Dynamic Clustering and Scaling Behavior of Active Particles under Confinement
Abstract
:1. Introduction
2. Methods and Models
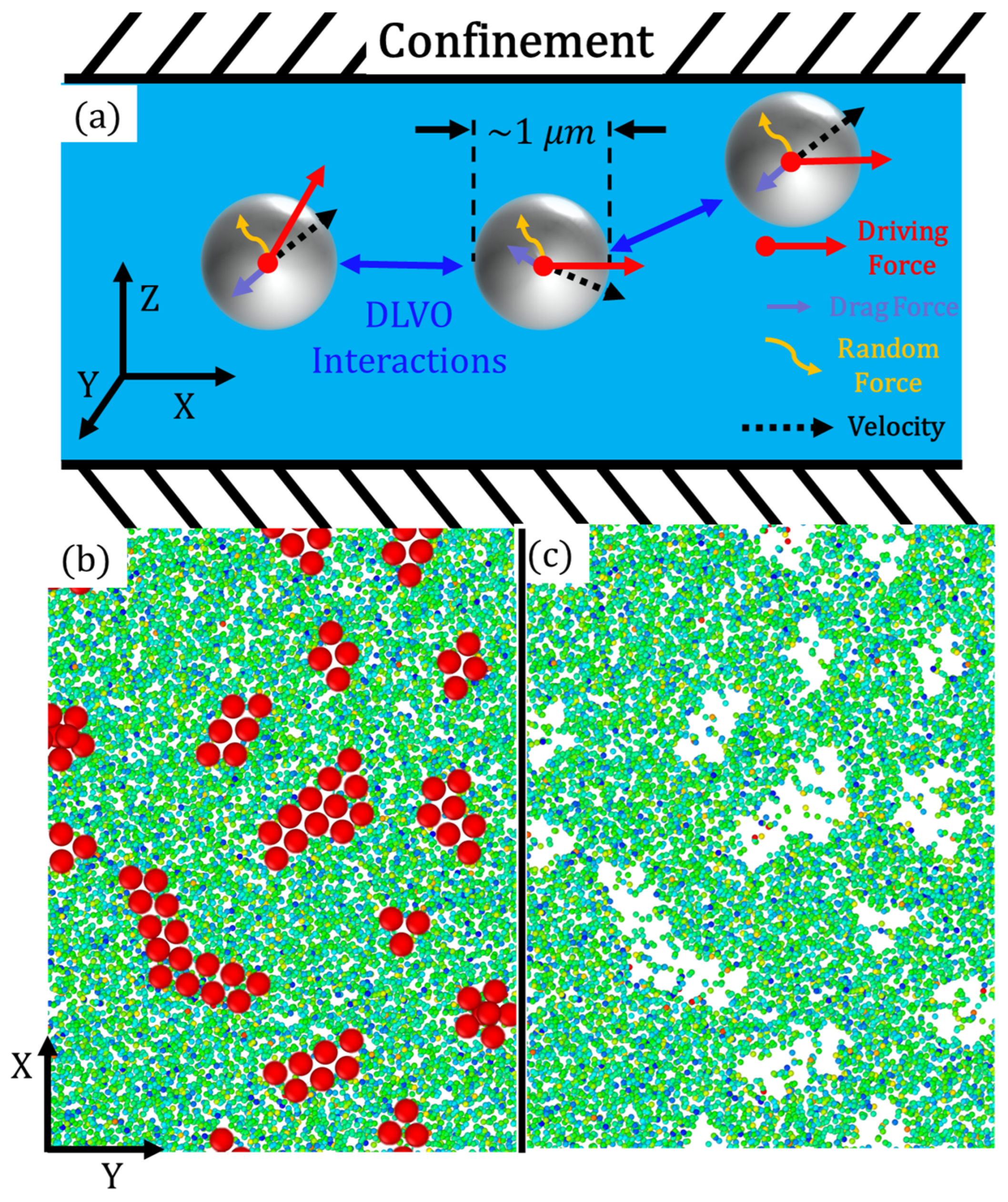
2.1. SRD—Explicit Hydrodynamic Solvent
2.2. Coarse-Grained Molecular Dynamics (CGMD) Active Particles and DLVO Potential
3. Results and Discussion
3.1. Dynamic Clustering Behavior
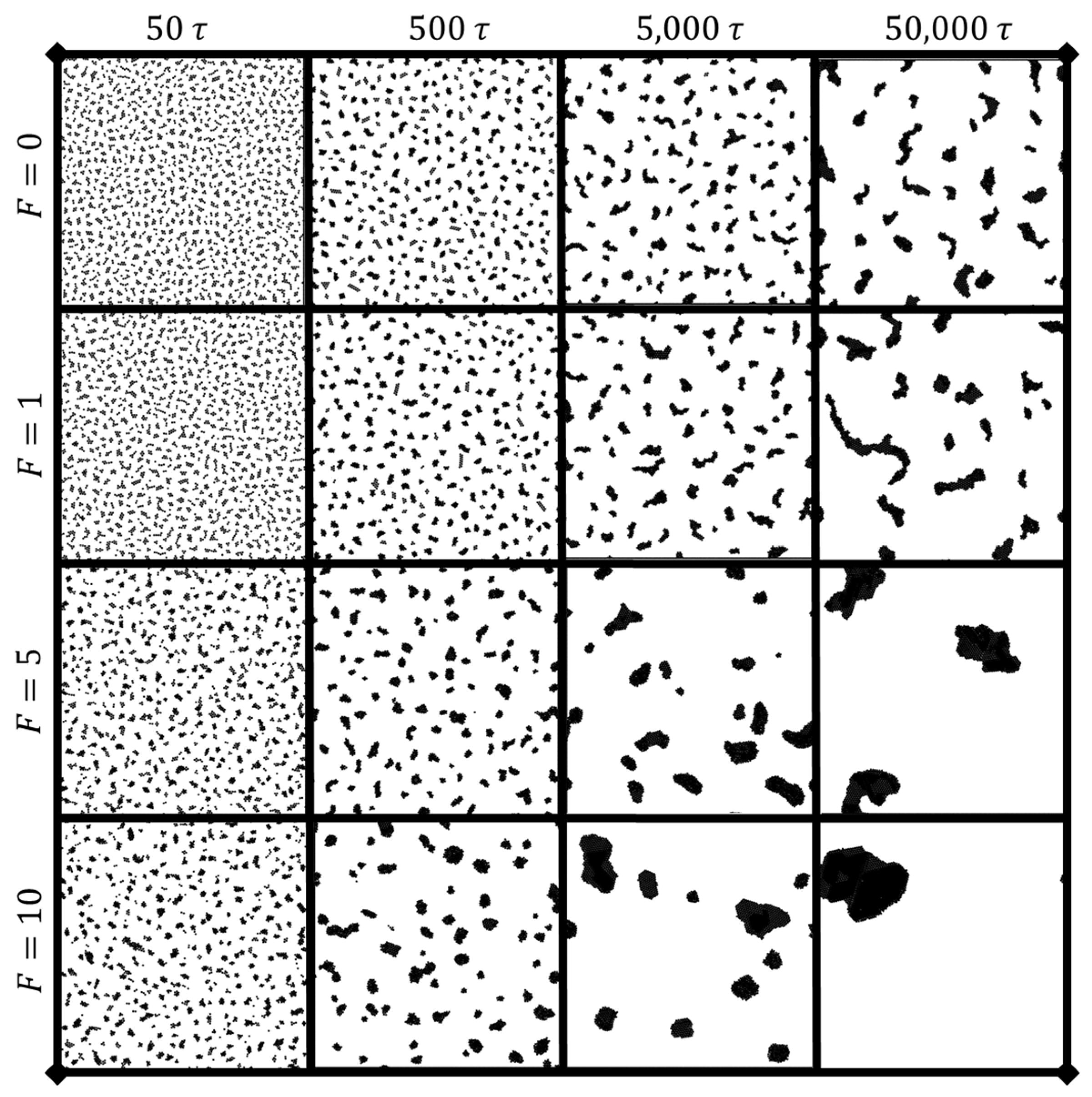
3.1.1. Time Evolution of Cluster Number and Cluster Size
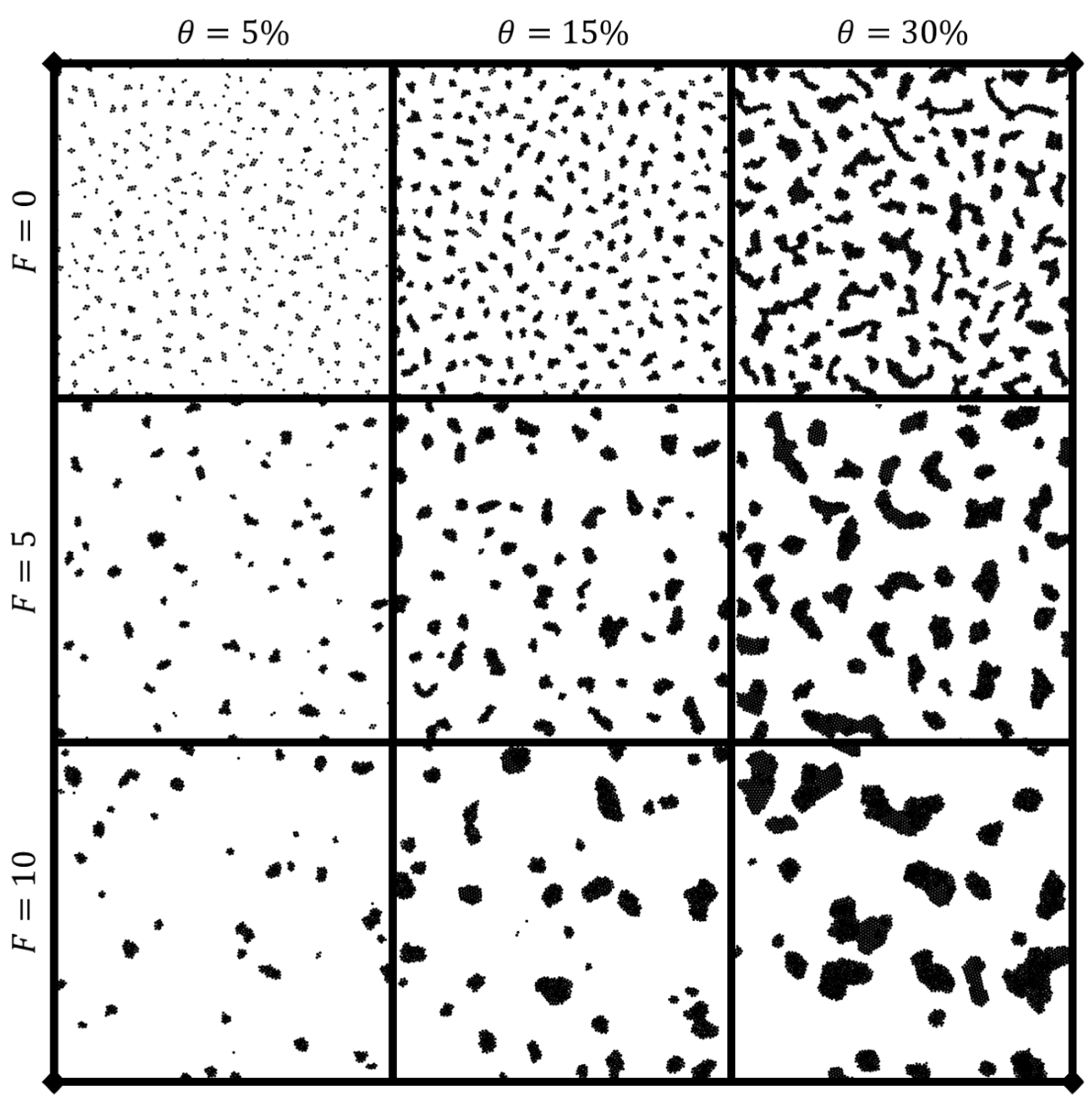
3.1.2. Cluster Dynamic Spatial Distribution
3.1.3. Phase Diagram of Dynamic Characteristics
3.2. Scaling Laws and Relationships
3.2.1. Particle Coverage-Governed Scaling
3.2.2. Driving Force-Governed Scaling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cui, X.L.; Li, J.; Ng, D.H.L.; Liu, J.; Liu, Y.; Yang, W.N. 3D hierarchical ACFs-based micromotors as efficient photo-Fentonlike catalysts. Carbon 2020, 158, 738–748. [Google Scholar] [CrossRef]
- Jurado-Sanchez, B.; Pacheco, M.; Rojo, J.; Escarpa, A. Magnetocatalytic Graphene Quantum Dots Janus Micromotors for Bacterial Endotoxin Detection. Angew. Chem. Int. Edit. 2017, 56, 6957–6961. [Google Scholar] [CrossRef] [PubMed]
- de Avila, B.E.F.; Lopez-Ramirez, M.A.; Baez, D.F.; Jodra, A.; Singh, V.V.; Kaufmann, K.; Wang, J. Aptamer-Modified Graphene-Based Catalytic Micromotors: Off-On Fluorescent Detection of Ricin. ACS Sens. 2016, 1, 217–221. [Google Scholar] [CrossRef]
- Cai, L.J.; Wang, H.; Yu, Y.R.; Bian, F.K.; Wang, Y.; Shi, K.Q.; Ye, F.F.; Zhao, Y.J. Stomatocyte structural color-barcode micromotors for multiplex assays. Natl. Sci. Rev. 2020, 7, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mou, F.Z.; Chen, C.R.; You, M.; Yin, Y.X.; Xu, L.L.; Guan, J.G. Light-controlled bubble propulsion of amorphous TiO2/Au Janus micromotors. Rsc. Adv. 2016, 6, 10697–10703. [Google Scholar] [CrossRef]
- Rojas, D.; Jurado-Sanchez, B.; Escarpa, A. “Shoot and Sense” Janus Micromotors-Based Strategy for the Simultaneous Degradation and Detection of Persistent Organic Pollutants in Food and Biological Samples. Anal. Chem. 2016, 88, 4153–4160. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Zhao, A.D.; Wang, F.M.; Ren, J.S.; Qu, X.G. Design of a plasmonic micromotor for enhanced photo-remediation of polluted anaerobic stagnant waters. Chem. Commun. 2016, 52, 5550–5553. [Google Scholar] [CrossRef]
- Delezuk, J.A.M.; Ramirez-Herrera, D.E.; de Avila, B.E.F.; Wang, J. Chitosan-based water-propelled micromotors with strong antibacterial activity. Nanoscale 2017, 9, 2195–2200. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Guix, M.; Schmidt, O.G. Wastewater Mediated Activation of Micromotors for Efficient Water Cleaning. Nano Lett. 2016, 16, 817–821. [Google Scholar] [CrossRef]
