Recent Advances in Nanopore Technology for Copper Detection and Their Potential Applications
Abstract
1. Introduction
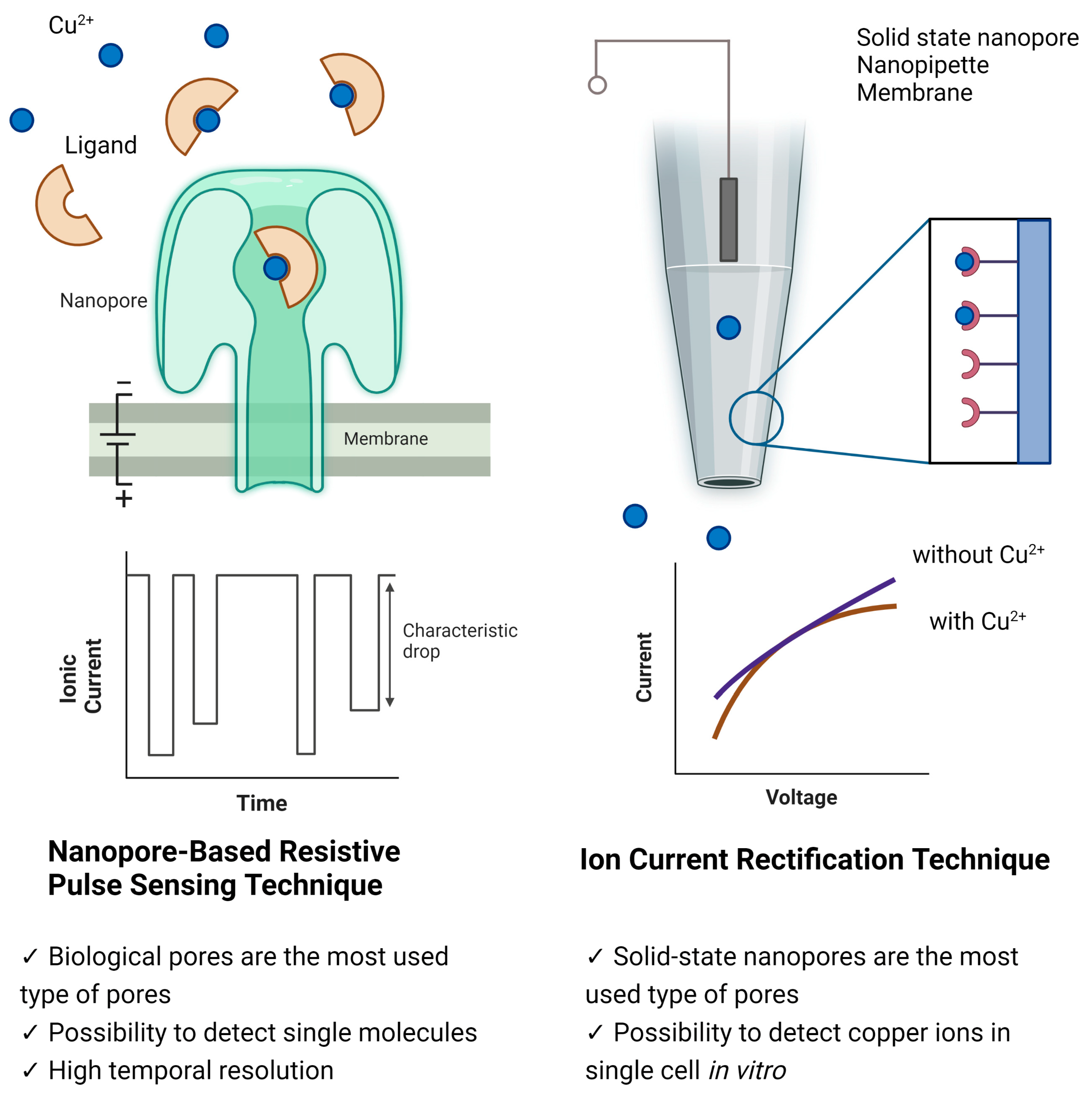
2. Techniques of Nanopore-Based Copper Detection
2.1. Nanopore-Based Resistive Pulse Sensing Technique
2.2. Ionic Current Rectification Technique
3. Applications of Nanopore-Based Sensors for Copper Detection
3.1. Nanopore-Based Resistive Pulse Sensing Technique
3.2. Ionic Current Rectification in Nanopores Technique
4. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| βCD | β-cyclodextrin |
| AAS | atomic absorption spectroscopy |
| AES | atomic emission spectroscopy |
| APTES | (3-aminopropyl)triethoxysilane |
| CMβCD | carboxymethyl-β-cyclodextrin |
| EDTA | ethylenediamine tetraacetic acid |
| ICP-MS | inductively coupled plasma mass spectrometry |
| ICR | ion current rectification |
| LOD | limit of detection |
| PGA | polyglutamic acid |
| PrP | prion peptide |
| RPS | resistive pulse sensing |
| TPPS | 5,10,15,20-tetrakis(4-sulfonatophenyl)-porphyrin |
References
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in Diseases and Treatments, and Copper-Based Anticancer Strategies. Med. Res. Rev. 2009, 30, 708–749. [Google Scholar] [CrossRef] [PubMed]
- Pavelková, M.; Vysloužil, J.; Kubová, K.; Vetchý, D. Biological Role of Copper as an Essential Trace Element in the Human Organism. Ceska Slov. Farm. 2018, 2018, 143–153. [Google Scholar]
- Gaggelli, E.; Kozlowski, H.; Valensin, D.; Valensin, G. Copper Homeostasis and Neurodegenerative Disorders (Alzheimer’s, Prion, and Parkinson’s Diseases and Amyotrophic Lateral Sclerosis). Chem. Rev. 2006, 106, 1995–2044. [Google Scholar] [CrossRef] [PubMed]
- Timoshenko, R.V.; Vaneev, A.N.; Savin, N.A.; Klyachko, N.L.; Parkhomenko, Y.N.; Salikhov, S.V.; Majouga, A.G.; Gorelkin, P.V.; Erofeev, A.S. Promising Approaches for Determination of Copper Ions in Biological Systems. Nanotechnol. Russ. 2020, 15, 121–134. [Google Scholar] [CrossRef]
- Wei, K.; Yao, F.; Kang, X.-F. Single-Molecule Porphyrin-Metal Ion Interaction and Sensing Application. Biosens. Bioelectron. 2018, 109, 272–278. [Google Scholar] [CrossRef]
- Gromadzka, G.; Tarnacka, B.; Flaga, A.; Adamczyk, A. Copper Dyshomeostasis in Neurodegenerative Diseases—Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 9259. [Google Scholar] [CrossRef]
- Witt, B.; Schaumlöffel, D.; Schwerdtle, T. Subcellular Localization of Copper—Cellular Bioimaging with Focus on Neurological Disorders. Int. J. Mol. Sci. 2020, 21, 2341. [Google Scholar] [CrossRef]
- Liddell, J.R.; Bush, A.I.; White, A.R. Copper in Brain and Neurodegeneration. In Encyclopedia of Inorganic and Bioinorganic Chemistry; John Wiley & Sons, Ltd.: Chichester, UK, 2004; pp. 1–21. [Google Scholar]
