Exploring Disulfiram’s Anticancer Potential: PLGA Nano-Carriers for Prolonged Drug Delivery and Potential Improved Therapeutic Efficacy
Abstract
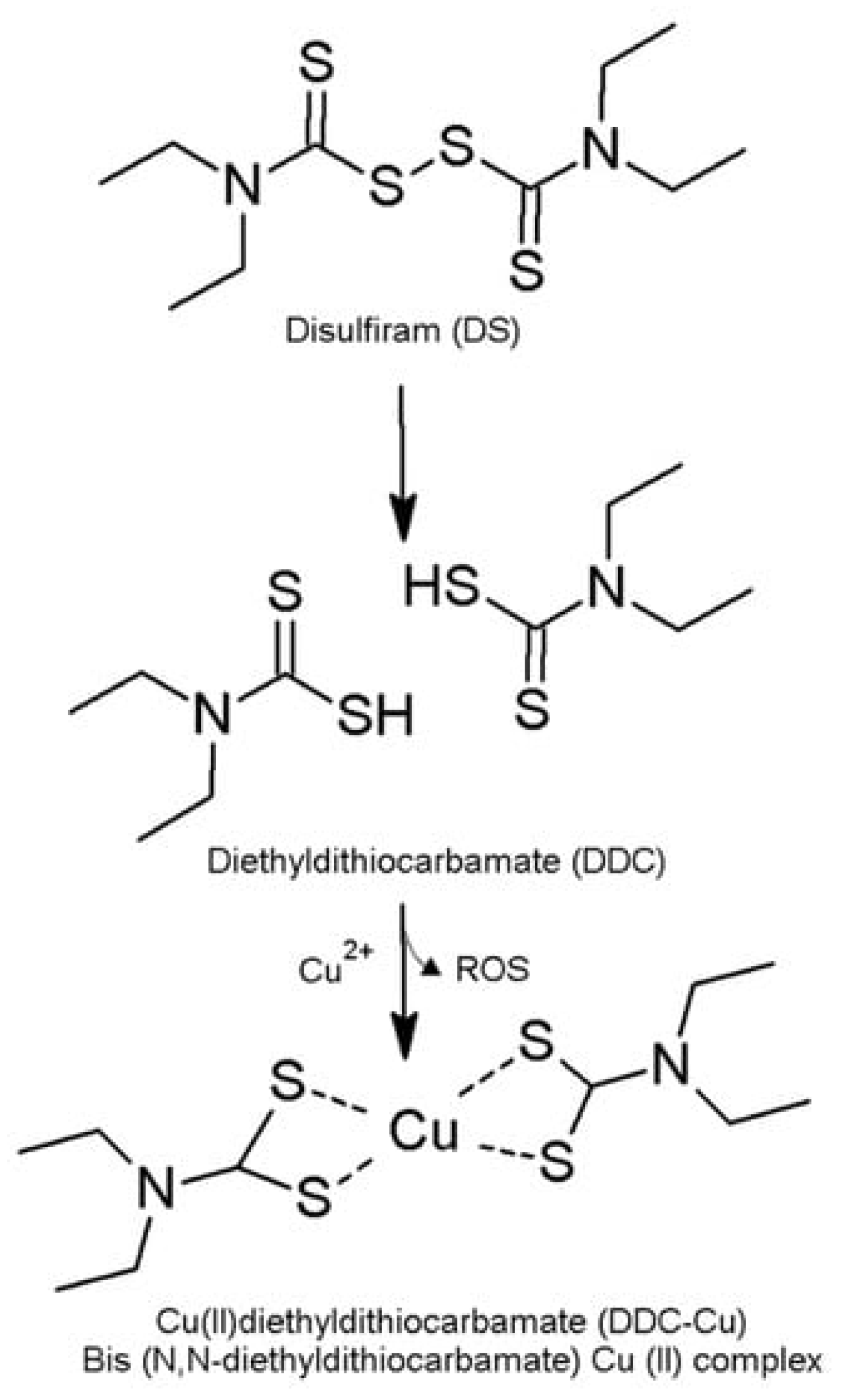
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Nanoparticle Method Development
Particle Size Reduction by High-Pressure Homogenisation (HPH)
Particle Size Reduction by Probe Sonication (PS)
2.2.2. Measurement of Particle Size, Polydispersity Index and Zeta Potential by Zeta Sizer
2.2.3. Freeze-Drying
2.2.4. Scanning Electron Microscopy
2.2.5. Encapsulation Efficiency
2.2.6. Cumulative Release Studies
2.2.7. Stability Studies in Horse Serum
2.2.8. HPLC Method
2.2.9. MTT Cytotoxicity Assay
2.2.10. Statistical Analysis
3. Results and Discussion
3.1. Characterisation of DS-Loaded NPs Prepared by DNP and SE Methods
3.1.1. Particle Size, Polydispersity Index, Zeta Potential and Surface Morphology
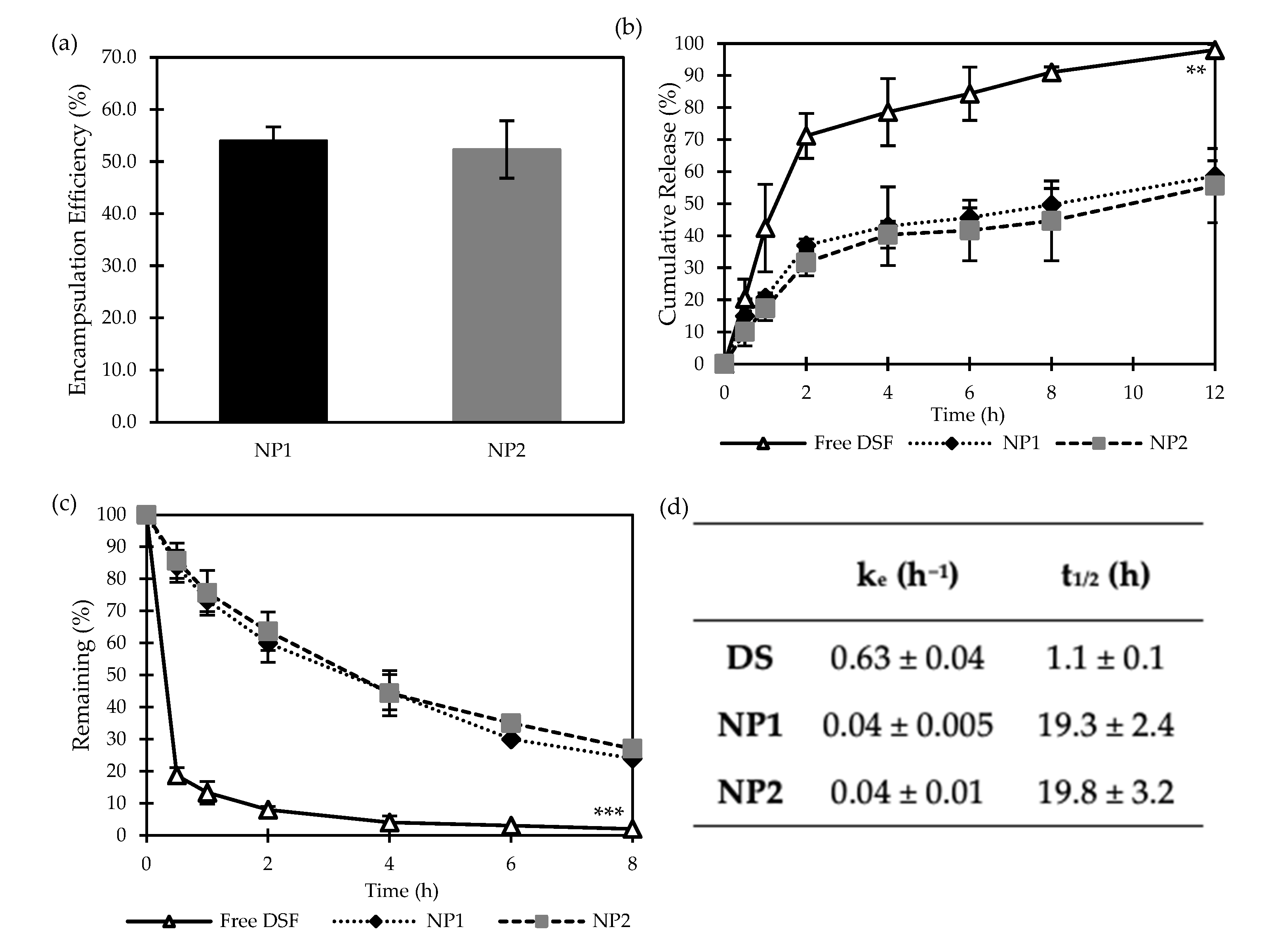
3.1.2. Encapsulation Efficiency and Cumulative Release Studies of DS-Loaded PLGA NPs
3.2. Disulfiram-Loaded PLGA PEGylated Nanoparticle Formulations by Direct Nanoprecipitation Method
3.2.1. Particle Size, Polydispersity Index, Zeta Potential and Surface Morphology
3.2.2. Encapsulation Efficiency and Cumulative Release Studies of DS-Loaded PLGA NPs
3.2.3. Stability Studies in Horse Serum
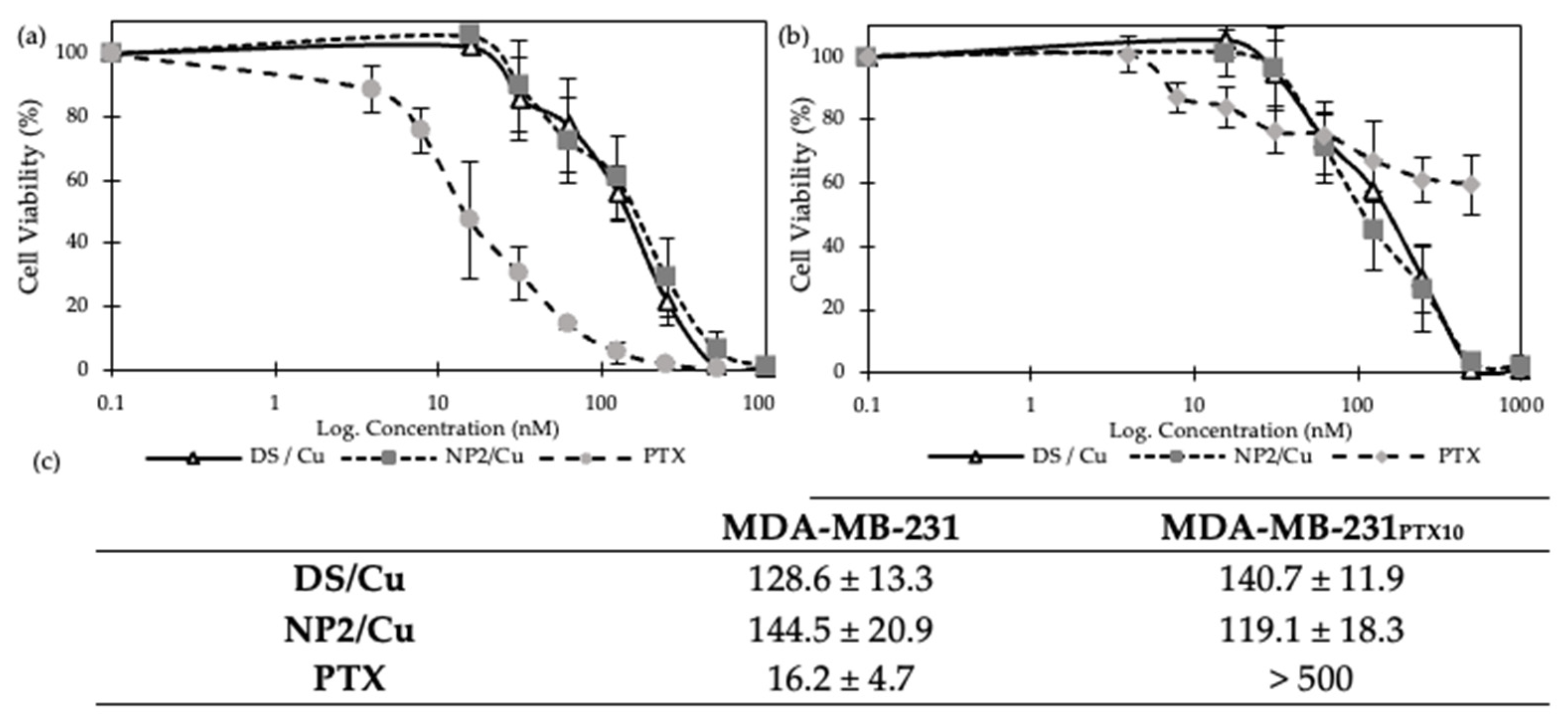
3.2.4. MTT Cytotoxicity Assay
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kashyap, D.; Pal, D.; Sharma, R.; Garg, V.K.; Goel, N.; Koundal, D.; Zaguia, A.; Koundal, S.; Belay, A. Global Increase in Breast Cancer Incidence: Risk Factors and Preventive Measures. BioMed Res. Int. 2022, 2022, 9605439. [Google Scholar] [CrossRef]
