Modified Bacteriophage for Tumor Detection and Targeted Therapy
Abstract
1. Introduction
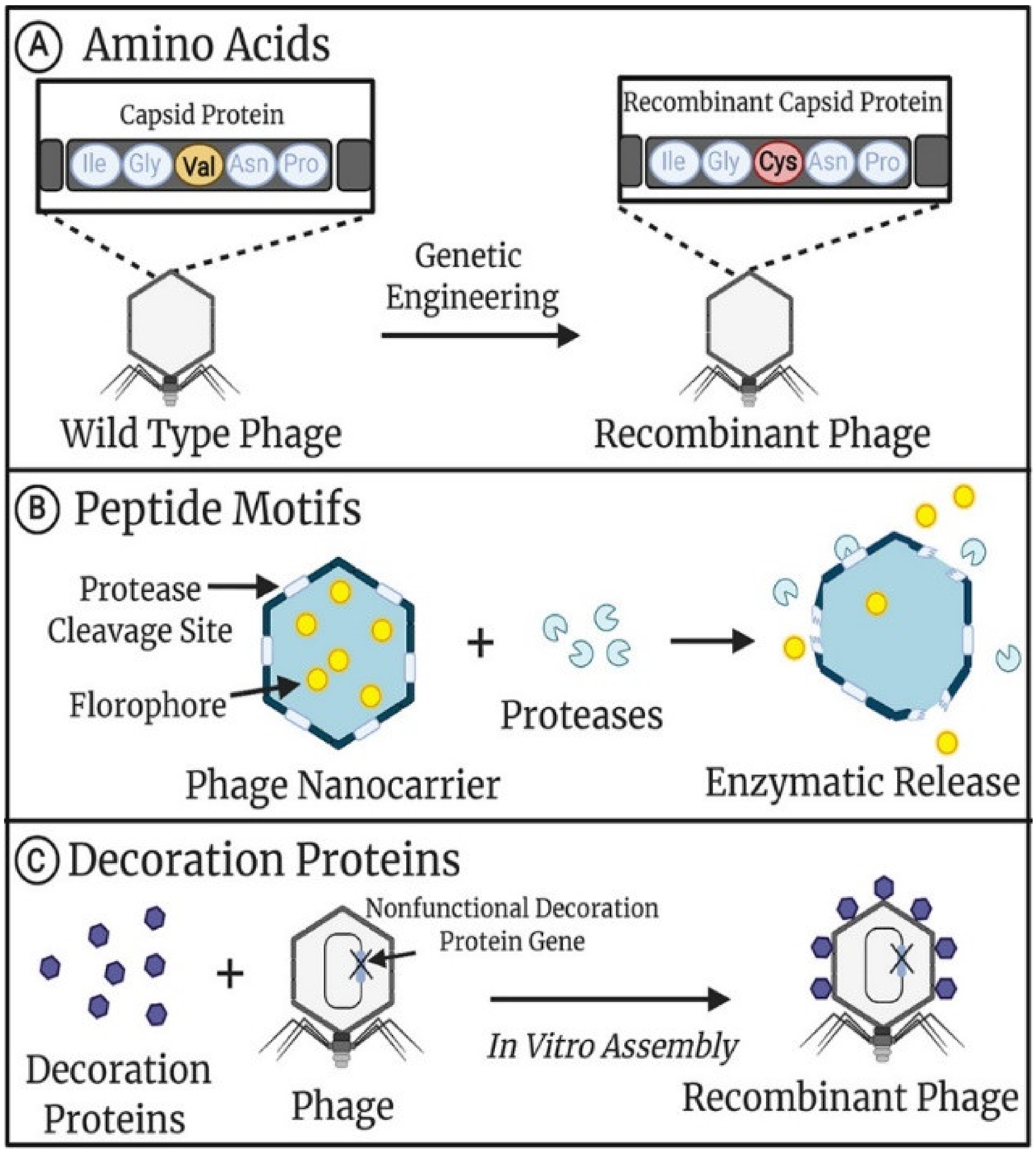
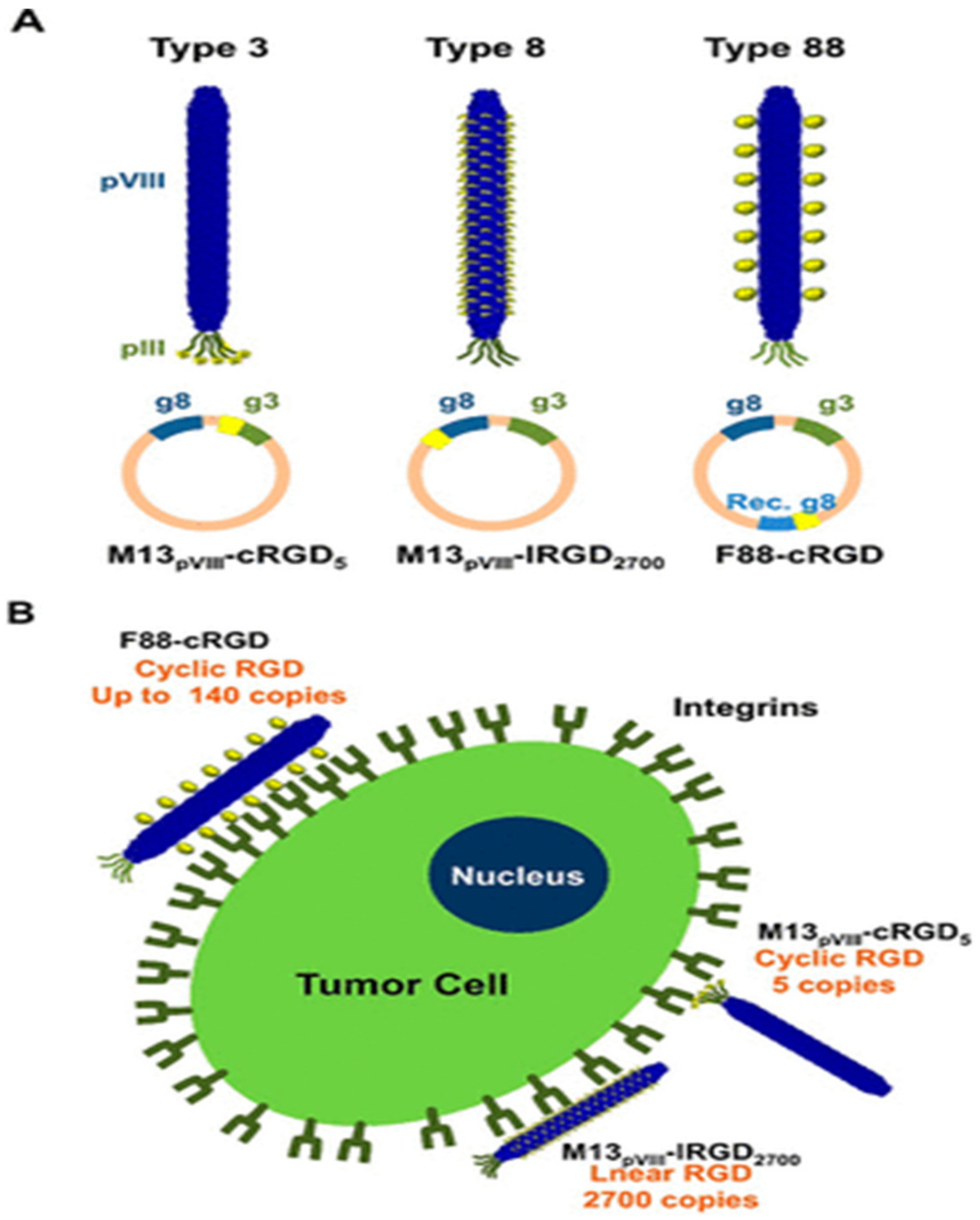
2. Phage Modification
2.1. Genetic Modification of Phage
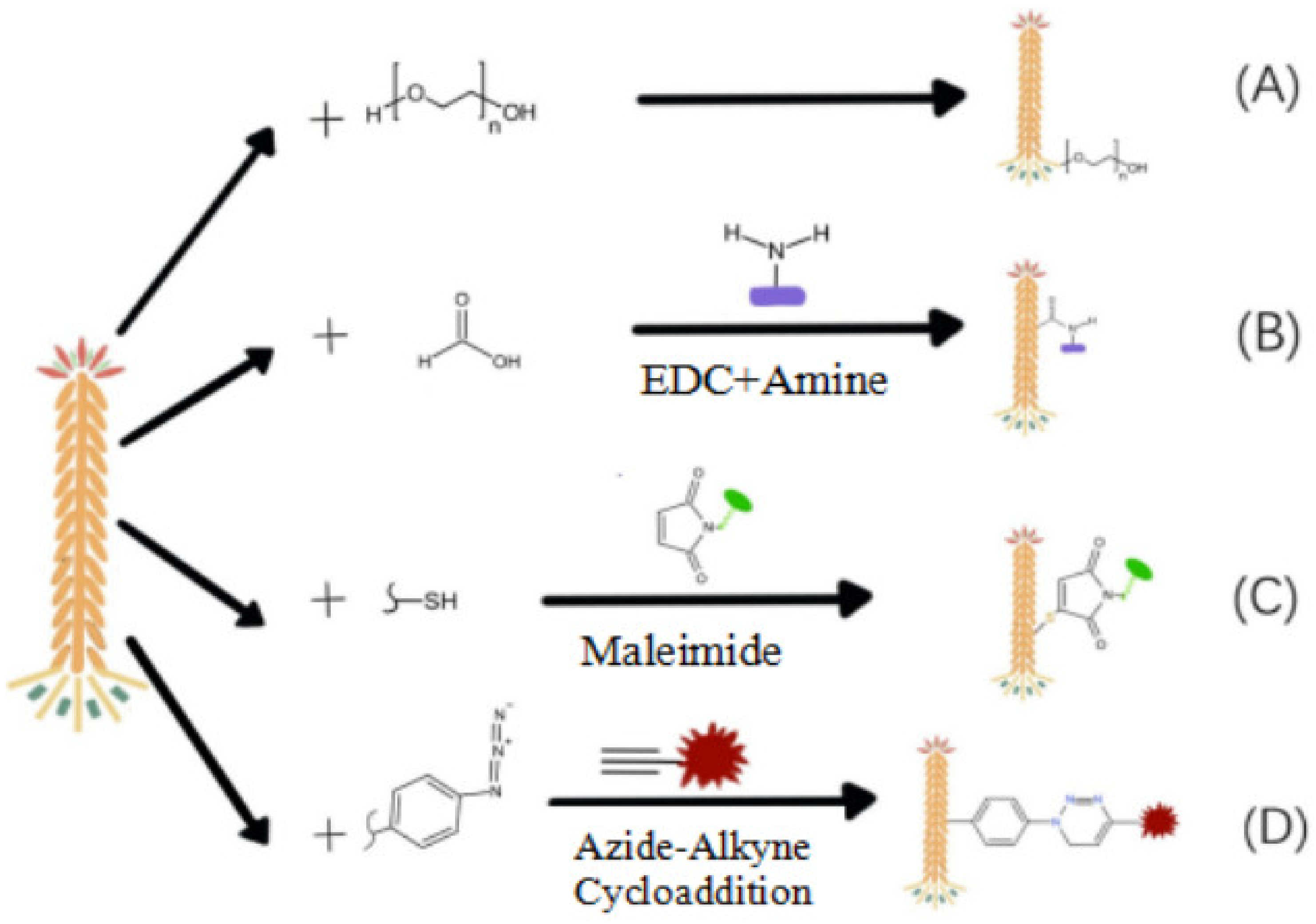
2.2. Chemical Modification of Phage
3. Modified Phage for Tumor Diagnosis
3.1. Tumor Diagnosis with Nanoparticles Modified Phage
3.2. Visual Diagnosis of Tumor Cell with Molecular Imaging Agent Modified Phage
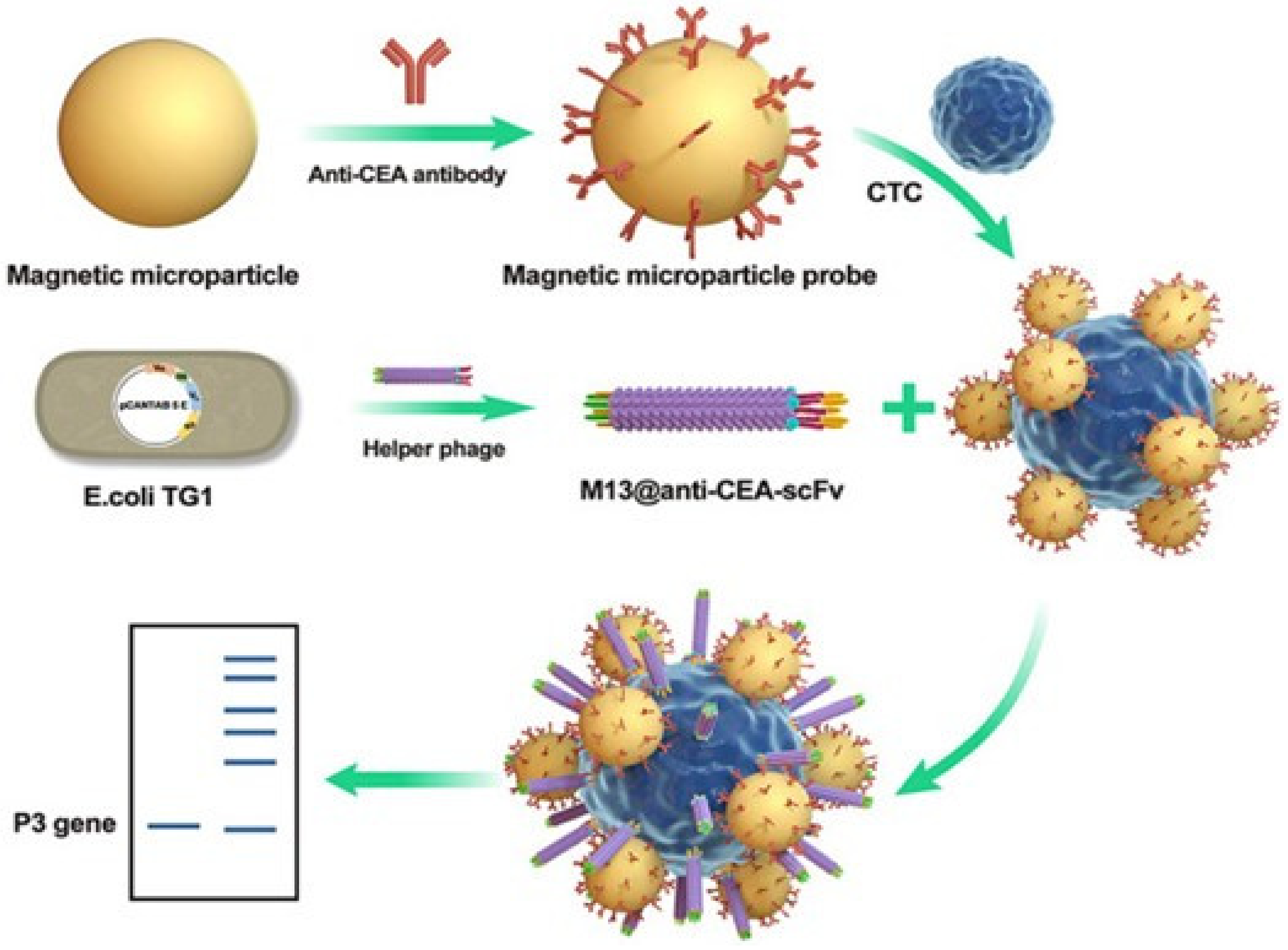
3.3. Tumor Diagnosis with Phage-Based Immuno-PCR
4. Modified Phage for Tumor Therapy
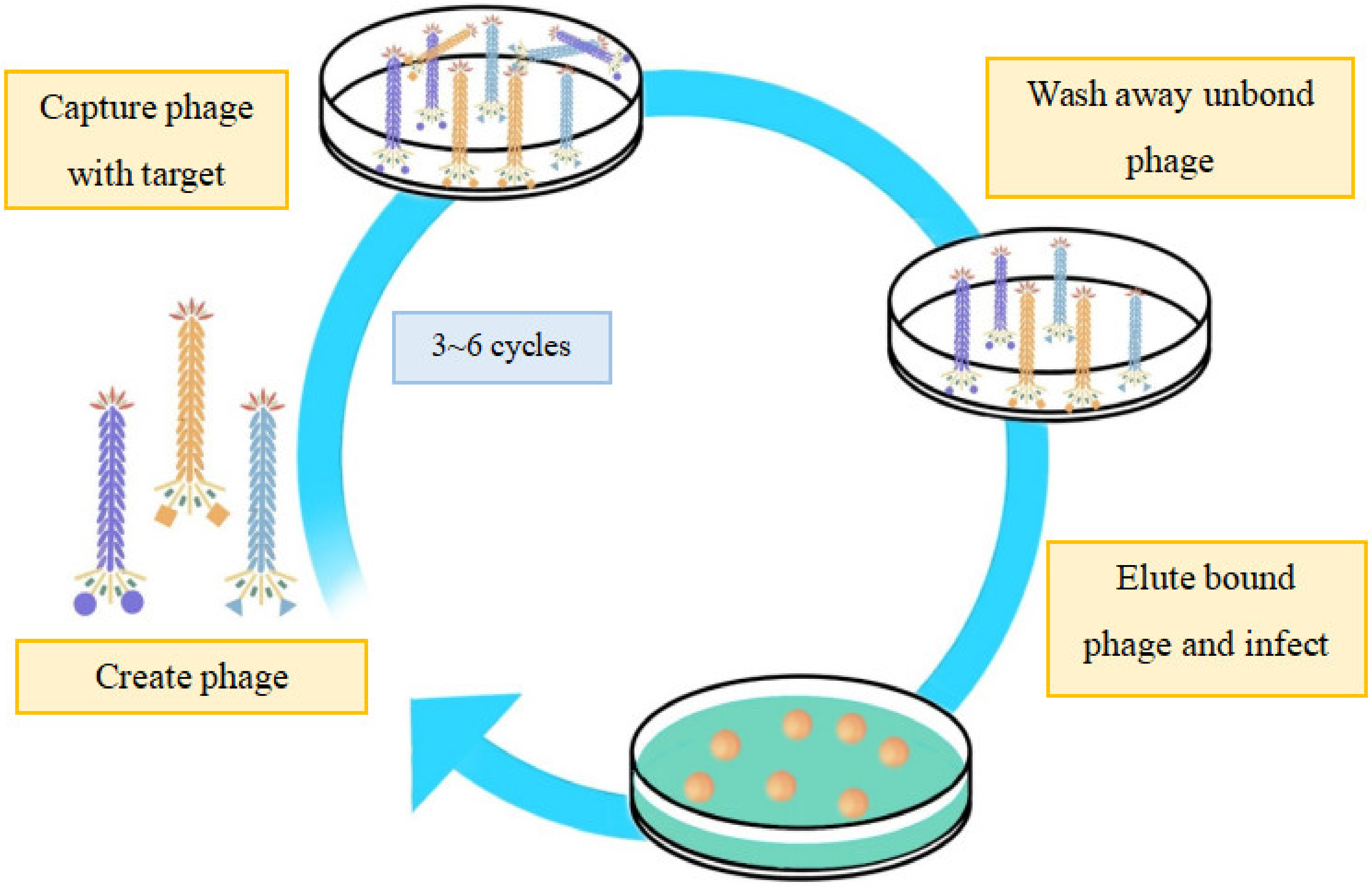
4.1. Screening of Novel Ligands and Short Peptides as Tumor-Targeting Drugs
4.2. Validation of Specific Tumor-Targeting Antibodies through Phage Display Technology
4.3. Targeted Delivery Vehicle for Chemotherapy Drugs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Tumor Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Tumors in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar]
- Roy, P.S.; Saikia, B.J. Tumor and Cure: A Critical Analysis. Indian J. Tumor 2016, 53, 441–442. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Weingart, S.N.; Zhang, L.; Sweeney, M.; Hassett, M. Chemotherapy medication errors. Lancet Oncol. 2018, 19, e191–e199. [Google Scholar]
- Behranvand, N.; Nasri, F.; Zolfaghari Emameh, R.; Khani, P.; Hosseini, A.; Garssen, J.; Falak, R. Chemotherapy: A Double-Edged Sword in Tumor Treatment. Tumor Immunol. Immunother. 2022, 71, 507–526. [Google Scholar]
- Connors, T.A.; Knox, R.J. Prodrugs in Tumor Chemotherapy. Stem. Cells 1995, 13, 501–511. [Google Scholar]
- Paczesny, J.; Bielec, K. Application of Bacteriophages in Nanotechnology. Nanomaterials 2020, 10, 1944. [Google Scholar]
- Abbaszadeh, F.; Leylabadlo, H.E.; Alinezhad, F.; Feizi, H.; Mobed, A.; Baghbanijavid, S.; Baghi, H.B. Bacteriophages: Cancer diagnosis, treatment, and future prospects. J. Pharm. Investig. 2021, 51, 23–34. [Google Scholar]
- Goracci, M.; Pignochino, Y.; Marchiò, S. Phage Display-Based Nanotechnology Applications in Tumor Immunotherapy. Molecules 2020, 25, E843. [Google Scholar]
- Weiss, G.A.; Sidhu, S.S. Design and Evolution of Artificial M13 Coat Proteins. J. Mol. Biol. 2000, 300, 213–219. [Google Scholar]
- Ackermann, H.-W. 5500 Phages Examined in the Electron Microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage Display. Chem. Rev. 1997, 97, 391–410. [Google Scholar]
- Karimi, M.; Mirshekari, H.; Moosavi Basri, S.M.; Bahrami, S.; Moghoofei, M.; Hamblin, M.R. Bacteriophages and Phage-Inspired Nanocarriers for Targeted Delivery of Therapeutic Cargos. Adv. Drug Deliv. Rev. 2016, 106, 45–62. [Google Scholar]
- Carmody, C.M.; Goddard, J.M.; Nugen, S.R. Bacteriophage Capsid Modification by Genetic and Chemical Methods. Bioconjug. Chem. 2021, 32, 466–481. [Google Scholar] [CrossRef]
- Pines, G.; Freed, E.F.; Winkler, J.D.; Gill, R.T. Bacterial Recombineering: Genome Engineering via Phage-Based Homologous Recombination. ACS Synth. Biol. 2015, 4, 1176–1185. [Google Scholar] [CrossRef]
- Hoess, R.H.; Ziese, M.; Sternberg, N. P1 Site-Specific Recombination: Nucleotide Sequence of the Recombining Sites. Proc. Natl. Acad. Sci. USA 1982, 79, 3398–3402. [Google Scholar]