- Kozlowski, H.; Janicka-Klos, A.; Brasun, J.; Gaggelli, E.; Valensin, D.; Valensin, G. Copper, Iron, and Zinc Ions Homeostasis and Their Role in Neurodegenerative Disorders (Metal Uptake, Transport, Distribution and Regulation). Coord. Chem. Rev. 2009, 253, 2665–2685. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. A DNAzyme Catalytic Beacon Sensor for Paramagnetic Cu2+ Ions in Aqueous Solution with High Sensitivity and Selectivity. J. Am. Chem. Soc. 2007, 129, 9838–9839. [Google Scholar] [CrossRef]
- Mohr, I.; Weiss, K.H. Biochemical Markers for the Diagnosis and Monitoring of Wilson Disease. Clin. Biochem. Rev. 2019, 40, 59–77. [Google Scholar] [CrossRef]
- Cotruvo, J.A., Jr.; Aron, A.T.; Ramos-Torres, K.M.; Chang, C.J. Synthetic Fluorescent Probes for Studying Copper in Biological Systems. Chem. Soc. Rev. 2015, 44, 4400–4414. [Google Scholar] [CrossRef] [PubMed]
- Siriwardhane, T.; Sulkanen, A.; Pathirathna, P.; Tremonti, A.; McElmurry, S.P.; Hashemi, P. Voltammetric Characterization of Cu(II) Complexation in Real-Time. Anal. Chem. 2016, 88, 7603–7608. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, L.; Liu, W.; Yu, Y.; Tian, Y. A Single Biosensor for Evaluating the Levels of Copper Ion and L -Cysteine in a Live Rat Brain with Alzheimer’s Disease. Angew. Chem. Int. Ed. 2015, 54, 14053–14056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, Y.; Zhao, F.; Shi, G.; Tian, Y. A Selective and Accurate Ratiometric Electrochemical Biosensor for Monitoring of Cu2+ Ions in a Rat Brain. Anal. Chem. 2015, 87, 2931–2936. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.J.; Yusof, N.A.; Alang Ahmad, S.A. Electrochemical Determination of Lead & Copper Ions Using Thiolated Calix [4]Arene-Modified Screen-Printed Carbon Electrode. Chemosensors 2021, 9, 157. [Google Scholar] [CrossRef]
- Ogra, Y.; Tanaka, Y.K.; Suzuki, N. Recent Advances in Copper Analyses by Inorganic Mass Spectrometry. J. Clin. Biochem. Nutr. 2022, 71, 2–6. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.-M.; Han, J. Fluorescent Chemosensors for Copper(II) Ion: Structure, Mechanism and Application. J. Photochem. Photobiol. C Photochem. Rev. 2017, 32, 78–103. [Google Scholar] [CrossRef]
- Corbini, G.; Dreassi, E.; Chiasserini, L.; Girolamo, M.M.; Mellace, P. Determination of Copper by AAS in Tear Fluid of Patients with Keratoconus. Anal. Biochem. 2021, 623, 114174. [Google Scholar] [CrossRef]
- Cao, Y.; Feng, J.; Tang, L.; Yu, C.; Mo, G.; Deng, B. A Highly Efficient Introduction System for Single Cell- ICP-MS and Its Application to Detection of Copper in Single Human Red Blood Cells. Talanta 2020, 206, 120174. [Google Scholar] [CrossRef]
- Chopra, T.; Sasan, S.; Devi, L.; Parkesh, R.; Kapoor, K.K. A Comprehensive Review on Recent Advances in Copper Sensors. Coord. Chem. Rev. 2022, 470, 214704. [Google Scholar] [CrossRef]
