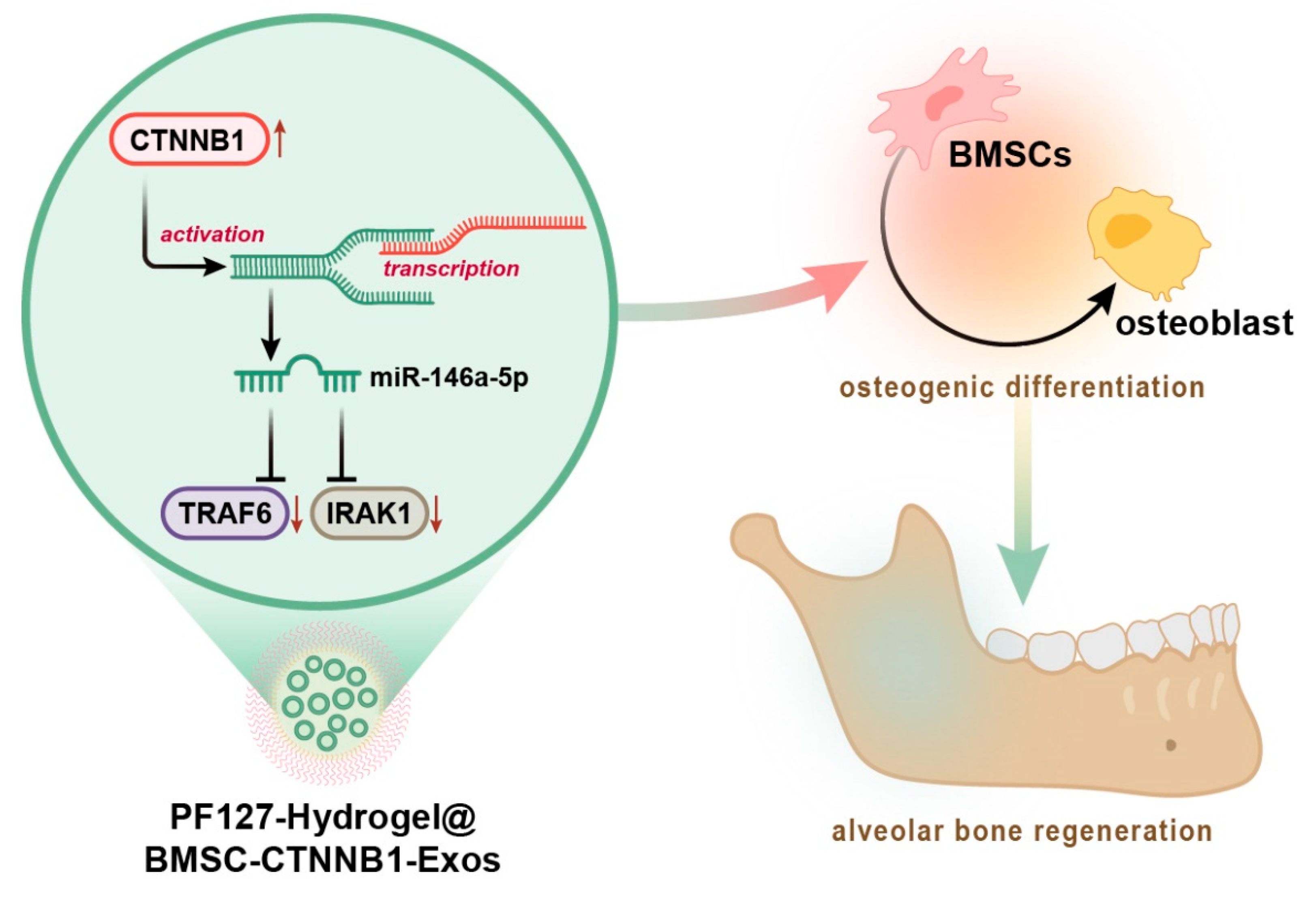
PF127 Hydrogel-Based Delivery of Exosomal CTNNB1 from Mesenchymal Stem Cells Induces Osteogenic Differentiation during the Repair of Alveolar Bone Defects
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
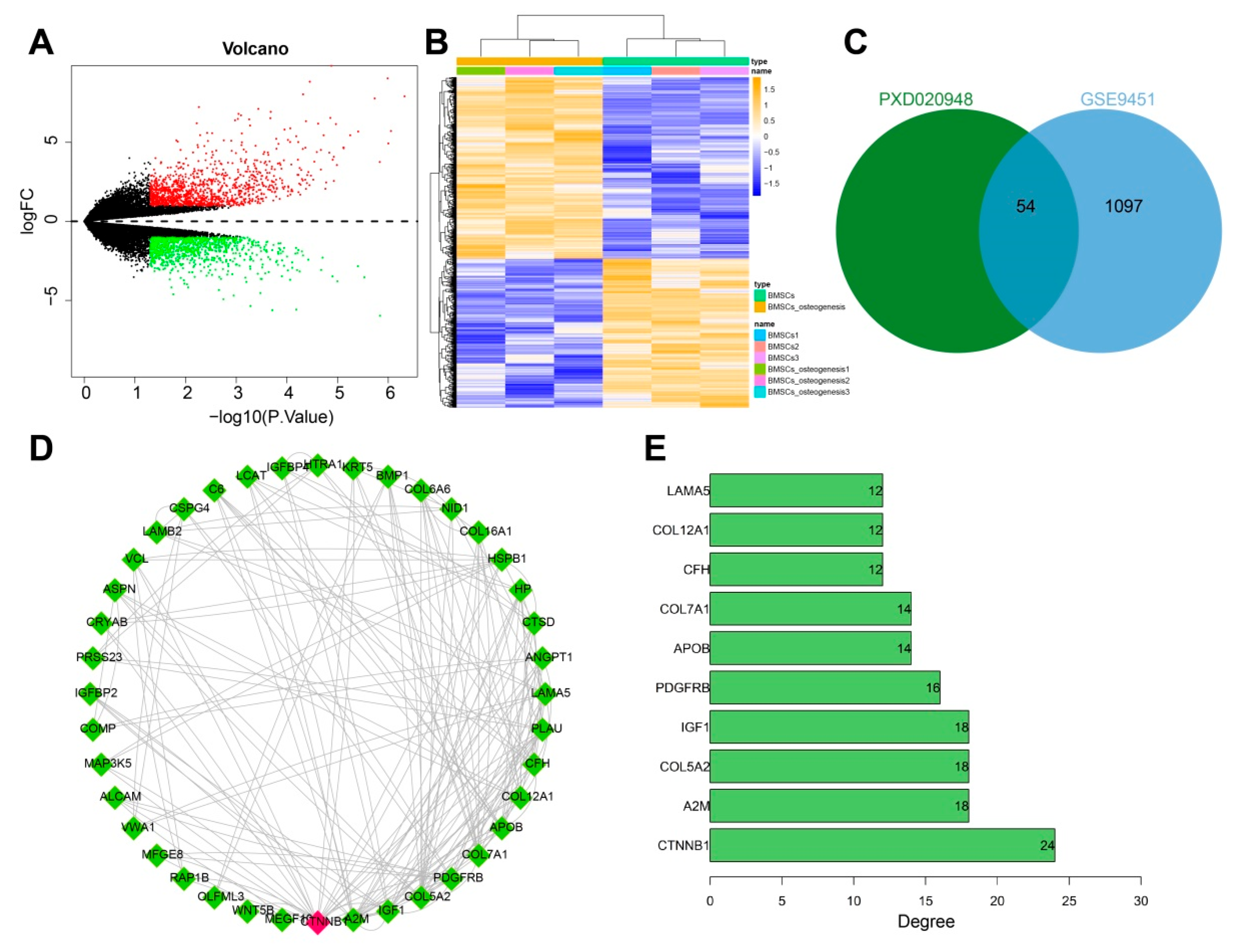
2.2. Bioinformatics Analysis
2.3. Culture of Rat BMSCs
2.4. Cell Grouping and Plasmid Transfection
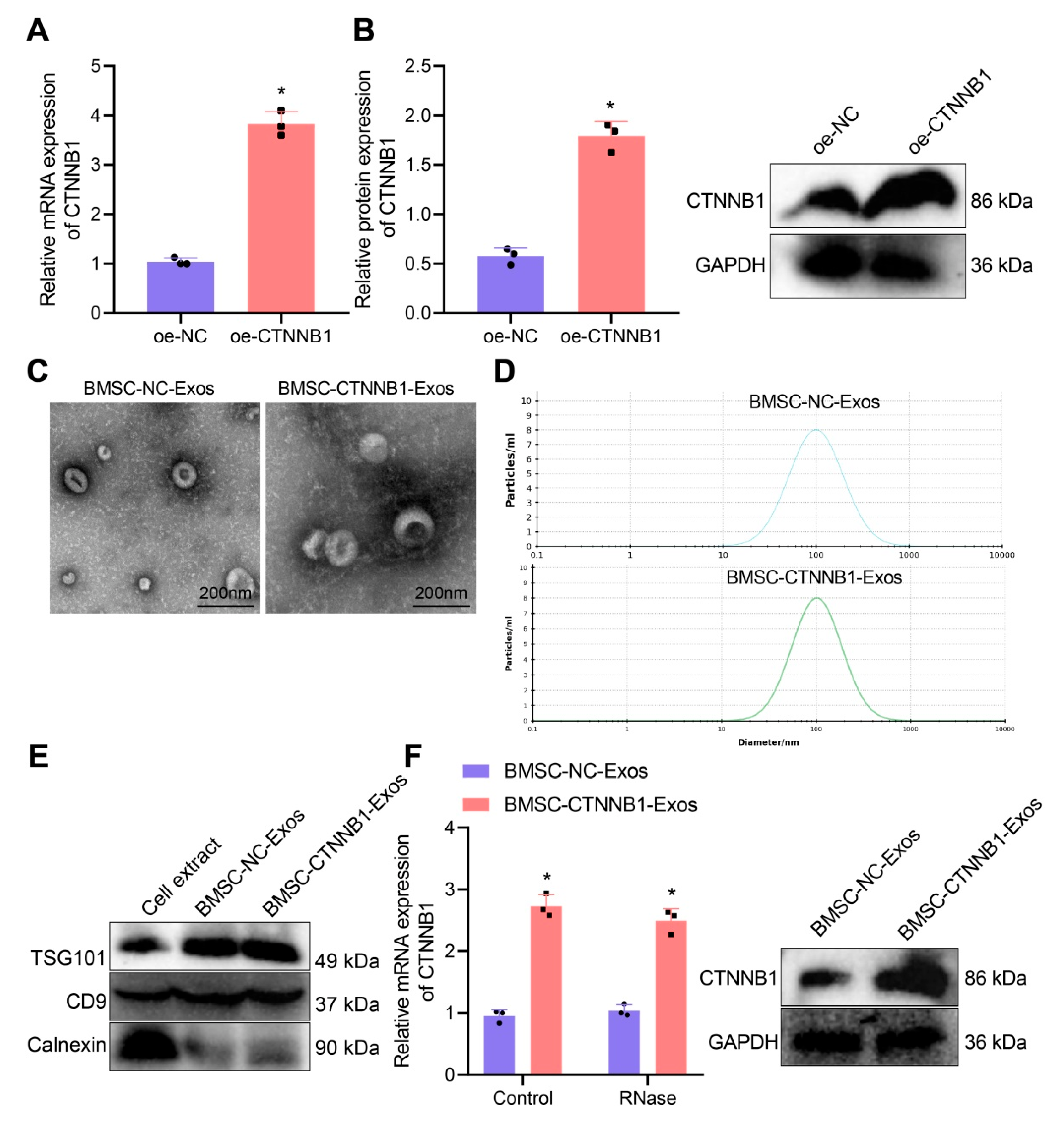
2.5. Isolation and Characterization of Exos from BMSCs
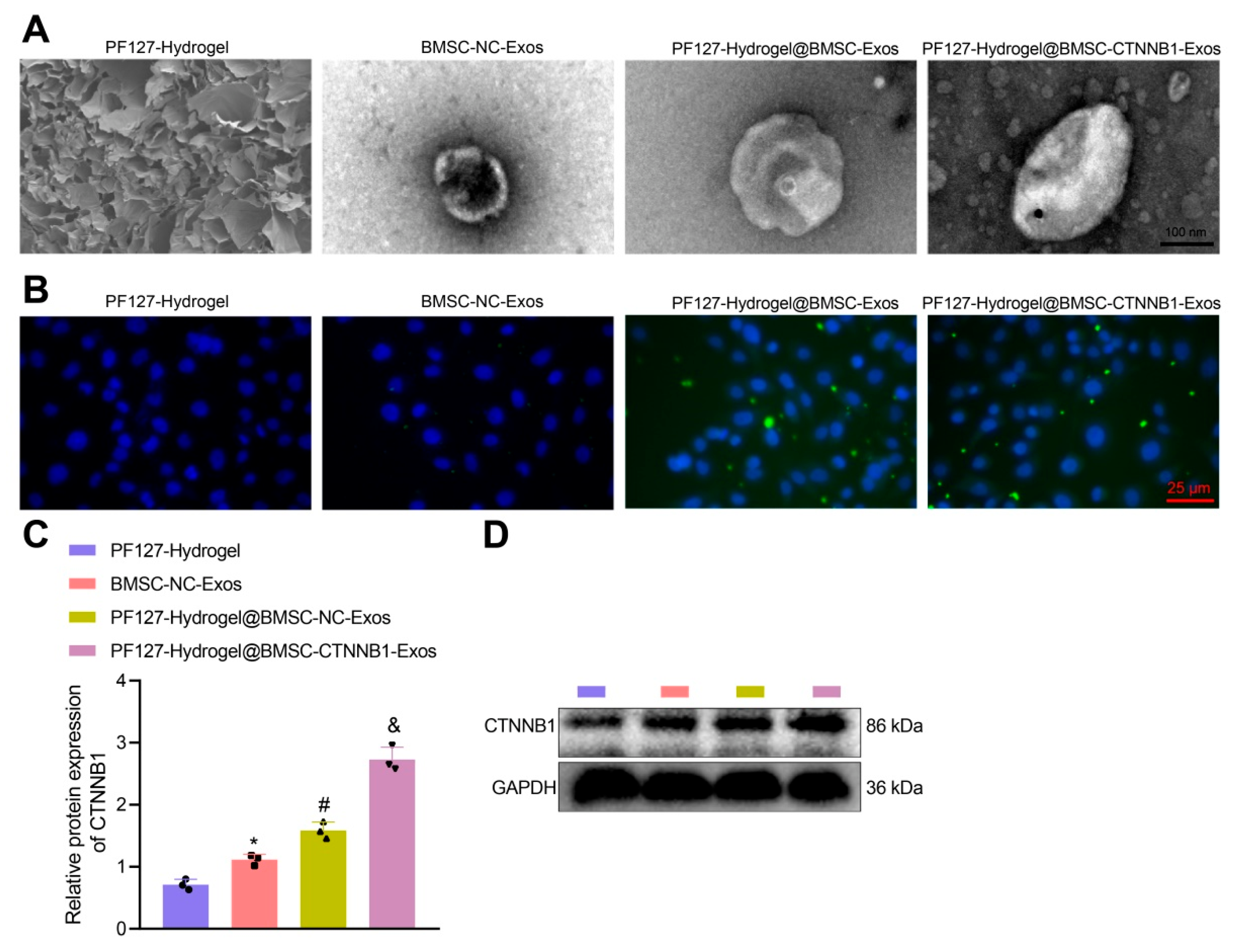
2.6. Construction of Composite Material of Hydrogel Loading with Exos
2.7. Uptake of Exos by BMSCs
2.8. Alkaline Phosphatase (ALP) Activity Assessment
2.9. Alizarin Red Staining
2.10. Chromatin Immunoprecipitation (ChIP)
2.11. Dual-Luciferase Reporter Gene Assay
2.12. RT-qPCR
2.13. Western Blot Analysis
2.14. Rat Model of Alveolar Bone Defects and In Vivo Experiment Protocols
2.15. Two-Photon Excited Fluorescence (TPEF) Imaging
2.16. Immunohistochemistry
2.17. Micro-CT Scanning of Newly Formed Bone Tissue
2.18. Statistical Analysis
3. Results and Discussion
3.1. CTNNB1 Might Be the Key Gene for BMSC-Exos to Promote the Differentiation of BMSCs into Osteoblasts
3.2. Characterization of BMSC−CTNNB1−Exos
3.3. PF127 Hydrogel Loaded with BMSC-Exos Efficiently Delivered CTNNB1 to BMSCs
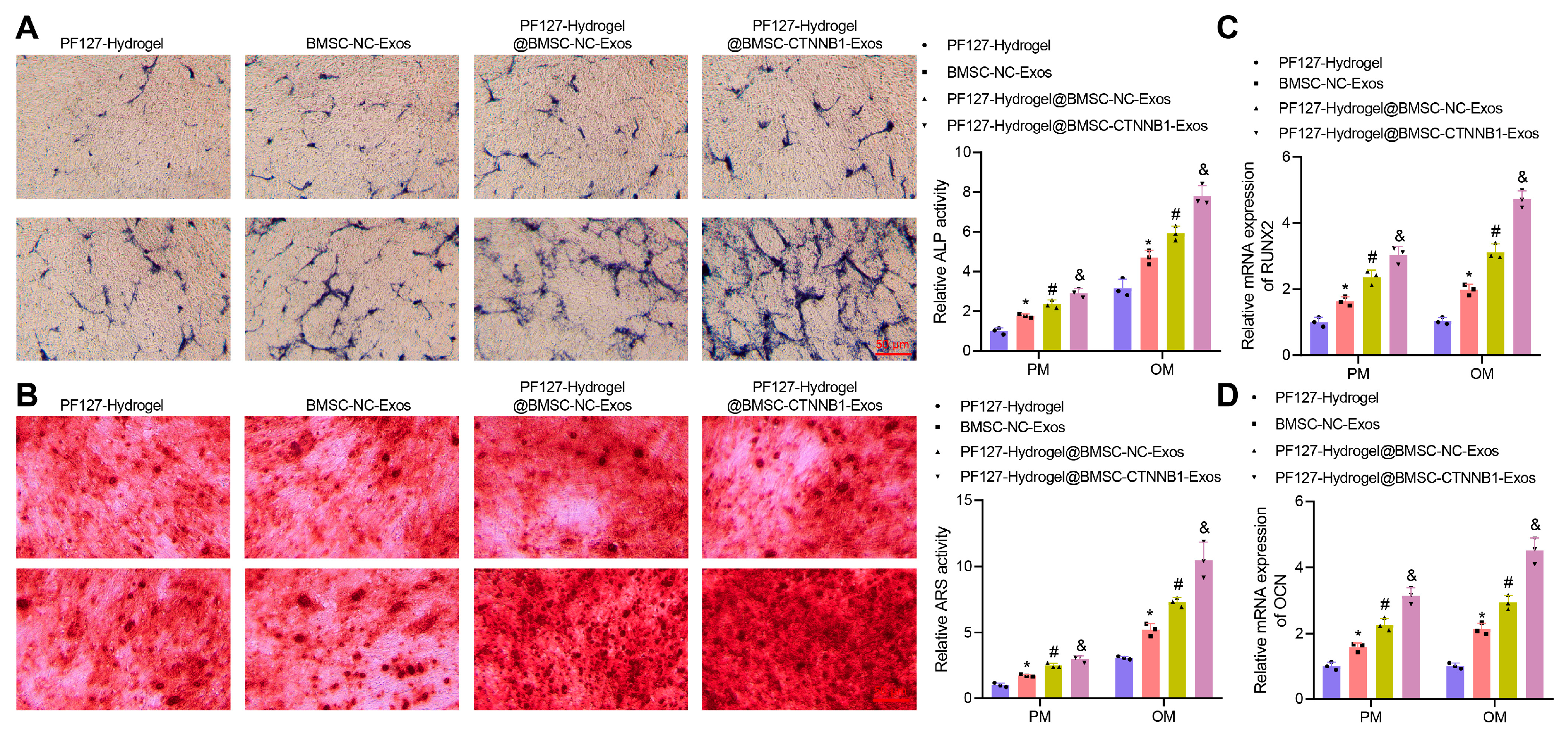
3.4. PF127 Hydrogel Loaded with BMSC-CTNNB1-Exos Promoted the Osteogenic Differentiation of BMSCs
3.5. CTNNB1 Activated the Transcription of miR-146a-5p and Further Targeted Inhibited IRAK1 and TRAF6 during Osteogenic Differentiation of BMSCs
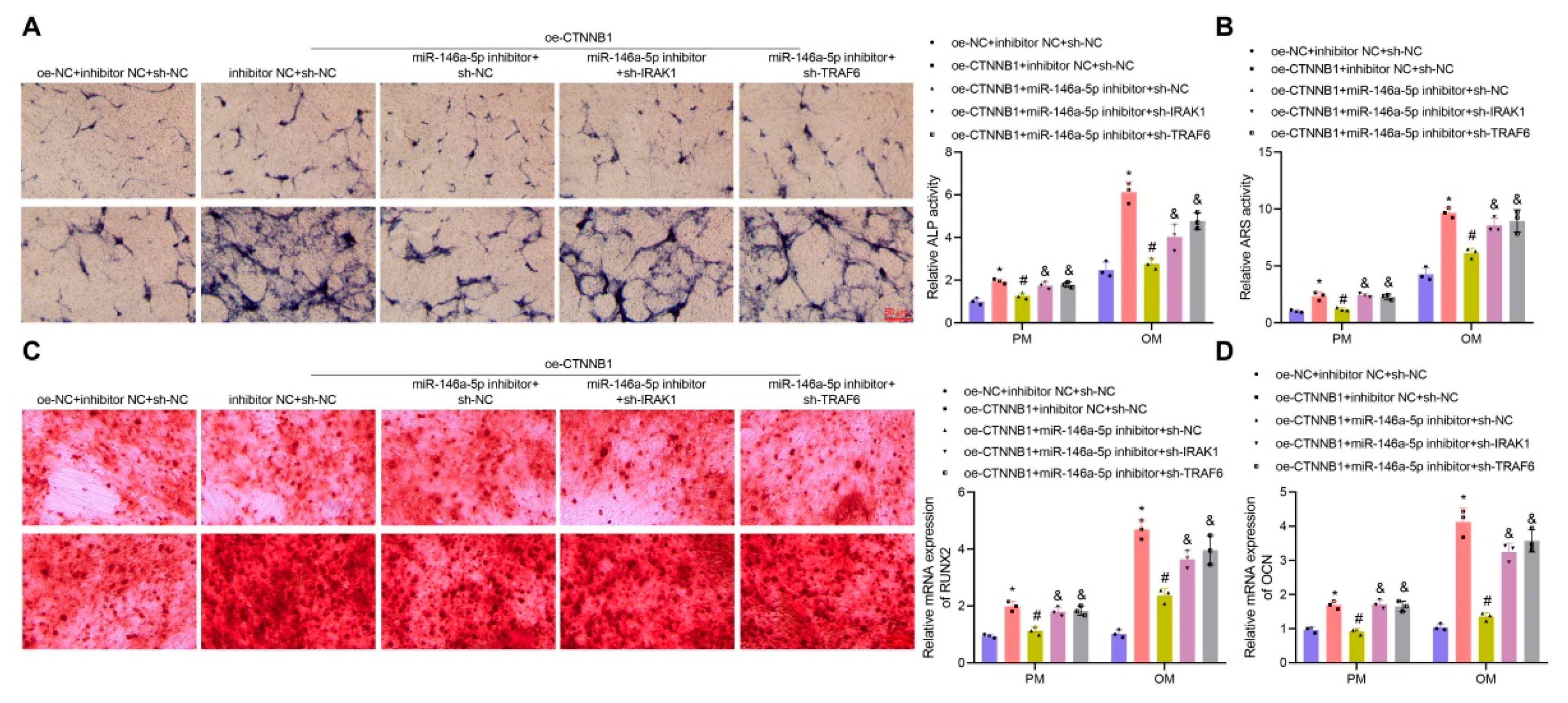
3.6. CTNNB1 Induced the Osteogenic Differentiation of BMSCs by Regulating the miR-146a-5p/IRAK1/TRAF6 Axis
3.7. PF127 Hydrogel Loaded with BMSC-CTNNB1-Exos Induced Alveolar Bone Regeneration in Rats