- Mahichi, F.; Synnott, A.J.; Yamamichi, K.; Osada, T.; Tanji, Y. Site-Specific Recombination of T2 Phage Using IP008 Long Tail Fiber Genes Provides a Targeted Method for Expanding Host Range While Retaining Lytic Activity. FEMS Microbiol. Lett. 2009, 295, 211–217. [Google Scholar]
- Marinelli, L.J.; Piuri, M.; Swigonová, Z.; Balachandran, A.; Oldfield, L.M.; van Kessel, J.C.; Hatfull, G.F. BRED: A Simple and Powerful Tool for Constructing Mutant and Recombinant Bacteriophage Genomes. PLoS ONE 2008, 3, e3957. [Google Scholar] [CrossRef]
- Richardson, C.D.; Ray, G.J.; DeWitt, M.A.; Curie, G.L.; Corn, J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016, 34, 339–344. [Google Scholar] [CrossRef]
- Haque, M.E.; Khan, F.; Chi, L.; Gurung, S.; Vadevoo, S.M.P.; Park, R.-W.; Kim, D.-K.; Kim, S.K.; Lee, B. A Phage Display-Identified Peptide Selectively Binds to Kidney Injury Molecule-1 (KIM-1) and Detects KIM-1-Overexpressing Tumors in Vivo. Cancer. Res. Treat. 2019, 51, 861–875. [Google Scholar] [CrossRef]
- Ran, Y.; Hu, H.; Zhou, Z.; Yu, L.; Sun, L.; Pan, J.; Liu, J.; Yang, Z. Profiling Tumor-Associated Autoantibodies for the Detection of Colon Tumor. Clin. Tumor Res. 2008, 14, 2696–2700. [Google Scholar]
- Wu, L.; Huang, T.; Yang, L.; Pan, J.; Zhu, S.; Yan, X. Sensitive and selective bacterial detection using tetracysteine-tagged phages in conjunction with biarsenical dye. Angew Chem. Int. Ed. Engl. 2011, 50, 5873–5877. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar]
- Kiro, R.; Shitrit, D.; Qimron, U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014, 11, 42–44. [Google Scholar]
- Lemay, M.L.; Horvath, P.; Moineau, S. The CRISPR-Cas app goes viral. Curr. Opin. Microbiol. 2017, 37, 103–109. [Google Scholar]
- Karikó, K. In vitro-Transcribed mRNA Therapeutics: Out of the Shadows and Into the Spotlight. Mol. Ther. 2019, 27, 691–692. [Google Scholar] [CrossRef]
- Chiem, K.; Lorenzo, M.M.; Rangel-Moreno, J.; Garcia-Hernandez, M.L.; Park, J.G.; Nogales, A.; Blasco, R.; Martínez-Sobrido, L. Bi-Reporter Vaccinia Virus for Tracking Viral Infections In Vitro and In Vivo. Microbiol. Spectr. 2021, 9, e0160121. [Google Scholar] [CrossRef]
- Kang, S.; Uchida, M.; O’Neil, A.; Li, R.; Prevelige, P.E.; Douglas, T. Implementation of P22 Viral Capsids as Nanoplatforms. Biomacromolecules 2010, 11, 2804–2809. [Google Scholar] [CrossRef]
- Tridgett, M.; Lloyd, J.R.; Kennefick, J.; Moore-Kelly, C.; Dafforn, T.R. Mutation of M13 Bacteriophage Major Coat Protein for Increased Conjugation to Exogenous Compounds. Bioconjug. Chem. 2018, 29, 1872–1875. [Google Scholar]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A Million Peptide Motifs for the Molecular Biologist. Mol. Cell 2014, 55, 161–169. [Google Scholar]
- Oślizło, A.; Miernikiewicz, P.; Piotrowicz, A.; Owczarek, B.; Kopciuch, A.; Figura, G.; Dąbrowska, K. Purification of Phage Display-Modified Bacteriophage T4 by Affinity Chromatography. BMC Biotechnol. 2011, 11, 59. [Google Scholar]
- Shivachandra, S.B.; Rao, M.; Janosi, L.; Sathaliyawala, T.; Matyas, G.R.; Alving, C.R.; Leppla, S.H.; Rao, V.B. In Vitro Binding of Anthrax Protective Antigen on Bacteriophage T4 Capsid Surface through Hoc-Capsid Interactions: A Strategy for Efficient Display of Large Full-Length Proteins. Virology 2006, 345, 190–198. [Google Scholar]