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2022, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Dewanjee, S.; Li, Y.; Jha, N.K.; Chen, Z.S.; Kumar, A.; Vishakha; Behl, T.; Jha, S.K.; Tang, H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer 2023, 22, 105. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Kim, D.; Ko, S.; Kim, A.; Mo, K.; Yoon, H. Breast Cancer Metastasis: Mechanisms and Therapeutic Implications. Int. J. Mol. Sci. 2022, 23, 6806. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Kaya, A.; Alves, D.; Ansari-Fard, N.; Tolaymat, I.; Arafat, B.; Najlah, M. Preparation and Characterization of Disulfiram and Beta Cyclodextrin Inclusion Complexes for Potential Application in the Treatment of SARS-CoV-2 via Nebulization. Molecules 2022, 27, 5600. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, X.; Lou, H.; Jiang, M.; Wu, X.; Qin, J.; Zhang, J.; Guan, X.; Li, W.; Zhang, W.; et al. Buffet-style Cu(II) for enhance disulfiram-based cancer therapy. J. Colloid Interface Sci. 2022, 624, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.K.; Seo, S.Y.; Kim, H.J.; Chie, E.K.; Kim, S.K.; Kim, I.H. Disulfiram, a Re-positioned Aldehyde Dehydrogenase Inhibitor, Enhances Radiosensitivity of Human Glioblastoma Cells In Vitro. Cancer Res. Treat. 2019, 51, 696–705. [Google Scholar] [CrossRef]
- Yip, N.C.; Fombon, I.S.; Liu, P.; Brown, S.; Kannappan, V.; Armesilla, A.L.; Xu, B.; Cassidy, J.; Darling, J.L.; Wang, W. Disulfiram modulated ROS–MAPK and NFκB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br. J. Cancer 2011, 104, 1564–1574. [Google Scholar] [CrossRef]
- Roudi, R.; Korourian, A.; Shariftabrizi, A.; Madjd, Z. Differential Expression of Cancer Stem Cell Markers ALDH1 and CD133 in Various Lung Cancer Subtypes. Cancer Investig. 2015, 33, 294–302. [Google Scholar] [CrossRef]