- Wybieralska, K.; Wajda, A. Removal of organic dyes from aqueous solutions with surfactant-modified magnetic nanoparticles. Pol. J. Chem. Technol. 2014, 16, 27–30. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Yeom, J. Pollutant-Degrading Multifunctional Micromotors. In Proceedings of the 12th IEEE International Conference on Nano/Micro Engineered and Molecular Systems, Los Angeles, CA, USA, 9–12 April 2017; pp. 16–20. [Google Scholar]
- Lee, C.S.; Gong, J.Y.; Oh, D.S.; Jeon, J.R.; Chang, Y.S. Zerovalent-Iron/Platinum Janus Micromotors with Spatially Separated Functionalities for Efficient Water Decontamination. ACS Appl. Nano Mater. 2018, 1, 768–776. [Google Scholar] [CrossRef]
- Uygun, M.; Singh, V.V.; Kaufmann, K.; Uygun, D.A.; de Oliveira, S.D.S.; Wang, J. Micromotor-Based Biomimetic Carbon Dioxide Sequestration: Towards Mobile Microscrubbers. Angew. Chem. Int. Edit. 2015, 54, 12900–12904. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.V.; Wang, J. Nano/micromotors for security/defense applications. A review. Nanoscale 2015, 7, 19377–19389. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.V.; Martin, A.; Kaufmann, K.; de Oliveira, S.D.S.; Wang, J. Zirconia/Graphene Oxide Hybrid Micromotors for Selective Capture of Nerve Agents. Chem. Mater. 2015, 27, 8162–8169. [Google Scholar] [CrossRef]
- Singh, V.V.; Jurado-Sanchez, B.; Sattayasamitsathit, S.; Orozco, J.; Li, J.X.; Galarnyk, M.; Fedorak, Y.; Wang, J. Multifunctional Silver-Exchanged Zeolite Micromotors for Catalytic Detoxification of Chemical and Biological Threats. Adv. Funct. Mater. 2015, 25, 2147–2155. [Google Scholar] [CrossRef]
- Singh, V.V.; Kaufmann, K.; Orozco, J.; Li, J.X.; Galarnyk, M.; Arya, G.; Wang, J. Micromotor-based on-off fluorescence detection of sarin and soman simulants. Chem. Commun. 2015, 51, 11190–11193. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Ajalloueian, F.; Boisen, A. Thread-Like Radical-Polymerization via Autonomously Propelled (TRAP) Bots. Adv. Mater. 2019, 31, 1901573. [Google Scholar] [CrossRef]
- Chang, X.C.; Chen, C.R.; Li, J.X.; Lu, X.L.; Liang, Y.Y.; Zhou, D.K.; Wang, H.C.; Zhang, G.Y.; Li, T.L.; Wang, J.; et al. Motile Micropump Based on Synthetic Micromotors for Dynamic Micropatterning. Acs. Appl. Mater. Inter. 2019, 11, 28507–28514. [Google Scholar] [CrossRef]
- Singh, V.V.; Soto, F.; Kaufmann, K.; Wang, J. Micromotor-Based Energy Generation. Angew. Chem. Int. Edit. 2015, 54, 6896–6899. [Google Scholar] [CrossRef]