- Grimsley, G.R.; Scholtz, J.M.; Pace, C.N. A Summary of the Measured PK Values of the Ionizable Groups in Folded Proteins. Protein Sci. 2009, 18, 247–251. [Google Scholar]
- Chan, A.O.-Y.; Ho, C.-M.; Chong, H.-C.; Leung, Y.-C.; Huang, J.-S.; Wong, M.-K.; Che, C.-M. Modification of N-Terminal α-Amino Groups of Peptides and Proteins Using Ketenes. J. Am. Chem. Soc. 2012, 134, 2589–2598. [Google Scholar]
- Zhao, X.; Cai, L.; Adogla, E.A.; Guan, H.; Lin, Y.; Wang, Q. Labeling of Enveloped Virus via Metabolic Incorporation of Azido Sugars. Bioconjug. Chem. 2015, 26, 1868–1872. [Google Scholar] [CrossRef]
- Huang, L.-L.; Lu, G.-H.; Hao, J.; Wang, H.; Yin, D.-L.; Xie, H.-Y. Enveloped Virus Labeling via Both Intrinsic Biosynthesis and Metabolic Incorporation of Phospholipids in Host Cells. Anal. Chem. 2013, 85, 5263–5270. [Google Scholar]
- Huang, L.-L.; Liu, K.; Zhang, Q.; Xu, J.; Zhao, D.; Zhu, H.; Xie, H.-Y. Integrating Two Efficient and Specific Bioorthogonal Ligation Reactions with Natural Metabolic Incorporation in One Cell for Virus Dual Labeling. Anal. Chem. 2017, 89, 11620–11627. [Google Scholar]
- Jin, X.; Newton, J.R.; Montgomery-Smith, S.; Smith, G. A Generalized Kinetic Model for Amine Modification of Proteins with Application to Phage Display. Biotechniques 2009, 46, 175–182. [Google Scholar]
- Zwick, M.B.; Shen, J.; Scott, J.K. Homodimeric Peptides Displayed by the Major Coat Protein of Filamentous Phage. J. Mol. Biol. 2000, 300, 307–320. [Google Scholar]
- Jencks, W.P. Studies on the Mechanism of Oxime and Semicarbazone Formation. In Organic Functional Group Analysis; Elsevier: Amsterdam, The Netherlands, 1968; pp. 120–130. [Google Scholar]
- Chen, F.J.; Zheng, M.; Nobile, V.; Gao, J. Fast and Cysteine-Specific Modification of Peptides, Proteins and Bacteriophage Using Chlorooximes. Chemistry 2022, 28, e202200058. [Google Scholar]
- Bar, H.; Yacoby, I.; Benhar, I. Killing cancer cells by targeted drug-carrying phage nanomedicines. BMC Biotechnol. 2008, 8, 37. [Google Scholar] [CrossRef]
- Niu, Z.; Bruckman, M.A.; Harp, B.; Mello, C.M.; Wang, Q. Bacteriophage M13 as a Scaffold for Preparing Conductive Polymeric Composite Fibers. Nano. Res. 2008, 1, 235–241. [Google Scholar]
- Korkmaz, N. Recombinant Bacteriophages as Gold Binding Bio-Templates. Colloids Surf. B Biointerfaces 2013, 112, 219–228. [Google Scholar] [CrossRef]
- Peng, H.; Borg, R.E.; Dow, L.P.; Pruitt, B.L.; Chen, I.A. Controlled Phage Therapy by Photothermal Ablation of Specific Bacterial Species Using Gold Nanorods Targeted by Chimeric Phages. Proc. Natl. Acad. Sci. USA 2020, 117, 1951–1961. [Google Scholar] [CrossRef]
- Avrameas, S.; Ternynck, T. The Cross-Linking of Proteins with Glutaraldehyde and Its Use for the Preparation of Immunoadsorbents. Immunochemistry 1969, 6, 53–66. [Google Scholar] [CrossRef]
- Hopwood, D.; Callen, C.R.; McCabe, M. The Reactions between Glutaraldehyde and Various Proteins. An Investigation of Their Kinetics. Histochem. J. 1970, 2, 137–150. [Google Scholar] [CrossRef]
- Kitov, P.I.; Vinals, D.F.; Ng, S.; Tjhung, K.F.; Derda, R. Rapid, Hydrolytically Stable Modification of Aldehyde-Terminated Proteins and Phage Libraries. J. Am. Chem. Soc. 2014, 136, 8149–8152. [Google Scholar]
- Sandman, K.E.; Benner, J.S.; Noren, C.J. Phage Display of Selenopeptides. J. Am. Chem. Soc. 2000, 122, 960–961. [Google Scholar]