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cheng, X.; Zhou, X.; Liu, C.; Xu, X. Oral Osteomicrobiology: The Role of Oral Microbiota in Alveolar Bone Homeostasis. Front. Cell. Infect. Microbiol. 2021, 11, 751503. [Google Scholar] [CrossRef]
- An, S.Y.; Lee, Y.J.; Neupane, S.; Jun, J.H.; Kim, J.Y.; Lee, Y.; Choi, K.S.; An, C.H.; Suh, J.Y.; Shin, H.I.; et al. Effects of vascular formation during alveolar bone process morphogenesis in mice. Histochem. Cell Biol. 2017, 148, 435–443. [Google Scholar] [CrossRef]
- Hathaway-Schrader, J.D.; Novince, C.M. Maintaining homeostatic control of periodontal bone tissue. Periodontol 2000 2021, 86, 157–187. [Google Scholar] [CrossRef]
- Ninomiya, T.; Hiraga, T.; Hosoya, A.; Ohnuma, K.; Ito, Y.; Takahashi, M.; Ito, S.; Asashima, M.; Nakamura, H. Enhanced bone-forming activity of side population cells in the periodontal ligament. Cell Transpl. 2014, 23, 691–701. [Google Scholar] [CrossRef]
- Holly, D.; Klein, M.; Mazreku, M.; Zamborsky, R.; Polak, S.; Danisovic, L.; Csobonyeiova, M. Stem Cells and Their Derivatives-Implications for Alveolar Bone Regeneration: A Comprehensive Review. Int. J. Mol. Sci. 2021, 22, 11746. [Google Scholar] [CrossRef] [PubMed]
- Pishavar, E.; Luo, H.; Naserifar, M.; Hashemi, M.; Toosi, S.; Atala, A.; Ramakrishna, S.; Behravan, J. Advanced Hydrogels as Exosome Delivery Systems for Osteogenic Differentiation of MSCs: Application in Bone Regeneration. Int. J. Mol. Sci. 2021, 22, 6203. [Google Scholar] [CrossRef] [PubMed]
- Baldino, L.; Cardea, S.; Reverchon, E. Nanostructured chitosan–gelatin hybrid aerogels produced by supercritical gel drying. Polym. Eng. Sci. 2017, 58, 1494–1499. [Google Scholar] [CrossRef]
- Kang, H.; Jiang, X.; Liu, Z.; Liu, F.; Yan, G.; Li, F. Biodegradable 3D Printed Scaffolds of Modified Poly (Trimethylene Carbonate) Composite Materials with Poly (L-Lactic Acid) and Hydroxyapatite for Bone Regeneration. Nanomaterials 2021, 11, 3215. [Google Scholar] [CrossRef]
