988–2994. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hernández, I.; Afseth, N.K.; López-Luke, T.; Contreras-Torres, F.F.; Wold, J.P.; Ornelas-Soto, N. Surface enhanced Raman spectroscopy of phenolic antioxidants: A systematic evaluation of ferulic acid, p-coumaric acid, caffeic acid and sinapic acid. Vib. Spectrosc. 2017, 89, 113–122. [Google Scholar] [CrossRef]
- Liu, C.; Hu, J.; Biswas, S.; Zhu, F.; Zhan, J.; Wang, G.; Tung, C.H.; Wang, Y. Surface-enhanced Raman scattering of phenols and catechols by a molecular analogue of titanium dioxide. Anal. Chem. 2020, 92, 5929–5936. [Google Scholar] [CrossRef] [PubMed]
- Cabalo, J.; Guicheteau, J.A.; Christesen, S. Toward understanding the influence of intermolecular interactions and molecular orientation on the chemical enhancement of SERS. J. Phys. Chem. A 2013, 117, 9028–9038. [Google Scholar] [CrossRef]
- Marshall, A.R.L.; Stokes, J.; Viscomi, F.N.; Proctor, J.E.; Gierschner, J.; Bouillard, J.S.G.; Adawi, A.M. Determining molecular orientation via single molecule SERS in a plasmonic nano-gap. Nanoscale 2017, 44, 17415–17421. [Google Scholar] [CrossRef]
- Weese, J. A reliable and fast method for the solution of Fredhol integral equations of the first kind based on Tikhonov regularization Comput. Phys. Commun. 1992, 69, 99–111. [Google Scholar] [CrossRef]
- Johnson, P.B.; Christy, R.W. Optical constants of the noble metals. Phys. Rev. B 1972, 12, 4370. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisini, G.; Scroccarello, A.; Ripanti, F.; Fasolato, C.; Cappelluti, F.; Capocefalo, A.; Della Pelle, F.; Compagnone, D.; Postorino, P. Nanoscale Surface-Enhanced Raman Spectroscopy Investigation of a Polyphenol-Based Plasmonic Nanovector. Nanomaterials 2023, 13, 377. https://doi.org/10.3390/nano13030377
Nisini G, Scroccarello A, Ripanti F, Fasolato C, Cappelluti F, Capocefalo A, Della Pelle F, Compagnone D, Postorino P. Nanoscale Surface-Enhanced Raman Spectroscopy Investigation of a Polyphenol-Based Plasmonic Nanovector. Nanomaterials. 2023; 13(3):377. https://doi.org/10.3390/nano13030377
Chicago/Turabian StyleNisini, Giacomo, Annalisa Scroccarello, Francesca Ripanti, Claudia Fasolato, Francesco Cappelluti, Angela Capocefalo, Flavio Della Pelle, Dario Compagnone, and Paolo Postorino. 2023. "Nanoscale Surface-Enhanced Raman Spectroscopy Investigation of a Polyphenol-Based Plasmonic Nanovector" Nanomaterials 13, no. 3: 377. https://doi.org/10.3390/nano13030377
APA StyleNisini, G., Scroccarello, A., Ripanti, F., Fasolato, C., Cappelluti, F., Capocefalo, A., Della Pelle, F., Compagnone, D., & Postorino, P. (2023). Nanoscale Surface-Enhanced Raman Spectroscopy Investigation of a Polyphenol-Based Plasmonic Nanovector. Nanomaterials, 13(3), 377. https://doi.org/10.3390/nano13030377