- Paranjpe, A.; Zhang, R.; Ali-Osman, F.; Bobustuc, G.C.; Srivenugopal, K.S. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis 2014, 35, 692–702. [Google Scholar] [CrossRef]
- Sharma, V.; Verma, V.; Lal, N.; Yadav, S.K.; Sarkar, S.; Mandalapu, D.; Porwal, K.; Rawat, T.; Maikhuri, J.P.; Rajender, S.; et al. Disulfiram and its novel derivative sensitize prostate cancer cells to the growth regulatory mechanisms of the cell by re-expressing the epigenetically repressed tumor suppressor—Estrogen receptor β. Mol. Carcinog. 2016, 55, 1843–1857. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; Wang, L.H.; Li, Y.; Fu, S.Y.; Xue, X.; Jia, L.N.; Yuan, X.Z.; Wang, Y.T.; Tang, X.; Yang, J.Y.; et al. Targeting ALDH2 with disulfiram/copper reverses the resistance of cancer cells to microtubule inhibitors. Exp. Cell Res. 2018, 362, 72–82. [Google Scholar] [CrossRef]
- Suliman, A.S.; Khoder, M.; Tolaymat, I.; Webster, M.; Alany, R.G.; Wang, W.; Elhissi, A.; Najlah, M. Cyclodextrin Diethyldithiocarbamate Copper II Inclusion Complexes: A Promising Chemotherapeutic Delivery System against Chemoresistant Triple Negative Breast Cancer Cell Lines. Pharmaceutics 2021, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Najlah, M.; Suliman, A.S.; Tolaymat, I.; Kurusamy, S.; Kannappan, V.; Elhissi, A.M.A.; Wang, W. Development of Injectable PEGylated Liposome Encapsulating Disulfiram for Colorectal Cancer Treatment. Pharmaceutics 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Sun, X.; Ma, L.; Li, C.; Xu, Z.; Ma, W.; Zhou, Y.; Zhao, Z.; Ma, D. Therapeutic effect of disulfiram inclusion complex embedded in hydroxypropyl-β-cyclodextrin on intracranial glioma-bearing male rats via intranasal route. Eur. J. Pharm. Sci. 2021, 156, 105590. [Google Scholar] [CrossRef] [PubMed]