- Almoshari, Y.; Ren, R.; Zhang, H.; Jia, Z.; Wei, X.; Chen, N.; Li, G.; Ryu, S.; Lele, S.M.; Reinhardt, R.A.; et al. GSK3 inhibitor-loaded osteotropic Pluronic hydrogel effectively mitigates periodontal tissue damage associated with experimental periodontitis. Biomaterials 2020, 261, 120293. [Google Scholar] [CrossRef]
- Toyota, A.; Shinagawa, R.; Mano, M.; Tokioka, K.; Suda, N. Regeneration in Experimental Alveolar Bone Defect Using Human Umbilical Cord Mesenchymal Stem Cells. Cell Transpl. 2021, 30, 963689720975391. [Google Scholar] [CrossRef]
- Veronesi, F.; Giavaresi, G.; Tschon, M.; Borsari, V.; Nicoli Aldini, N.; Fini, M. Clinical use of bone marrow, bone marrow concentrate, and expanded bone marrow mesenchymal stem cells in cartilage disease. Stem Cells Dev. 2013, 22, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Chung, V.H.; Chen, A.Y.; Jeng, L.B.; Kwan, C.C.; Cheng, S.H.; Chang, S.C. Engineered autologous bone marrow mesenchymal stem cells: Alternative to cleft alveolar bone graft surgery. J. Craniofac. Surg. 2012, 23, 1558–1563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guo, S.; Shi, W.; Liu, Q.; Huo, F.; Wu, Y.; Tian, W. Bone Marrow Mesenchymal Stem Cell-Derived Small Extracellular Vesicles Promote Periodontal Regeneration. Tissue Eng. Part A 2021, 27, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Valenta, T.; Hausmann, G.; Basler, K. The many faces and functions of beta-catenin. EMBO J. 2012, 31, 2714–2736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Y.; Xiang, L.; Wang, Z.; Xiao, G.G.; Ju, D. Diosgenin protects against alveolar bone loss in ovariectomized rats via regulating long non-coding RNAs. Exp. Ther. Med. 2018, 16, 3939–3950. [Google Scholar] [CrossRef]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280. [Google Scholar] [CrossRef]