- Rotariu, O.; Udrea, L.E.; Strachan, N.J.C.; Heys, S.D.; Schofield, A.C.; Gilbert, F.J.; Badescu, V. Studies of magnetic carrier particles capture for blood vessel embolization. J. Optoelectron. Adv. Mater. 2006, 8, 1758–1760. [Google Scholar]
- Udrea, L.E.; Rotariu, O.; Popa, M.I. Magnetic Support Nanoparticles for the Targeting of Drugs: Physical Characterization and Manipulation in Magnetic Field. Sci. Study Res.-Chem. C 2006, 7, 151–156. [Google Scholar]
- Alexiou, C.; Tietze, R.; Schreiber, E.; Lyer, S. Nanomedicine. Bundesgesundheitsbla 2010, 53, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, R.; Tosi, G.; Grabrucker, A.M. Emerging Use of Nanotechnology in the Treatment of Neurological Disorders. Curr. Pharm. Design 2015, 21, 3111–3130. [Google Scholar] [CrossRef] [PubMed]
- Talelli, M.; Aires, A.; Marciello, M. Protein-modified Magnetic Nanoparticles for Biomedical Applications. Curr. Org. Chem. 2016, 20, 1252–1261. [Google Scholar] [CrossRef]
- Choi, A.; Seo, K.D.; Kim, D.W.; Kim, B.C.; Kim, D.S. Recent advances in engineering microparticles and their nascent utilization in biomedical delivery and diagnostic applications. Lab. Chip. 2017, 17, 591–613. [Google Scholar] [CrossRef] [PubMed]
- Karshalev, E.; de Avila, B.E.F.; Beltran-Gastelum, M.; Angsantikul, P.; Tang, S.S.; Mundaca-Uribe, R.; Zhang, F.Y.; Zhao, J.; Zhang, L.F.; Wang, J. Micromotor Pills as a Dynamic Oral Delivery Platform. ACS Nano 2018, 12, 8397–8405. [Google Scholar] [CrossRef] [PubMed]
- Bing, X.M.; Zhang, X.L.; Li, J.; Ng, D.H.L.; Yang, W.N.; Yang, J. 3D hierarchical tubular micromotors with highly selective recognition and capture for antibiotics. J. Mater. Chem. A 2020, 8, 2809–2819. [Google Scholar] [CrossRef]
- Yu, Y.R.; Guo, J.H.; Wang, Y.T.; Shao, C.M.; Wang, Y.; Zhao, Y.J. Bioinspired Helical Micromotors as Dynamic Cell Microcarriers. ACS Appl. Mater. Inter. 2020, 12, 16097–16103. [Google Scholar] [CrossRef]
- Tang, S.S.; Zhang, F.Y.; Gong, H.; Wei, F.N.; Zhuang, J.; Karshalev, E.; de Avila, B.E.F.; Huang, C.Y.; Zhou, Z.D.; Li, Z.X.; et al. Enzyme-powered Janus platelet cell robots for active and targeted drug delivery. Sci. Robot. 2020, 5, eaba6137. [Google Scholar] [CrossRef]
- Jiang, W.S.; Ma, L.; Xu, X.B. Recent progress on the design and fabrication of micromotors and their biomedical applications. Bio.-Des. Manuf. 2018, 1, 225–236. [Google Scholar] [CrossRef]
- Cui, Q.H.; Le, T.H.; Lin, Y.J.; Miao, Y.B.; Sung, I.T.; Tsai, W.B.; Chan, H.Y.; Lin, Z.H.; Sung, H.W. A self-powered battery-driven drug delivery device that can function as a micromotor and galvanically actuate localized payload release. Nano. Energy 2019, 66, 104120. [Google Scholar] [CrossRef]