- Zeng, X.; Xiang, Y.; Liu, Q.; Wang, L.; Ma, Q.; Ma, W.; Zeng, D.; Yin, Y.; Wang, D. Nanopore Technology for the Application of Protein Detection. Nanomaterials 2021, 11, 1942. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The Potential and Challenges of Nanopore Sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Hui, J.; Mao, H. Nanopore Technology and Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef] [PubMed]
- Duan, R.; Xia, F.; Jiang, L. Constructing Tunable Nanopores and Their Application in Drug Delivery. ACS Nano 2013, 7, 8344–8349. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Liang, Y.; Yang, J. Nanopore Detection Assisted DNA Information Processing. Nanomaterials 2022, 12, 3135. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, W.; Razaghi, R.; Busan, S.; Weeks, K.M.; Timp, W.; Smibert, P. Direct Detection of RNA Modifications and Structure Using Single-Molecule Nanopore Sequencing. Cell Genom. 2022, 2, 100097. [Google Scholar] [CrossRef]
- Lucas, F.L.R.; Versloot, R.C.A.; Yakovlieva, L.; Walvoort, M.T.C.; Maglia, G. Protein Identification by Nanopore Peptide Profiling. Nat. Commun. 2021, 12, 5795. [Google Scholar] [CrossRef]
- Afshar Bakshloo, M.; Kasianowicz, J.J.; Pastoriza-Gallego, M.; Mathé, J.; Daniel, R.; Piguet, F.; Oukhaled, A. Nanopore-Based Protein Identification. J. Am. Chem. Soc. 2022, 144, 2716–2725. [Google Scholar] [CrossRef]
- Ren, R.; Sun, M.; Goel, P.; Cai, S.; Kotov, N.A.; Kuang, H.; Xu, C.; Ivanov, A.P.; Edel, J.B. Single-Molecule Binding Assay Using Nanopores and Dimeric NP Conjugates. Adv. Mater. 2021, 33, 2103067. [Google Scholar] [CrossRef]
- Roozbahani, G.M.; Chen, X.; Zhang, Y.; Wang, L.; Guan, X. Nanopore Detection of Metal Ions: Current Status and Future Directions. Small Methods 2020, 4, 2000266. [Google Scholar] [CrossRef]
- Liang, L.; Qin, F.; Wang, S.; Wu, J.; Li, R.; Wang, Z.; Ren, M.; Liu, D.; Wang, D.; Astruc, D. Overview of the Materials Design and Sensing Strategies of Nanopore Devices. Coord. Chem. Rev. 2023, 478, 214998. [Google Scholar] [CrossRef]
- Wang, G.; Wang, L.; Han, Y.; Zhou, S.; Guan, X. Nanopore Detection of Copper Ions Using a Polyhistidine Probe. Biosens. Bioelectron. 2014, 53, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Fang, Z.; Zheng, X.; Xi, D. Nanopore-Based Strategy for Sensing of Copper(II) Ion and Real-Time Monitoring of a Click Reaction. ACS Sens. 2019, 4, 1323–1328. [Google Scholar] [CrossRef]
- Hu, P.; Wang, Y.; Zhang, Y.; Jin, Y. Glass Nanopore Detection of Copper Ions in Single Cells Based on Click Chemistry. Anal. Chem. 2022, 94, 14273–14279. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Mayer, S.F.; Cao, C.; Dal Peraro, M. Biological Nanopores for Single-Molecule Sensing. iScience 2022, 25, 104145. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gooding, J.J. The Application of Single Molecule Nanopore Sensing for Quantitative Analysis. Chem. Soc. Rev. 2022, 51, 3862–3885. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, W.; Song, L.; Wang, X.; Sun, W.; Song, P.; Ashraf, G.; Liu, B.; Zhao, Y.-D. Recent Advances in Ionic Current Rectification Based Nanopore Sensing: A Mini-Review. Sens. Actuators Rep. 2021, 3, 100042. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Yu, L.; Li, Y.; Qian, G.; Chang, S. Nanopipettes: A Potential Tool for DNA Detection. Analyst 2019, 144, 5037–5047. [Google Scholar] [CrossRef]
- Varongchayakul, N.; Song, J.; Meller, A.; Grinstaff, M.W. Single-Molecule Protein Sensing in a Nanopore: A Tutorial. Chem. Soc. Rev. 2018, 47, 8512–8524. [Google Scholar] [CrossRef]
- Yu, R.; Ying, Y.; Gao, R.; Long, Y. Confined Nanopipette Sensing: From Single Molecules, Single Nanoparticles, to Single Cells. Angew. Chem. Int. Ed. 2019, 58, 3706–3714. [Google Scholar] [CrossRef]
- Shi, W.; Friedman, A.K.; Baker, L.A. Nanopore Sensing. Anal. Chem. 2017, 89, 157–188. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, L.; Bouchet-Spinelli, A.; Raillon, C.; Buhot, A. Sensing with Nanopores and Aptamers: A Way Forward. Sensors 2020, 20, 4495. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Zhou, S.; Roozbahani, G.M.; Zhang, Y.; Wang, D.; Guan, X. Displacement Chemistry-Based Nanopore Analysis of Nucleic Acids in Complicated Matrices. Chem. Commun. 2018, 54, 13977–13980. [Google Scholar] [CrossRef] [PubMed]
- Rauf, S.; Zhang, L.; Ali, A.; Liu, Y.; Li, J. Label-Free Nanopore Biosensor for Rapid and Highly Sensitive Cocaine Detection in Complex Biological Fluids. ACS Sens. 2017, 2, 227–234. [Google Scholar] [CrossRef]
- Ji, Z.; Jordan, M.; Jayasinghe, L.; Guo, P. Insertion of Channel of Phi29 DNA Packaging Motor into Polymer Membrane for High-Throughput Sensing. Nanomedicine 2020, 25, 102170. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, Z. Fabrication and Applications of Solid-State Nanopores. Sensors 2019, 19, 1886. [Google Scholar] [CrossRef]
- Magi, A.; Semeraro, R.; Mingrino, A.; Giusti, B.; D’Aurizio, R. Nanopore Sequencing Data Analysis: State of the Art, Applications and Challenges. Brief. Bioinform. 2017, 19, 1256–1272. [Google Scholar] [CrossRef]
- Acharya, A.; Prajapati, J.D.; Kleinekathöfer, U. Atomistic Simulation of Molecules Interacting with Biological Nanopores: From Current Understanding to Future Directions. J. Phys. Chem. B 2022, 126, 3995–4008. [Google Scholar] [CrossRef]
- Vlassiouk, I.; Smirnov, S.; Siwy, Z. Nanofluidic Ionic Diodes. Comparison of Analytical and Numerical Solutions. ACS Nano 2008, 2, 1589–1602. [Google Scholar] [CrossRef]
- Siwy, Z.; Heins, E.; Harrell, C.C.; Kohli, P.; Martin, C.R. Conical-Nanotube Ion-Current Rectifiers: The Role of Surface Charge. J. Am. Chem. Soc. 2004, 126, 10850–10851. [Google Scholar] [CrossRef] [PubMed]