- Qiu, Z.; Lin, S.; Hu, X.; Zeng, J.; Xiao, T.; Ke, Z.; Lv, H. Involvement of miR-146a-5p/neurogenic locus notch homolog protein 1 in the proliferation and differentiation of STRO-1 (+) human dental pulp stem cells. Eur. J. Oral. Sci. 2019, 127, 294–303. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, X.; Zhang, R.; Xie, Y.; Feng, Z.; Li, F.; Han, J.; Sun, H.; Ouyang, Q.; Hua, S.; et al. Human umbilical cord mesenchymal stem cell-derived exosomal miR-146a-5p reduces microglial-mediated neuroinflammation via suppression of the IRAK1/TRAF6 signaling pathway after ischemic stroke. Aging 2021, 13, 3060–3079. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Zhang, D.; Chen, T.; Hu, A.; Han, X. Glycemic fluctuation exacerbates inflammation and bone loss and alters microbiota profile around implants in diabetic mice with experimental peri-implantitis. Int. J. Implant Dent. 2021, 7, 79. [Google Scholar] [CrossRef]
- Lasser, C.; Eldh, M.; Lotvall, J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012, 59, e3037. [Google Scholar]
- Liao, Z.; Luo, R.; Li, G.; Song, Y.; Zhan, S.; Zhao, K.; Hua, W.; Zhang, Y.; Wu, X.; Yang, C. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 2019, 9, 4084–4100. [Google Scholar] [CrossRef]
- Pritchard, A.; Tousif, S.; Wang, Y.; Hough, K.; Khan, S.; Strenkowski, J.; Chacko, B.K.; Darley-Usmar, V.M.; Deshane, J.S. Lung Tumor Cell-Derived Exosomes Promote M2 Macrophage Polarization. Cells 2020, 9, 1303. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- King, G.N.; King, N.; Cruchley, A.T.; Wozney, J.M.; Hughes, F.J. Recombinant human bone morphogenetic protein-2 promotes wound healing in rat periodontal fenestration defects. J. Dent. Res. 1997, 76, 1460–1470. [Google Scholar] [CrossRef] [PubMed]
- Yong, K.W.; Choi, J.R.; Choi, J.Y.; Cowie, A.C. Recent Advances in Mechanically Loaded Human Mesenchymal Stem Cells for Bone Tissue Engineering. Int. J. Mol. Sci. 2020, 21, 5816. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A.I.; Correa, D. The MSC: An injury drugstore. Cell Stem Cell 2011, 9, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Lin, D.; Zhao, H.; Chen, L.; Cai, B.; Lin, K.; Shen, S.G. Optimized BMSC-derived osteoinductive exosomes immobilized in hierarchical scaffold via lyophilization for bone repair through Bmpr2/Acvr2b competitive receptor-activated Smad pathway. Biomaterials 2021, 272, 120718. [Google Scholar] [CrossRef]
- Luo, Z.W.; Li, F.X.; Liu, Y.W.; Rao, S.S.; Yin, H.; Huang, J.; Chen, C.Y.; Hu, Y.; Zhang, Y.; Tan, Y.J.; et al. Aptamer-functionalized exosomes from bone marrow stromal cells target bone to promote bone regeneration. Nanoscale 2019, 11, 20884–20892. [Google Scholar] [CrossRef]
- Hung, K.S.; Chen, M.S.; Lan, W.C.; Cho, Y.C.; Saito, T.; Huang, B.H.; Tsai, H.Y.; Hsieh, C.C.; Ou, K.L.; Lin, H.Y. Three-Dimensional Printing of a Hybrid Bioceramic and Biopolymer Porous Scaffold for Promoting Bone Regeneration Potential. Materials 2022, 15, 1971. [Google Scholar] [CrossRef]
- Xiang, L.; Zheng, J.; Zhang, M.; Ai, T.; Cai, B. FOXQ1 promotes the osteogenic differentiation of bone mesenchymal stem cells via Wnt/beta-catenin signaling by binding with ANXA2. Stem Cell Res. Ther. 2020, 11, 403. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wan, S.; Yang, M. Circ_0067680 expedites the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells through miR-4429/CTNNB1/Wnt/beta-catenin pathway. Biol. Direct 2021, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, Z.; Pan, D.; Li, H.; Shen, J. Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomes Combined Pluronic F127 Hydrogel Promote Chronic Diabetic Wound Healing and Complete Skin Regeneration. Int. J. Nanomed. 2020, 15, 5911–5926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Rong, Y.; Luo, C.; Cui, W. Bone marrow mesenchymal stem cell-derived exosomes prevent osteoarthritis by regulating synovial macrophage polarization. Aging 2020, 12, 25138–25152. [Google Scholar] [CrossRef]