- Chen, S.; Lovell, S.; Lee, S.; Fellner, M.; Mace, P.D.; Bogyo, M. Identification of Highly Selective Covalent Inhibitors by Phage Display. Nat. Biotechnol. 2021, 39, 490–498. [Google Scholar] [CrossRef]
- Zheng, M.; Chen, F.-J.; Li, K.; Reja, R.M.; Haeffner, F.; Gao, J. Lysine-Targeted Reversible Covalent Ligand Discovery for Proteins via Phage Display. J. Am. Chem. Soc. 2022, 144, 15885–15893. [Google Scholar] [CrossRef]
- Tookmanian, E.M.; Fenlon, E.E.; Brewer, S.H. Synthesis and Protein Incorporation of Azido-Modified Unnatural Amino Acids. RSC. Adv. 2015, 5, 1274–1281. [Google Scholar] [CrossRef]
- Mukai, Y.; Yoshioka, Y.; Tsutsumi, Y. Phage Display and PEGylation of Therapeutic Proteins. Comb. Chem. High. Throughput. Screen. 2005, 8, 145–152. [Google Scholar] [CrossRef]
- Özdemir, C.; Güner, A. Solubility profiles of poly(ethylene glycol)/solvent systems, i: Qualitative comparison of solubility parameter approaches. Eur. Polym. J. 2007, 43, 3068–3093. [Google Scholar] [CrossRef]
- Bugelski, P.J.; Capocasale, R.J.; Makropoulos, D.; Marshall, D.; Fisher, P.W.; Lu, J.; Achuthanandam, R.; Spinka-Doms, T.; Kwok, D.; Graden, D.; et al. CNTO 530: Molecular Pharmacology in Human UT-7EPO Cells and Pharmacokinetics and Pharmacodynamics in Mice. J. Biotechnol. 2008, 134, 171–180. [Google Scholar]
- Ning, L.; He, B.; Zhou, P.; Derda, R.; Huang, J. Molecular Design of Peptide-Fc Fusion Drugs. Curr. Drug. Metab. 2019, 20, 203–208. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Song, S.; Yin, L.; Sun, D.; Gu, J. Recent Advances in the Bioanalytical Methods of Polyethylene Glycols and PEGylated Pharmaceuticals. J. Sep. Sci. 2020, 43, 1978–1997. [Google Scholar] [CrossRef]
- Tagliavini, V.; Honisch, C.; Serratì, S.; Azzariti, A.; Bonchio, M.; Ruzza, P.; Carraro, M. Enhancing the Biological Activity of Polyoxometalate-Peptide Nano-Fibrils by Spacer Design. RSC. Adv. 2021, 11, 4952–4957. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Tsutsumi, Y.; Yoshioka, Y.; Nishibata, T.; Kobayashi, K.; Okamoto, T.; Mukai, Y.; Shimizu, T.; Nakagawa, S.; Nagata, S.; et al. Site-Specific PEGylation of a Lysine-Deficient TNF-Alpha with Full Bioactivity. Nat. Biotechnol. 2003, 21, 546–552. [Google Scholar] [CrossRef]
- Kim, K.-P.; Cha, J.-D.; Jang, E.-H.; Klumpp, J.; Hagens, S.; Hardt, W.-D.; Lee, K.-Y.; Loessner, M.J. PEGylation of Bacteriophages Increases Blood Circulation Time and Reduces T-Helper Type 1 Immune Response. Microb. Biotechnol. 2008, 1, 247–257. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Gao, J.; Ouyang, H.; He, Y.; Fu, Z. PEGylated Ni Single-Atom Catalysts as Ultrasensitive Electrochemiluminescent Probes with Favorable Aqueous Dispersibility for Assaying Drug-Resistant Pathogens. Anal. Chem. 2022, 94, 14047–14053. [Google Scholar] [CrossRef]
- Wang, X.; Yang, T.; Zhang, X.; Chen, M.; Wang, J. In situ growth of gold nanoparticles on Hg2+-binding M13 phages for mercury sensing. Nanoscale 2017, 9, 16728–16734. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yang, T.; Wang, S.Y.; Du, K.D.; Chen, M.L.; Wang, J.H. M13 phage as network frame for the quantification of Pb2+ based on the Pb2+-induced in-situ growth of gold nanoparticles. Anal. Chim. Acta. 2019, 1073, 72–78. [Google Scholar]