- Li, M.L.; Brinkmann, M.; Pagonabarraga, I.; Seemann, R.; Fleury, J.B. Spatiotemporal control of cargo delivery performed by programmable self-propelled Janus droplets. Commun. Phys. 2018, 1, 23. [Google Scholar] [CrossRef]
- Dong, Y.; Yi, C.; Yang, S.S.; Wang, J.; Chen, P.; Liu, X.; Du, W.; Wang, S.; Liu, B.F. A substrate-free graphene oxide-based micromotor for rapid adsorption of antibiotics. Nanoscale 2019, 11, 4562–4570. [Google Scholar] [CrossRef]
- Yoon, M.; Borrmann, P.; Tomanek, D. Targeted medication delivery using magnetic nanostructures. J. Phys.-Condens. Mat. 2007, 19, 086210. [Google Scholar] [CrossRef]
- Li, J.; Ji, F.; Ng, D.H.L.; Liu, J.; Bing, X.M.; Wang, P. Bioinspired Pt-free molecularly imprinted hydrogel-based magnetic Janus micromotors for temperature-responsive recognition and adsorption of erythromycin in water. Chem. Eng. J. 2019, 369, 611–620. [Google Scholar] [CrossRef]
- Tietze, R.; Lyer, S.; Schreiber, E.; Mann, J.; Durr, S.; Alexiou, C. Local Cancer Therapy with Magnetic Nanoparticles. Else. Kroner. Fresen. S 2011, 2, 154–164. [Google Scholar]
- Hu, J.N.; Huang, S.W.; Zhu, L.; Huang, W.J.; Zhao, Y.P.; Jin, K.L.; ZhuGe, Q.C. Tissue Plasminogen Activator-Porous Magnetic Microrods for Targeted Thrombolytic Therapy after Ischemic Stroke. ACS Appl. Mater. Inter. 2018, 10, 32988–32997. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.M.; Fan, X.J.; Meng, X.H.; Song, J.M.; Chen, W.N.; Sun, L.N.; Xie, H. Magnetic biohybrid micromotors with high maneuverability for efficient drug loading and targeted drug delivery. Nanoscale 2019, 11, 18382–18392. [Google Scholar] [CrossRef]
- Fridjonsson, E.O.; Seymour, J.D. Colloid particle transport in a microcapillary: NMR study of particle and suspending fluid dynamics. Chem. Eng. Sci. 2016, 153, 165–173. [Google Scholar] [CrossRef]
- Udrea, L.E.; Strachan, N.J.C.; Badescu, V.; Rotariu, O. An in vitro study of magnetic particle targeting in small blood vessels. Phys. Med. Biol. 2006, 51, 4869–4881. [Google Scholar] [CrossRef]
- Wang, J.W. Anomalous Diffusion of Active Brownian Particles in Crystalline Phases. IOP Conf. Ser. Earth Environ. 2019, 237, 052005. [Google Scholar] [CrossRef]
- Fodor, E.; Nemoto, T.; Vaikuntanathan, S. Dissipation controls transport and phase transitions in active fluids: Mobility, diffusion and biased ensembles. New J. Phys. 2020, 22, 013052. [Google Scholar] [CrossRef]
- Maloney, R.C.; Liao, G.J.; Klapp, S.H.L.; Hall, C.K. Clustering and phase separation in mixtures of dipolar and active particles. Soft. Matter. 2020, 16, 3779–3791. [Google Scholar] [CrossRef] [PubMed]