- Ma, S.; Wu, J.; Hu, H.; Mu, Y.; Zhang, L.; Zhao, Y.; Bian, X.; Jing, W.; Wei, P.; Zhao, B.; et al. Novel fusion peptides deliver exosomes to modify injectable thermo-sensitive hydrogels for bone regeneration. Mater. Today Bio 2022, 13, 100195. [Google Scholar] [CrossRef]
- Diniz, I.M.; Chen, C.; Xu, X.; Ansari, S.; Zadeh, H.H.; Marques, M.M.; Shi, S.; Moshaverinia, A. Pluronic F-127 hydrogel as a promising scaffold for encapsulation of dental-derived mesenchymal stem cells. J. Mater. Sci. Mater. Med. 2015, 26, 153. [Google Scholar] [CrossRef]
- Venkatesh, M.P.; Kumar, T.M.; Avinash, B.S.; Kumar, G.S. Development, in vitro and in vivo evaluation of novel injectable smart gels of azithromycin for chronic periodontitis. Curr. Drug Deliv. 2013, 10, 188–197. [Google Scholar] [CrossRef]
- Nakao, Y.; Fukuda, T.; Zhang, Q.; Sanui, T.; Shinjo, T.; Kou, X.; Chen, C.; Liu, D.; Watanabe, Y.; Hayashi, C.; et al. Exosomes from TNF-alpha-treated human gingiva-derived MSCs enhance M2 macrophage polarization and inhibit periodontal bone loss. Acta Biomater. 2021, 122, 306–324. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Wang, R.; Song, Z.; Shen, Z.; Zhao, Y.; Huang, S.; Lin, Z. Exosomes Secreted by Stem Cells from Human Exfoliated Deciduous Teeth Promote Alveolar Bone Defect Repair through the Regulation of Angiogenesis and Osteogenesis. ACS Biomater. Sci. Eng. 2019, 5, 3561–3571. [Google Scholar] [CrossRef]
- Wang, L.; Xu, W.; Chen, Y.; Wang, J. Alveolar bone repair of rhesus monkeys by using BMP-2 gene and mesenchymal stem cells loaded three-dimensional printed bioglass scaffold. Sci. Rep. 2019, 9, 18175. [Google Scholar] [CrossRef]
- Chen, N.; Ren, R.; Wei, X.; Mukundan, R.; Li, G.; Xu, X.; Zhao, G.; Zhao, Z.; Lele, S.M.; Reinhardt, R.A.; et al. Thermoresponsive Hydrogel-Based Local Delivery of Simvastatin for the Treatment of Periodontitis. Mol. Pharm. 2021, 18, 1992–2003. [Google Scholar] [CrossRef]