- Woermann, D. Electrochemical Transport Properties of a Cone-Shaped Nanopore: High and Low Electrical Conductivity States Depending on the Sign of an Applied Electrical Potential Difference. Phys. Chem. Chem. Phys. 2003, 5, 1853–1858. [Google Scholar] [CrossRef]
- Momotenko, D.; Girault, H.H. Scan-Rate-Dependent Ion Current Rectification and Rectification Inversion in Charged Conical Nanopores. J. Am. Chem. Soc. 2011, 133, 14496–14499. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.; Zhang, K.; Jiang, Y.; Yu, P.; Mao, L. Ion Current Rectification: From Nanoscale to Microscale. Sci. China Chem. 2019, 62, 1346–1359. [Google Scholar] [CrossRef]
- Duleba, D.; Johnson, R.P. Sensing with Ion Current Rectifying Solid-State Nanopores. Curr. Opin. Electrochem. 2022, 34, 100989. [Google Scholar] [CrossRef]
- Ma, W.; Xie, W.; Tian, R.; Zeng, X.; Liang, L.; Hou, C.; Huo, D.; Wang, D. An Ultrasensitive Aptasensor of SARS-CoV-2 N Protein Based on Ion Current Rectification with Nanopipettes. Sens. Actuators B Chem. 2023, 377, 133075. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tsutsui, M.; Zhou, Y.; Miao, X.-S. Solid-State Nanopore Systems: From Materials to Applications. NPG Asia Mater. 2021, 13, 48. [Google Scholar] [CrossRef]
- Braha, O.; Gu, L.-Q.; Zhou, L.; Lu, X.; Cheley, S.; Bayley, H. Simultaneous Stochastic Sensing of Divalent Metal Ions. Nat. Biotechnol. 2000, 18, 1005–1007. [Google Scholar] [CrossRef]
- Guo, Y.; Jian, F.; Kang, X. Nanopore Sensor for Copper Ion Detection Using a Polyamine Decorated β-Cyclodextrin as the Recognition Element. RSC Adv. 2017, 7, 15315–15320. [Google Scholar] [CrossRef]
- Wang, L.; Yao, F.; Kang, X. Nanopore Single-Molecule Analysis of Metal Ion–Chelator Chemical Reaction. Anal. Chem. 2017, 89, 7958–7965. [Google Scholar] [CrossRef]
- Qi, Z.-L.; Cheng, Y.-H.; Xu, Z.; Chen, M.-L. Recent Advances in Porphyrin-Based Materials for Metal Ions Detection. Int. J. Mol. Sci. 2020, 21, 5839. [Google Scholar] [CrossRef] [PubMed]
- Pyrzynska, K.; Kilian, K.; Pęgier, M. Porphyrins as Chelating Agents for Molecular Imaging in Nuclear Medicine. Molecules 2022, 27, 3311. [Google Scholar] [CrossRef] [PubMed]
- Mayne, L.J.; Christie, S.D.R.; Platt, M. A Tunable Nanopore Sensor for the Detection of Metal Ions Using Translocation Velocity and Biphasic Pulses. Nanoscale 2016, 8, 19139–19147. [Google Scholar] [CrossRef] [PubMed]
- Actis, P.; McDonald, A.; Beeler, D.; Vilozny, B.; Millhauser, G.; Pourmand, N. Copper Sensing with a Prion Protein Modified Nanopipette. RSC Adv. 2012, 2, 11638. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, V.O.; Fokin, V.V.; Finn, M.G. Mechanism of the Ligand-Free CuI-Catalyzed Azide–Alkyne Cycloaddition Reaction. Angew. Chem. Int. Ed. 2005, 44, 2210–2215. [Google Scholar] [CrossRef]