- Najlah, M.; Ahmed, Z.; Iqbal, M.; Wang, Z.; Tawari, P.; Wang, W.; McConville, C. Development and characterisation of disulfiram-loaded PLGA nanoparticles for the treatment of non-small cell lung cancer. Eur. J. Pharm. Biopharm. 2017, 112, 224–233. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.; Chen, W.; Li, F. Old wine in new bottles: Advanced drug delivery systems for disulfiram-based cancer therapy. J. Control Release 2020, 319, 352–359. [Google Scholar] [CrossRef]
- Wang, Z.; Tan, J.; McConville, C.; Kannappan, V.; Tawari, P.E.; Brown, J.; Ding, J.; Armesilla, A.L.; Irache, J.M.; Mei, Q.B.; et al. Poly lactic-co-glycolic acid controlled delivery of disulfiram to target liver cancer stem-like cells. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 641–657. [Google Scholar] [CrossRef]
- Madala, H.R.; Punganuru, S.R.; Ali-Osman, F.; Zhang, R.; Srivenugopal, K.S. Brain- and brain tumor-penetrating disulfiram nanoparticles: Sequence of cytotoxic events and efficacy in human glioma cell lines and intracranial xenografts. Oncotarget 2018, 9, 3459–3482. [Google Scholar] [CrossRef]
- Song, W.; Tang, Z.; Lei, T.; Wen, X.; Wang, G.; Zhang, D.; Deng, M.; Tang, X.; Chen, X. Stable loading and delivery of disulfiram with mPEG-PLGA/PCL mixed nanoparticles for tumor therapy. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 377–386. [Google Scholar] [CrossRef]
- Kannappan, V.; Liu, Y.; Wang, Z.; Azar, K.; Kurusamy, S.; Kilari, R.S.; Armesilla, A.L.; Morris, M.R.; Najlah, M.; Liu, P.; et al. PLGA–Nano-Encapsulated Disulfiram Inhibits Hypoxia-Induced NF-κB, Cancer Stem Cells, and Targets Glioblastoma In Vitro and In Vivo. Mol. Cancer Ther. 2022, 21, 1273–1284. [Google Scholar] [CrossRef]