- Zhang, C.; Li, T.; Zhou, C.; Huang, L.; Li, Y.; Wang, H.; Duan, P.; Zou, S.; Mei, L. Parathyroid hormone increases alveolar bone homoeostasis during orthodontic tooth movement in rats with periodontitis via crosstalk between STAT3 and beta-catenin. Int. J. Oral. Sci. 2020, 12, 38. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, F.; Huang, L.; Zhang, H.; Li, B.; Zhou, W.; Li, X.; Du, Y.; Pan, Y.; Wang, R. Human amnion-derived mesenchymal stem cells promote osteogenic differentiation of lipopolysaccharide-induced human bone marrow mesenchymal stem cells via ANRIL/miR-125a/APC axis. Stem Cell Res. Ther. 2021, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Tang, X.; Li, Q.; Wang, H.; Sun, H.; Tian, J. Mesenchymal stem cell extracellular vesicles-derived microRNA-194-5p delays the development of intervertebral disc degeneration by targeting TRAF6. Regen. Ther. 2022, 19, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ji, Z.; Xu, Z.; Du, Z.; Wang, Z. Overexpression of miR-146a-5p Ameliorates Inflammation and Autophagy in TLCs-Induced AR42J Cell Model of Acute Pancreatitis by Inhibiting IRAK1/TRAF6/NF-kappaB Pathway. Ann. Clin. Lab. Sci. 2022, 52, 416–425. [Google Scholar] [PubMed]
- Sun, X.; Cui, S.; Fu, X.; Liu, C.; Wang, Z.; Liu, Y. MicroRNA-146-5p promotes proliferation, migration and invasion in lung cancer cells by targeting claudin-12. Cancer Biomark. 2019, 25, 89–99. [Google Scholar] [CrossRef]
- Zhai, M.; Zhu, Y.; Yang, M.; Mao, C. Human Mesenchymal Stem Cell Derived Exosomes Enhance Cell-Free Bone Regeneration by Altering Their miRNAs Profiles. Adv. Sci. 2020, 7, 2001334. [Google Scholar] [CrossRef]
- Deng, L.; Qing, W.; Lai, S.; Zheng, J.; Liu, C.; Huang, H.; Peng, P.; Mu, Y. Differential Expression Profiling of microRNAs in Human Placenta-Derived Mesenchymal Stem Cells Cocultured with Grooved Porous Hydroxyapatite Scaffolds. DNA Cell Biol. 2022, 41, 292–304. [Google Scholar] [CrossRef]