- Ringgaard, L.; Melander, F.; Eliasen, R.; Henriksen, J.R.; Jølck, R.I.; Engel, T.B.; Bak, M.; Fliedner, F.P.; Kristensen, K.; Elema, D.R.; et al. Tumor Repolarization by an Advanced Liposomal Drug Delivery System Provides a Potent New Approach for Chemo-Immunotherapy. Sci. Adv. 2020, 6, eaba5628. [Google Scholar] [CrossRef]
- Chen, F.; Tang, F.; Yang, C.-T.; Zhao, X.; Wang, J.; Thierry, B.; Bansal, V.; Dai, J.; Zhou, X. Fast and Highly Sensitive Detection of Pathogens Wreathed with Magnetic Nanoparticles Using Dark-Field Microscopy. ACS Sens. 2018, 3, 2175–2181. [Google Scholar]
- Xiao, L.; Wei, L.; He, Y.; Yeung, E.S. Single Molecule Biosensing Using Color Coded Plasmon Resonant Metal Nanoparticles. Anal. Chem. 2010, 82, 6308–6314. [Google Scholar]
- Hu, L.Y.; Kelly, K.A.; Sutcliffe, J.L. High-Throughput Approaches to the Development of Molecular Imaging Agents. Mol. Imaging. Biol. 2017, 19, 163–182. [Google Scholar]
- Yang, A.; Yang, L.; Liu, W.; Li, Z.; Xu, H.; Yang, X. Tumor Necrosis Factor Alpha Blocking Peptide Loaded PEG-PLGA Nanoparticles: Preparation and in Vitro Evaluation. Int. J. Pharm. 2007, 331, 123–132. [Google Scholar]
- Li, K.; Chen, Y.; Li, S.; Nguyen, H.G.; Niu, Z.; You, S.; Mello, C.M.; Lu, X.; Wang, Q. Chemical Modification of M13 Bacteriophage and Its Application in Tumor Cell Imaging. Bioconjug. Chem. 2010, 21, 1369–1377. [Google Scholar] [CrossRef]
- Maisano, D.; Mimmi, S.; Dattilo, V.; Marino, F.; Gentile, M.; Vecchio, E.; Fiume, G.; Nisticò, N.; Aloisio, A.; de Santo, M.P.; et al. A novel phage display based platform for exosome diversity characterization. Nanoscale 2022, 14, 2998–3003. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, S.; Niu, G.; Chen, K.; Yan, Y.; Liu, Z.; Chen, X. Phage display peptide probes for imaging early response to bevacizumab treatment. Amino Acids 2011, 41, 1103–1112. [Google Scholar] [CrossRef]
- Lomakin, Y.A.; Kaminskaya, A.N.; Stepanov, A.V.; Shmidt, A.A.; Gabibov, A.G.; Belogurov, A.A. Probing Surface Membrane Receptors Using Engineered Bacteriophage Bioconjugates. Bioconjug. Chem. 2019, 30, 1500–1506. [Google Scholar]
- Ishina, I.A.; Filimonova, I.N.; Zakharova, M.Y.; Ovchinnikova, L.A.; Mamedov, A.E.; Lomakin, Y.A.; Belogurov, A.A. Exhaustive Search of the Receptor Ligands by the CyCLOPS (Cytometry Cell-Labeling Operable Phage Screening) Technique. Int. J. Mol. Sci. 2020, 21, E6258. [Google Scholar]
- Morley, A.A. Digital PCR: A Brief History. Biomol. Detect. Quantif. 2014, 1, 1–2. [Google Scholar]
- Kanagal-Shamanna, R. Digital PCR: Principles and Applications. Methods. Mol. Biol. 2016, 1392, 43–50. [Google Scholar]
- Niemeyer, C.M.; Adler, M.; Wacker, R. Immuno-PCR: High Sensitivity Detection of Proteins by Nucleic Acid Amplification. Trends Biotechnol. 2005, 23, 208–216. [Google Scholar]
- Brasino, M.; Cha, J.N. Real-Time Femtomolar Detection of Tumor Biomarkers from Photoconjugated Antibody-Phage Constructs. Analyst 2016, 142, 91–97. [Google Scholar] [CrossRef]
- Kim, H.-J.; McCoy, M.; Gee, S.J.; González-Sapienza, G.G.; Hammock, B.D. Noncompetitive Phage Anti-Immunocomplex Real-Time Polymerase Chain Reaction for Sensitive Detection of Small Molecules. Anal. Chem. 2011, 83, 246–253. [Google Scholar]