- Elnawasany, S.; Haggag, Y.A.; Shalaby, S.M.; Soliman, N.A.; Saadany, A.A.; Ibrahim, M.A.A.; Badria, F. Anti-cancer effect of nano-encapsulated boswellic acids, curcumin and naringenin against HepG-2 cell line. BMC Complement. Med. Ther. 2023, 23, 270. [Google Scholar] [CrossRef] [PubMed]
- Bouaouina, H.; Desrumaux, A.; Loisel, C.; Legrand, J. Functional properties of whey proteins as affected by dynamic high-pressure treatment. Int. Dairy J. 2006, 16, 275–284. [Google Scholar] [CrossRef]
- Sharma, S.; Sahni, J.K.; Ali, J.; Baboota, S. Effect of high-pressure homogenization on formulation of TPGS loaded nanoemulsion of rutin—Pharmacodynamic and antioxidant studies. Drug Deliv. 2015, 22, 541–551. [Google Scholar] [CrossRef]
- O’Donnell, P.B.; McGinity, J.W. Preparation of microspheres by the solvent evaporation technique. Adv. Drug Deliv. Rev. 1997, 28, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Fasehee, H.; Zarrinrad, G.; Tavangar, S.M.; Ghaffari, S.H.; Faghihi, S. The inhibitory effect of disulfiram encapsulated PLGA NPs on tumor growth: Different administration routes. Mater. Sci. Eng. C 2016, 63, 587–595. [Google Scholar] [CrossRef]
- Ou, A.T.; Zhang, J.X.; Fang, Y.f.; Wang, R.; Tang, X.P.; Zhao, P.F.; Zhao, Y.G.; Zhang, M.; Huang, Y.Z. Disulfiram-loaded lactoferrin nanoparticles for treating inflammatory diseases. Acta Pharmacol. Sin. 2021, 42, 1913–1920. [Google Scholar] [CrossRef]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent Advances in Repurposing Disulfiram and Disulfiram Derivatives as Copper-Dependent Anticancer Agents. Front. Mol. Biosci. 2021, 8, 41316. [Google Scholar] [CrossRef]