- Czirok, A.; Vicsek, T. Collective behavior of interacting self-propelled particles. Phys. A 2000, 281, 17–29. [Google Scholar] [CrossRef]
- Deutsch, A.; Theraulaz, G.; Vicsek, T. Collective motion in biological systems. Interface Focus 2012, 2, 689–692. [Google Scholar] [CrossRef]
- Mehes, E.; Vicsek, T. Collective motion of cells: From experiments to models. Integr. Biol. 2014, 6, 831–854. [Google Scholar] [CrossRef]
- Ferdinandy, B.; Ozogany, K.; Vicsek, T. Collective motion of groups of self-propelled particles following interacting leaders. Phys. A 2017, 479, 467–477. [Google Scholar] [CrossRef]
- Czirok, A.; Vicsek, M.; Vicsek, T. Collective motion of organisms in three dimensions. Phys. A 1999, 264, 299–304. [Google Scholar] [CrossRef]
- Czirok, A.; Barabasi, A.L.; Vicsek, T. Collective motion of self-propelled particles: Kinetic phase transition in one dimension. Phys. Rev. Lett. 1999, 82, 209–212. [Google Scholar] [CrossRef]
- Sherman, Z.M.; Swan, J.W. Transmutable Colloidal Crystals and Active Phase Separation via Dynamic, Directed Self-Assembly with Toggled External Fields. ACS Nano 2019, 13, 764–771. [Google Scholar] [CrossRef]
- Alapan, Y.; Yigit, B.; Beker, O.; Demirors, A.F.; Sitti, M. Shape-encoded dynamic assembly of mobile micromachines. Nat. Mater. 2019, 18, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Soheilian, R.; Abdi, H.; Maloney, C.E.; Erb, R.M. Assembling particle clusters with incoherent 3D magnetic fields. J. Colloid Interf. Sci. 2018, 513, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Mallory, S.A.; Valeriani, C.; Cacciuto, A. An Active Approach to Colloidal Self-Assembly. Annu. Rev. Phys. Chem. 2018, 69, 59–79. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.Y.; Wurger, A.; Lintuvuori, J.S. Hydrodynamic self-assembly of active colloids: Chiral spinners and dynamic crystals. Soft. Matter. 2019, 15, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Gaspard, P.; Kapral, R. Active Matter, Microreversibility, and Thermodynamics. Res. China 2020, 2020, 9739231. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Adhikari, R. Generalized Stokes laws for active colloids and their applications. J. Phys. Commun. 2018, 2, 025025. [Google Scholar] [CrossRef]
- Theurkauff, I.; Cottin-Bizonne, C.; Palacci, J.; Ybert, C.; Bocquet, L. Dynamic Clustering in Active Colloidal Suspensions with Chemical Signaling. Phys. Rev. Lett. 2012, 108, 268303. [Google Scholar] [CrossRef]
- Laskar, A.; Manna, R.K.; Shklyaev, O.E.; Balazs, A.C. Modeling the biomimetic self-organization of active objects in fluids. Nano Today 2019, 29, 100804. [Google Scholar] [CrossRef]