- Nebra, N.; García-Álvarez, J. Recent Progress of Cu-Catalyzed Azide-Alkyne Cycloaddition Reactions (CuAAC) in Sustainable Solvents: Glycerol, Deep Eutectic Solvents, and Aqueous Media. Molecules 2020, 25, 2015. [Google Scholar] [CrossRef]
- Yáñez-Sedeño, P.; González-Cortés, A.; Campuzano, S.; Pingarrón, J.M. Copper(I)-Catalyzed Click Chemistry as a Tool for the Functionalization of Nanomaterials and the Preparation of Electrochemical (Bio)Sensors. Sensors 2019, 19, 2379. [Google Scholar] [CrossRef]
- Zheng, W.; Li, H.; Chen, W.; Zhang, J.; Wang, N.; Guo, X.; Jiang, X. Rapid Detection of Copper in Biological Systems Using Click Chemistry. Small 2018, 14, 1703857. [Google Scholar] [CrossRef]
- Qin, Y.; Li, M.; Yang, Y.; Gao, Z.; Zhang, H.; Zhao, J. A Unimolecular DNA Fluorescent Probe for Determination of Copper Ions Based on Click Chemistry. RSC Adv. 2020, 10, 6017–6021. [Google Scholar] [CrossRef]
- Wei, X.; Wang, X.; Zhang, Z.; Luo, Y.; Wang, Z.; Xiong, W.; Jain, P.K.; Monnier, J.R.; Wang, H.; Hu, T.Y.; et al. A Click Chemistry Amplified Nanopore Assay for Ultrasensitive Quantification of HIV-1 P24 Antigen in Clinical Samples. Nat. Commun. 2022, 13, 6852. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, T.; Sheng, Y.; Liu, L.; Wu, H.-C. Enhanced Sensitivity in Nanopore Sensing of Cancer Biomarkers in Human Blood via Click Chemistry. Small 2019, 15, 1804078. [Google Scholar] [CrossRef]
- Zhang, Y.; Takahashi, Y.; Hong, S.P.; Liu, F.; Bednarska, J.; Goff, P.S.; Novak, P.; Shevchuk, A.; Gopal, S.; Barozzi, I.; et al. High-Resolution Label-Free 3D Mapping of Extracellular PH of Single Living Cells. Nat. Commun. 2019, 10, 5610. [Google Scholar] [CrossRef]
- Actis, P.; Vilozny, B.; Seger, R.A.; Li, X.; Jejelowo, O.; Rinaudo, M.; Pourmand, N. Voltage-Controlled Metal Binding on Polyelectrolyte-Functionalized Nanopores. Langmuir 2011, 27, 6528–6533. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; He, H.; Xu, X.; Jin, Y. Single Glass Nanopore-Based Regenerable Sensing Platforms with a Non-Immobilized Polyglutamic Acid Probe for Selective Detection of Cupric Ions. Anal. Chim. Acta 2015, 889, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Li, Z.; Chen, J.; Hou, Y.; Zhang, J.; Sun, R.; Bu, Z.; Wang, L.; Wang, M.; Chen, X.; et al. Tannic Acid Modified Single Nanopore with Multivalent Metal Ions Recognition and Ultra-Trace Level Detection. Nano Today 2020, 33, 100868. [Google Scholar] [CrossRef]
- Vaneev, A.N.; Timoshenko, R.V.; Gorelkin, P.V.; Klyachko, N.L.; Korchev, Y.E.; Erofeev, A.S. Nano- and Microsensors for In Vivo Real-Time Electrochemical Analysis: Present and Future Perspectives. Nanomaterials 2022, 12, 3736. [Google Scholar] [CrossRef]
- Hsu, J.-P.; Chen, Y.-C.; Wu, C.-T. Detection of the Trace Level of Heavy Metal Ions by PH-Regulated Conical Nanochannels. J. Taiwan Inst. Chem. Eng. 2020, 109, 145–152. [Google Scholar] [CrossRef]