- Lanz, J.; Biniaz-Harris, N.; Kuvaldina, M.; Jain, S.; Lewis, K.; Fallon, B.A. Disulfiram: Mechanisms, Applications, and Challenges. Antibiotics 2023, 12, 524. [Google Scholar] [CrossRef]



| Formulation * | PLGA | mPEG2k-PLGA | DS | Total |
|---|---|---|---|---|
| NP1E | 10 | -- | -- | 10 |
| NP1 | 9 | -- | 1 | 10 |
| NP2E | 9.5 | 0.5 | -- | 10 |
| NP2 | 8.5 | 0.5 | 1 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumbuya, I.; Pereira, A.M.; Tolaymat, I.; Al Dalaty, A.; Arafat, B.; Webster, M.; Pierscionek, B.; Khoder, M.; Najlah, M. Exploring Disulfiram’s Anticancer Potential: PLGA Nano-Carriers for Prolonged Drug Delivery and Potential Improved Therapeutic Efficacy. Nanomaterials 2024, 14, 1133. https://doi.org/10.3390/nano14131133
Dumbuya I, Pereira AM, Tolaymat I, Al Dalaty A, Arafat B, Webster M, Pierscionek B, Khoder M, Najlah M. Exploring Disulfiram’s Anticancer Potential: PLGA Nano-Carriers for Prolonged Drug Delivery and Potential Improved Therapeutic Efficacy. Nanomaterials. 2024; 14(13):1133. https://doi.org/10.3390/nano14131133
Chicago/Turabian StyleDumbuya, Ibrahim, Ana Maria Pereira, Ibrahim Tolaymat, Adnan Al Dalaty, Basel Arafat, Matt Webster, Barbara Pierscionek, Mouhamad Khoder, and Mohammad Najlah. 2024. "Exploring Disulfiram’s Anticancer Potential: PLGA Nano-Carriers for Prolonged Drug Delivery and Potential Improved Therapeutic Efficacy" Nanomaterials 14, no. 13: 1133. https://doi.org/10.3390/nano14131133
APA StyleDumbuya, I., Pereira, A. M., Tolaymat, I., Al Dalaty, A., Arafat, B., Webster, M., Pierscionek, B., Khoder, M., & Najlah, M. (2024). Exploring Disulfiram’s Anticancer Potential: PLGA Nano-Carriers for Prolonged Drug Delivery and Potential Improved Therapeutic Efficacy. Nanomaterials, 14(13), 1133. https://doi.org/10.3390/nano14131133