- Theers, M.; Westphal, E.; Qi, K.; Winkler, R.G.; Gompper, G. Clustering of microswimmers: Interplay of shape and hydrodynamics. Soft Matter. 2018, 14, 8590–8603. [Google Scholar] [CrossRef]
- Kanso, E.; Michelin, S. Phoretic and hydrodynamic interactions of weakly confined autophoretic particles. J. Chem. Phys. 2019, 150, 044902. [Google Scholar] [CrossRef]
- Popescu, M.N.; Uspal, W.E.; Eskandari, Z.; Tasinkevych, M.; Dietrich, S. Effective squirmer models for self-phoretic chemically active spherical colloids. Eur. Phys. J. E 2018, 41, 145. [Google Scholar] [CrossRef]
- Kuron, M.; Kreissl, P.; Holm, C. Toward Understanding of Self-Electrophoretic Propulsion under Realistic Conditions: From Bulk Reactions to Confinement Effects. Accounts Chem. Res. 2018, 51, 2998–3005. [Google Scholar] [CrossRef] [PubMed]
- Popescu, M.N.; Uspal, W.E.; Dominguez, A.; Dietrich, S. Effective Interactions between Chemically Active Colloids and Interfaces. Acc. Chem. Res. 2018, 51, 2991–2997. [Google Scholar] [CrossRef]
- Zottl, A.; Stark, H. Emergent behavior in active colloids. J. Phys.-Condens Mat. 2016, 28, 253001. [Google Scholar] [CrossRef]
- Delfau, J.B.; Molina, J.; Sano, M. Collective behavior of strongly confined suspensions of squirmers. Epl.-Europhys. Lett. 2016, 114, 24001. [Google Scholar] [CrossRef]
- Thutupalli, S.; Geyer, D.; Singh, R.; Adhikari, R.; Stone, H.A. Flow-induced phase separation of active particles is controlled by boundary conditions. Proc. Natl. Acad. Sci. USA 2018, 115, 5403–5408. [Google Scholar] [CrossRef]
- Theillard, M.; Alonso-Matilla, R.; Saintillan, D. Geometric control of active collective motion. Soft Matter 2017, 13, 363–375. [Google Scholar] [CrossRef]
- Lei, L.J.; Wang, S.; Zhang, X.Y.; Lai, W.J.; Wu, J.Y.; Gao, Y.X. Phoretic self-assembly of active colloidal molecules*. Chin. Phys. B 2021, 30, 056112. [Google Scholar] [CrossRef]
- Soto, R.; Golestanian, R. Self-Assembly of Catalytically Active Colloidal Molecules: Tailoring Activity through Surface Chemistry. Phys. Rev. Lett. 2014, 112, 068301. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, J.; Granick, S. Directed Self-Assembly Pathways of Active Colloidal Clusters. Angew. Chem. Int. Edit. 2016, 55, 5166–5169. [Google Scholar] [CrossRef]
- Ebbens, S.J.; Gregory, D.A. Catalytic Janus Colloids: Controlling Trajectories of Chemical Microswimmers. Acc. Chem. Res. 2018, 51, 1931–1939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lei, H.; Atzberger, P.J. Fluctuating hydrodynamic methods for fluid-structure interactions in confined channel geometries. Appl. Math Mech.-Engl. 2018, 39, 125–152. [Google Scholar] [CrossRef]