- Müller, L.K.; Duznovic, I.; Tietze, D.; Weber, W.; Ali, M.; Stein, V.; Ensinger, W.; Tietze, A.A. Ultrasensitive and Selective Copper(II) Detection: Introducing a Bioinspired and Robust Sensor. Chem.—A Eur. J. 2020, 26, 8511–8517. [Google Scholar] [CrossRef]
- Zhao, X.-P.; Wang, S.-S.; Younis, M.R.; Xia, X.-H.; Wang, C. Asymmetric Nanochannel–Ionchannel Hybrid for Ultrasensitive and Label-Free Detection of Copper Ions in Blood. Anal. Chem. 2018, 90, 896–902. [Google Scholar] [CrossRef]



| Analyte | Type of Nanopore | LOD | Linear Range | Sensing Principe | Used Ligand | Refs. |
|---|---|---|---|---|---|---|
| Cu2+ | α-hemolysin | 40 nM | - | RPS | Histidine-peptide | [33] |
| Cu2+/Cu+ | α-hemolysin | 67 pM | RPS | DNA | [34] | |
| Cu2+ | α-hemolysin | 12 nM | 0.08–20 µM | RPS | Polyamine-decorated cyclodextrins | [60] |
| Cu2+ | α-hemolysin | - | - | RPS | Carboxymethyl-β-cyclodextrin | [61] |
| Cu2+ | α-hemolysin | 16 nM | 0.03–1.0 µM | RPS | TPPS | [62] |
| Cu2+ | α-hemolysin | 1 ppm | - | RPS | Modified silicon dioxide nanoparticles | [64] |
| Cu2+ | α-hemolysin | - | - | RPS | Prion peptide | [65] |
| Cu2+ | Glass nanopipette | - | 4–100 µM | ICR | Chitosan and poly(acrylic acid) | [74] |
| Cu2+ | Glass Nanopipette | 1.05 µM | 7.5–60 µM | ICR | PGA | [75] |
| Cu2+ | Glass Nanopipette | - | ~1–40 µM | ICR | Thiolated ssDNA with an alkynyl end, azide-end single-stranded DNA | [35] |
| Cu2+ | PET nanochannel | - | 10–500 µM | ICR | 18-crown-6 | [78] |
| Cu2+ | PET nanochannel | 18 nM (Fluorescence quenching) | 10 fM–0.1 μM (ICR technique) 1–100 μM (Fluorescence quenching) | ICR and Fluorescence quenching | 5/6-FAM-Dap-β-Ala-His fluorescent peptide | [79] |
| Cu2+ | Nanochannel array of porous anodic alumina | 0.1 fM | - | ICR | PGA | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vaneev, A.N.; Timoshenko, R.V.; Gorelkin, P.V.; Klyachko, N.L.; Erofeev, A.S. Recent Advances in Nanopore Technology for Copper Detection and Their Potential Applications. Nanomaterials 2023, 13, 1573. https://doi.org/10.3390/nano13091573
Vaneev AN, Timoshenko RV, Gorelkin PV, Klyachko NL, Erofeev AS. Recent Advances in Nanopore Technology for Copper Detection and Their Potential Applications. Nanomaterials. 2023; 13(9):1573. https://doi.org/10.3390/nano13091573
Chicago/Turabian StyleVaneev, Alexander N., Roman V. Timoshenko, Petr V. Gorelkin, Natalia L. Klyachko, and Alexander S. Erofeev. 2023. "Recent Advances in Nanopore Technology for Copper Detection and Their Potential Applications" Nanomaterials 13, no. 9: 1573. https://doi.org/10.3390/nano13091573
APA StyleVaneev, A. N., Timoshenko, R. V., Gorelkin, P. V., Klyachko, N. L., & Erofeev, A. S. (2023). Recent Advances in Nanopore Technology for Copper Detection and Their Potential Applications. Nanomaterials, 13(9), 1573. https://doi.org/10.3390/nano13091573