- Hou, J.; Shen, J.; Zhao, N.; Yang, C.-T.; Thierry, B.; Zhou, X.; Zhu, J.; Mao, C. Detection of a Single Circulating Tumor Cell Using a Genetically Engineered Antibody-like Phage Nanofiber Probe. Mater. Today Adv. 2021, 12, 100168. [Google Scholar] [CrossRef]
- Manivannan, A.C.; Dhandapani, R.; Velmurugan, P.; Thangavelu, S.; Paramasivam, R.; Ragunathan, L.; Saravanan, M. Phage in Tumor Treatment—Biology of Therapeutic Phage and Screening of Tumor Targeting Peptide. Expert Opin. Drug. Deliv. 2022, 19, 873–882. [Google Scholar] [CrossRef]
- Dmitrieva, M.D.; Voitova, A.A.; Dymova, M.A.; Richter, V.A.; Kuligina, E.V. Tumor-Targeting Peptides Search Strategy for the Delivery of Therapeutic and Diagnostic Molecules to Tumor Cells. Int. J. Mol. Sci. 2020, 22, E314. [Google Scholar]
- Wu, L.P.; Ahmadvand, D.; Su, J.; Hall, A.; Tan, X.; Farhangrazi, Z.S.; Moghimi, S.M. Crossing the Blood-Brain-Barrier with Nanoligand Drug Carriers Self-Assembled from a Phage Display Peptide. Nat. Commun. 2019, 10, 4635. [Google Scholar] [CrossRef]
- Saw, P.E.; Song, E.-W. Phage Display Screening of Therapeutic Peptide for Tumor Targeting and Therapy. Protein Cell 2019, 10, 787–807. [Google Scholar] [CrossRef]
- Veeranarayanan, S.; Azam, A.H.; Kiga, K.; Watanabe, S.; Cui, L. Bacteriophages as Solid Tumor Theragnostic Agents. Int. J. Mol. Sci. 2021, 23, 402. [Google Scholar]
- Pranjol, M.Z.I.; Hajitou, A. Bacteriophage-Derived Vectors for Targeted Tumor Gene Therapy. Viruses 2015, 7, 268–284. [Google Scholar]
- Aquino, C.; Sarkar, M. One Bead-One Compound (OBOC) Peptidomimetic-Encoded Library Synthesis via Split-and-Pool Methods. Methods Mol. Biol. 2022, 2541, 105–120. [Google Scholar]
- Houghten, R.A.; Pinilla, C.; Blondelle, S.E.; Appel, J.R.; Dooley, C.T.; Cuervo, J.H. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature 1991, 354, 84–86. [Google Scholar] [CrossRef]
- Lam, K.S.; Sroka, T.; Chen, M.L.; Zhao, Y.; Lou, Q.; Wu, J.; Zhao, Z.G. Application of “One-Bead One-Compound” Combinatorial Library Methods in Signal Transduction Research. Life Sci. 1998, 62, 1577–1583. [Google Scholar]
- Lam, K.S.; Salmon, S.E.; Hersh, E.M.; Hruby, V.J.; Kazmierski, W.M.; Knapp, R.J. A New Type of Synthetic Peptide Library for Identifying Ligand-Binding Activity. Nature 1991, 354, 82–84. [Google Scholar]
- Aina, O.H.; Liu, R.; Sutcliffe, J.L.; Marik, J.; Pan, C.-X.; Lam, K.S. From Combinatorial Chemistry to Tumor-Targeting Peptides. Mol. Pharm. 2007, 4, 631–651. [Google Scholar]
- Liu, R.; Marik, J.; Lam, K.S. A Novel Peptide-Based Encoding System for “One-Bead One-Compound” Peptidomimetic and Small Molecule Combinatorial Libraries. J. Am. Chem. Soc. 2002, 124, 7678–7680. [Google Scholar] [CrossRef]
- Hao, D.; Ma, B.; He, C.; Liu, R.; Farmer, D.L.; Lam, K.S.; Wang, A. Surface modification of polymeric electrospun scaffolds via a potent and high-affinity integrin α4β1 ligand improved the adhesion, spreading and survival of human chorionic villus-derived mesenchymal stem cells: A new insight for fetal tissue engineering. J. Mater. Chem. B 2020, 8, 1649–1659. [Google Scholar]
- Tanaka, S.; Sugimachi, K.; Yamashita, Y.I.; Ohga, T.; Shirabe, K.; Shimada, M.; Wands, J.R.; Sugimachi, K. Tie2 vascular endothelial receptor expression and function in hepatocellular carcinoma. Hepatology 2002, 35, 861–867. [Google Scholar]