- Zhang, J.; Luijten, E.; Grzybowski, B.A.; Granick, S. Active colloids with collective mobility status and research opportunities. Chem. Soc. Rev. 2017, 46, 5551–5569. [Google Scholar] [CrossRef] [PubMed]
- Shaebani, M.R.; Wysocki, A.; Winkler, R.G.; Gompper, G.; Rieger, H. Computational models for active matter. Nat. Rev. Phys. 2020, 2, 181–199. [Google Scholar] [CrossRef]
- Winkler, A.; Winter, D.; Chaudhuri, P.; Statt, A.; Virnau, P.; Horbach, J.; Binder, K. Computer simulations of structure, dynamics, and phase behavior of colloidal fluids in confined geometry and under shear. Eur. Phys. J.-Spec. Top. 2013, 222, 2787–2801. [Google Scholar] [CrossRef]
- Puertas, A.M.; Voigtmann, T. Microrheology of colloidal systems. J. Phys.-Condens Mat. 2014, 26, 243101. [Google Scholar] [CrossRef]
- Tierno, P. Recent advances in anisotropic magnetic colloids: Realization, assembly and applications. Phys. Chem. Chem. Phys. 2014, 16, 23515–23528. [Google Scholar] [CrossRef]
- Foss, D.R.; Brady, J.F. Structure, diffusion and rheology of Brownian suspensions by Stokesian Dynamics simulation. J. Fluid Mech. 2000, 407, 167–200. [Google Scholar] [CrossRef]
- Malevanets, A.; Kapral, R. Mesoscopic model for solvent dynamics. J. Chem. Phys. 1999, 110, 8605–8613. [Google Scholar] [CrossRef]
- Hoskins, P.R.; Hardman, D. Three-dimensional imaging and computational modelling for estimation of wall stresses in arteries. Brit. J. Radiol. 2009, 82, S3–S17. [Google Scholar] [CrossRef]
- Plimpton, S. Fast Parallel Algorithms for Short-Range Molecular-Dynamics. J. Comput. Phys. 1995, 117, 1039. [Google Scholar] [CrossRef]
- Lin, M.Y.; Lindsay, H.M.; Weitz, D.A.; Klein, R.; Ball, R.C.; Meakin, P. Universal Diffusion-Limited Colloid Aggregation. J. Phys.-Condens Mat. 1990, 2, 3093–3113. [Google Scholar] [CrossRef]
- Meakin, P. Models for Colloidal Aggregation. Annu. Rev. Phys. Chem. 1988, 39, 237–267. [Google Scholar] [CrossRef]
- Bostrom, M.; Deniz, V.; Franks, G.V.; Ninham, B.W. Extended DLVO theory: Electrostatic and non-electrostatic forces in oxide suspensions. Adv. Colloid Interface Sci. 2006, 123, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Hecht, M.; Harting, J.; Ihle, T.; Herrmann, H.J. Simulation of claylike colloids. Phys. Rev. E 2005, 72, 011408. [Google Scholar] [CrossRef] [PubMed]
- Padding, J.T.; Louis, A.A. Interplay between hydrodynamic and Brownian fluctuations in sedimenting colloidal suspensions. Phys. Rev. E 2008, 77, 011402. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.G.; Family, F.; Lam, P.M. Dynamic Scaling of the Island-Size Distribution and Percolation in a Model of Submonolayer Molecular-Beam Epitaxy. Phys. Rev. B 1994, 50, 8781–8797. [Google Scholar] [CrossRef]