- Li, X.; Liao, J.; Su, X.; Li, W.; Bi, Z.; Wang, J.; Su, Q.; Huang, H.; Wei, Y.; Gao, Y.; et al. Human urine-derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR-146a-5p which targets IRAK1. Theranostics 2020, 10, 9561–9578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Le, X.; Zheng, S.; Zhang, K.; He, J.; Liu, M.; Tu, C.; Rao, W.; Du, H.; Ouyang, Y.; et al. MicroRNA-146a-5p-modified human umbilical cord mesenchymal stem cells enhance protection against diabetic nephropathy in rats through facilitating M2 macrophage polarization. Stem Cell Res. Ther. 2022, 13, 171. [Google Scholar] [CrossRef]
- Han, Y.; Yu, C.; Yu, Y. Astragalus polysaccharide alleviates alveolar bone destruction by regulating local osteoclastogenesis during periodontitis. J. Appl. Biomed. 2021, 19, 97–104. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Zhou, Q.; Zhang, H.; Zhao, N.; Liao, L. PF127 Hydrogel-Based Delivery of Exosomal CTNNB1 from Mesenchymal Stem Cells Induces Osteogenic Differentiation during the Repair of Alveolar Bone Defects. Nanomaterials 2023, 13, 1083. https://doi.org/10.3390/nano13061083
He L, Zhou Q, Zhang H, Zhao N, Liao L. PF127 Hydrogel-Based Delivery of Exosomal CTNNB1 from Mesenchymal Stem Cells Induces Osteogenic Differentiation during the Repair of Alveolar Bone Defects. Nanomaterials. 2023; 13(6):1083. https://doi.org/10.3390/nano13061083
Chicago/Turabian StyleHe, Longlong, Qin Zhou, Hengwei Zhang, Ningbo Zhao, and Lifan Liao. 2023. "PF127 Hydrogel-Based Delivery of Exosomal CTNNB1 from Mesenchymal Stem Cells Induces Osteogenic Differentiation during the Repair of Alveolar Bone Defects" Nanomaterials 13, no. 6: 1083. https://doi.org/10.3390/nano13061083
APA StyleHe, L., Zhou, Q., Zhang, H., Zhao, N., & Liao, L. (2023). PF127 Hydrogel-Based Delivery of Exosomal CTNNB1 from Mesenchymal Stem Cells Induces Osteogenic Differentiation during the Repair of Alveolar Bone Defects. Nanomaterials, 13(6), 1083. https://doi.org/10.3390/nano13061083