- Wu, X.; Li, Z.; Yao, M.; Wang, H.; Qu, S.; Chen, X.; Li, J.; Sun, Y.; Xu, Y.; Gu, J. Identification and characterization of a novel peptide ligand of Tie2 for targeting gene therapy. Acta Biochim. Biophys. Sin. 2008, 40, 217–225. [Google Scholar] [CrossRef]
- Ayat, H.; Burrone, O.R.; Sadghizadeh, M.; Jahanzad, E.; Rastgou, N.; Moghadasi, S.; Arbabi, M. Isolation of ScFv Antibody Fragments against HER2 and CEA Tumor Antigens from Combinatorial Antibody Libraries Derived from Tumor Patients. Biologicals 2013, 41, 345–354. [Google Scholar]
- Lin, H.; Zhang, H.; Wang, J.; Lu, M.; Zheng, F.; Wang, C.; Tang, X.; Xu, N.; Chen, R.; Zhang, D.; et al. A Novel Human Fab Antibody for Trop2 Inhibits Breast Tumor Growth in Vitro and in Vivo. Int. J. Tumor 2014, 134, 1239–1249. [Google Scholar]
- Han, L.; Xia, H.; Yin, L.; Petrenko, V.A.; Liu, A. Selected landscape phage probe as selective recognition interface for sensitive total prostate-specific antigen immunosensor. Biosens. Bioelectron. 2018, 106, 1–6. [Google Scholar]
- Frenzel, A.; Schirrmann, T.; Hust, M. Phage Display-Derived Human Antibodies in Clinical Development and Therapy. MAbs 2016, 8, 1177–1194. [Google Scholar]
- Kumar, R.; Parray, H.A.; Shrivastava, T.; Sinha, S.; Luthra, K. Phage Display Antibody Libraries: A Robust Approach for Generation of Recombinant Human Monoclonal Antibodies. Int. J. Biol. Macromol. 2019, 135, 907–918. [Google Scholar]
- Luqmani, Y.A. Mechanisms of Drug Resistance in Tumor Chemotherapy. Med. Princ. Pract. 2005, 14, 35–48. [Google Scholar]
- Hamad, I.; Moghimi, S.M. Critical Issues in Site-Specific Targeting of Solid Tumours: The Carrier, the Tumour Barriers and the Bioavailable Drug. Expert Opin. Drug Deliv. 2008, 5, 205–219. [Google Scholar]
- Petrenko, V.A.; Jayanna, P.K. Phage Protein-Targeted Tumor Nanomedicines. FEBS Lett. 2014, 588, 341–349. [Google Scholar]
- Loi, M.; Di Paolo, D.; Soster, M.; Brignole, C.; Bartolini, A.; Emionite, L.; Sun, J.; Becherini, P.; Curnis, F.; Petretto, A.; et al. Novel Phage Display-Derived Neuroblastoma-Targeting Peptides Potentiate the Effect of Drug Nanocarriers in Preclinical Settings. J. Control Release 2013, 170, 233–241. [Google Scholar]
- Choi, D.S.; Jin, H.-E.; Yoo, S.Y.; Lee, S.-W. Cyclic RGD Peptide Incorporation on Phage Major Coat Proteins for Improved Internalization by HeLa Cells. Bioconjug. Chem. 2014, 25, 216–223. [Google Scholar]
- Nagano, K.; Tsutsumi, Y. Development of Novel Drug Delivery Systems Using Phage Display Technology for Clinical Application of Protein Drugs. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2016, 92, 156–166. [Google Scholar]
- Hölig, P.; Bach, M.; Völkel, T.; Nahde, T.; Hoffmann, S.; Müller, R.; Kontermann, R.E. Novel RGD Lipopeptides for the Targeting of Liposomes to Integrin-Expressing Endothelial and Melanoma Cells. Protein. Eng. Des. Sel. 2004, 17, 433–441. [Google Scholar] [CrossRef]
- Wang, T.; D’Souza, G.G.M.; Bedi, D.; Fagbohun, O.A.; Potturi, L.P.; Papahadjopoulos-Sternberg, B.; Petrenko, V.A.; Torchilin, V.P. Enhanced Binding and Killing of Target Tumor Cells by Drug-Loaded Liposomes Modified with Tumor-Specific Phage Fusion Coat Protein. Nanomedicine 2010, 5, 563–574. [Google Scholar] [CrossRef]
- Mudd, G.E.; Scott, H.; Chen, L.; van Rietschoten, K.; Ivanova-Berndt, G.; Dzionek, K.; Brown, A.; Watcham, S.; White, L.; Park, P.U.; et al. Discovery of BT8009: A Nectin-4 Targeting Bicycle Toxin Conjugate for the Treatment of Tumor. J. Med. Chem. 2022, 65, 14337–14347. [Google Scholar] [CrossRef]
- Zhu, S.; Qian, L.; Hong, M.; Zhang, L.; Pei, Y.; Jiang, Y. RGD-Modified PEG-PAMAM-DOX Conjugate: In Vitro and in Vivo Targeting to Both Tumor Neovascular Endothelial Cells and Tumor Cells. Adv. Mater. 2011, 23, H84–H89. [Google Scholar]
- Longo, N.; Harding, C.O.; Burton, B.K.; Grange, D.K.; Vockley, J.; Wasserstein, M.; Rice, G.M.; Dorenbaum, A.; Neuenburg, J.K.; Musson, D.G.; et al. Single-Dose, Subcutaneous Recombinant Phenylalanine Ammonia Lyase Conjugated with Polyethylene Glycol in Adult Patients with Phenylketonuria: An Open-Label, Multicentre, Phase 1 Dose-Escalation Trial. Lancet 2014, 384, 37–44. [Google Scholar]
- Abello, N.; Kerstjens, H.A.M.; Postma, D.S.; Bischoff, R. Selective Acylation of Primary Amines in Peptides and Proteins. J. Proteome Res. 2007, 6, 4770–4776. [Google Scholar]
| Reference No. | Species of Phage | Modified Site | Modified Material | Diagnosis Target |
|---|---|---|---|---|
| 68 | M13 | P8 protein | Folic acid and fluorescent molecules | Human KB cancer cell |
| 69 | M13 | P3 protein | Random peptides with a disulfide constrained loop | Exosome of Eμ-myc tumor |
| 70 | M13 | P3 protein | Random 12-mer peptides | LS174T colorectal cancer cell |
| 71 | M13K07 | Biotinylated P3 protein | PE-Cy7-Strept | Raji and Raji-FL cell |
| 72 | fADL-1e | 3FLAG or hemagglutinin | B-cell receptor peptide ligand | Raji and Raji-FL cell |
| 76 | Fd derived filamentous phage | P3 protein with antibody binding peptide and pBPA | Mouse IgG1 clone Mab1 raised against recombinant TNFα/IL-6/IL-1β | TNFα/IL-6/IL-1β |
| 78 | M13 | P3 | Anti-CEA-scFv | Circulating tumor cell |
| Reference No. | Species of Phage | Modified Site | Modified Material | Target |
|---|---|---|---|---|
| 93 | M13 | P3 protein | Peptide ligand and polyethylenimine | Tyrosine kinase with immunoglobulin and epidermal growth factor homology domain-2 |
| 94 | M13 | P3 protein | HER2 and CEA anti-scFv | HER2 and CEA antigen |
| 95 | M13 | P3 protein | Anti-Trop2 Fab antibody | Human trophoblastic cell surface antigen 2 |
| 96 | f8/8 phage | P8 protein | Fusion peptide | Total prostate-specific antigen |
| 97 | Dyax Fab phage display library | P3 protein | Anti-IgG1 Fab | Vascular endothelial growth factor receptor |
| 98 | Dyax Fab phage display library | P3 protein | Recombinant human IgG1 mAb | Metastatic non-small cell lung tumor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Y.; Wang, J.; Li, Y.; Yang, C.-T.; Zhou, X. Modified Bacteriophage for Tumor Detection and Targeted Therapy. Nanomaterials 2023, 13, 665. https://doi.org/10.3390/nano13040665
Shen Y, Wang J, Li Y, Yang C-T, Zhou X. Modified Bacteriophage for Tumor Detection and Targeted Therapy. Nanomaterials. 2023; 13(4):665. https://doi.org/10.3390/nano13040665
Chicago/Turabian StyleShen, Yuanzhao, Jingyu Wang, Yuting Li, Chih-Tsung Yang, and Xin Zhou. 2023. "Modified Bacteriophage for Tumor Detection and Targeted Therapy" Nanomaterials 13, no. 4: 665. https://doi.org/10.3390/nano13040665
APA StyleShen, Y., Wang, J., Li, Y., Yang, C.-T., & Zhou, X. (2023). Modified Bacteriophage for Tumor Detection and Targeted Therapy. Nanomaterials, 13(4), 665. https://doi.org/10.3390/nano13040665