- Weitz, D.A.; Huang, J.S.; Lin, M.Y.; Sung, J. Dynamics of Diffusion-Limited Kinetic Aggregation. Phys. Rev. Lett. 1984, 53, 1657–1660. [Google Scholar] [CrossRef]
- Rycroft, C.H.; Wu, C.H.; Yu, Y.; Kamrin, K. Reference map technique for incompressible fluid-structure interaction. J. Fluid Mech. 2020, 898, A9. [Google Scholar] [CrossRef]
- Theillard, M.; Saintillan, D. Computational mean-field modeling of confined active fluids. J. Comput. Phys. 2019, 397, 108841. [Google Scholar] [CrossRef]
- Tateno, M.; Tanaka, H. Numerical prediction of colloidal phase separation by direct computation of Navier-Stokes equation. NPJ Comput. Mater. 2019, 5, 40. [Google Scholar] [CrossRef]
- Howard, M.P.; Nikoubashman, A.; Palmer, J.C. Modeling hydrodynamic interactions in soft materials with multiparticle collision dynamics. Curr. Opin Chem. Eng. 2019, 23, 34–43. [Google Scholar] [CrossRef]
- Mirabello, G.; Ianiro, A.; Bomans, P.H.H.; Yoda, T.; Arakaki, A.; Friedrich, H.; de With, G.; Sommerdijk, N.A.J.M. Crystallization by particle attachment is a colloidal assembly process. Nat. Mater. 2020, 19, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Riest, J.; Winkler, R.G.; Gompper, G.; Dhont, J.K.G.; Nagele, G. Clustering and dynamics of particles in dispersions with competing interactions: Theory and simulation. Soft Matter 2018, 14, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Cates, M.E. Hydrodynamically Interrupted Droplet Growth in Scalar Active Matter. Phys. Rev. Lett. 2019, 123, 148005. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Mukherjee, S.; Dhar, J.; Ghosal, S.; Chakraborty, S. The effect of the finite size of ions and Debye layer overspill on the screened Coulomb interactions between charged flat plates. Electrophoresis 2020, 41, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Duan, W.T.; Ahmed, S.; Sen, A.; Mallouk, T.E. From One to Many: Dynamic Assembly and Collective Behavior of Self-Propelled Colloidal Motors. Accounts Chem. Res. 2015, 48, 1938–1946. [Google Scholar] [CrossRef]
- Padding, J.T.; Louis, A.A. Hydrodynamic interactions and Brownian forces in colloidal suspensions: Coarse-graining over time and length scales. Phys. Rev. E 2006, 74, 031402. [Google Scholar] [CrossRef]
- Yu, N.; Lou, X.; Chen, K.; Yang, M.C. Phototaxis of active colloids by self-thermophoresis. Soft Matter 2019, 15, 408–414. [Google Scholar] [CrossRef]
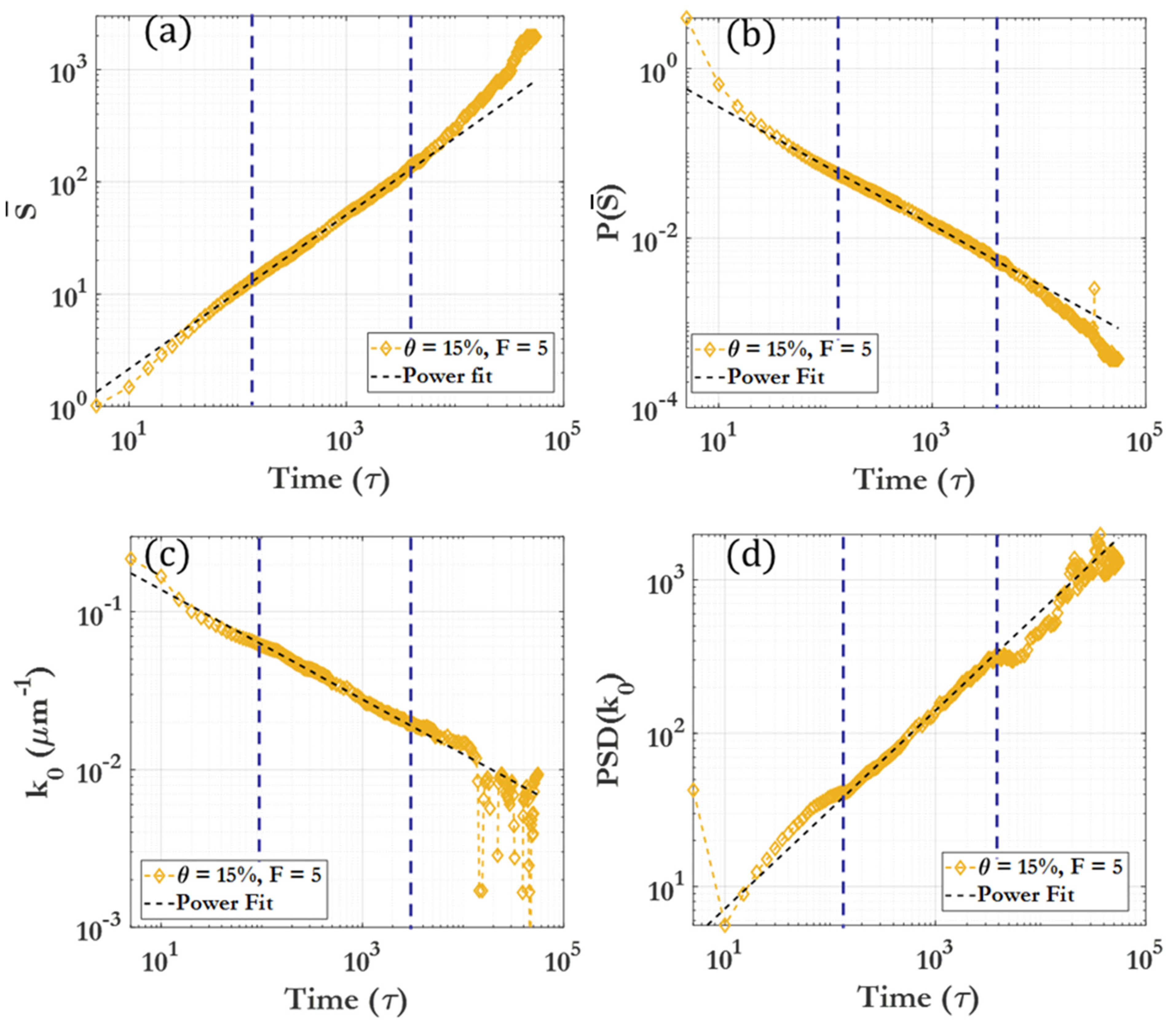
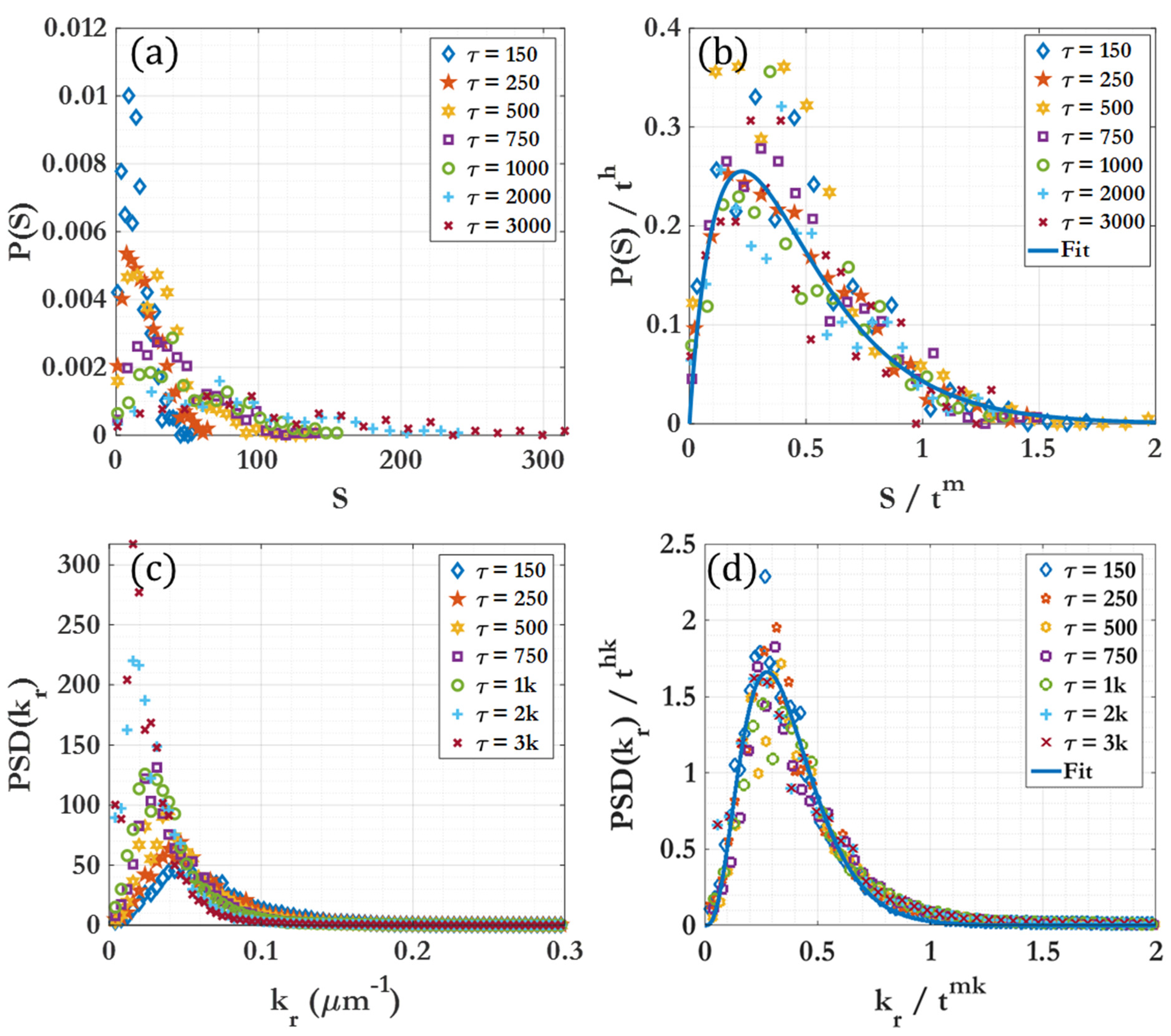




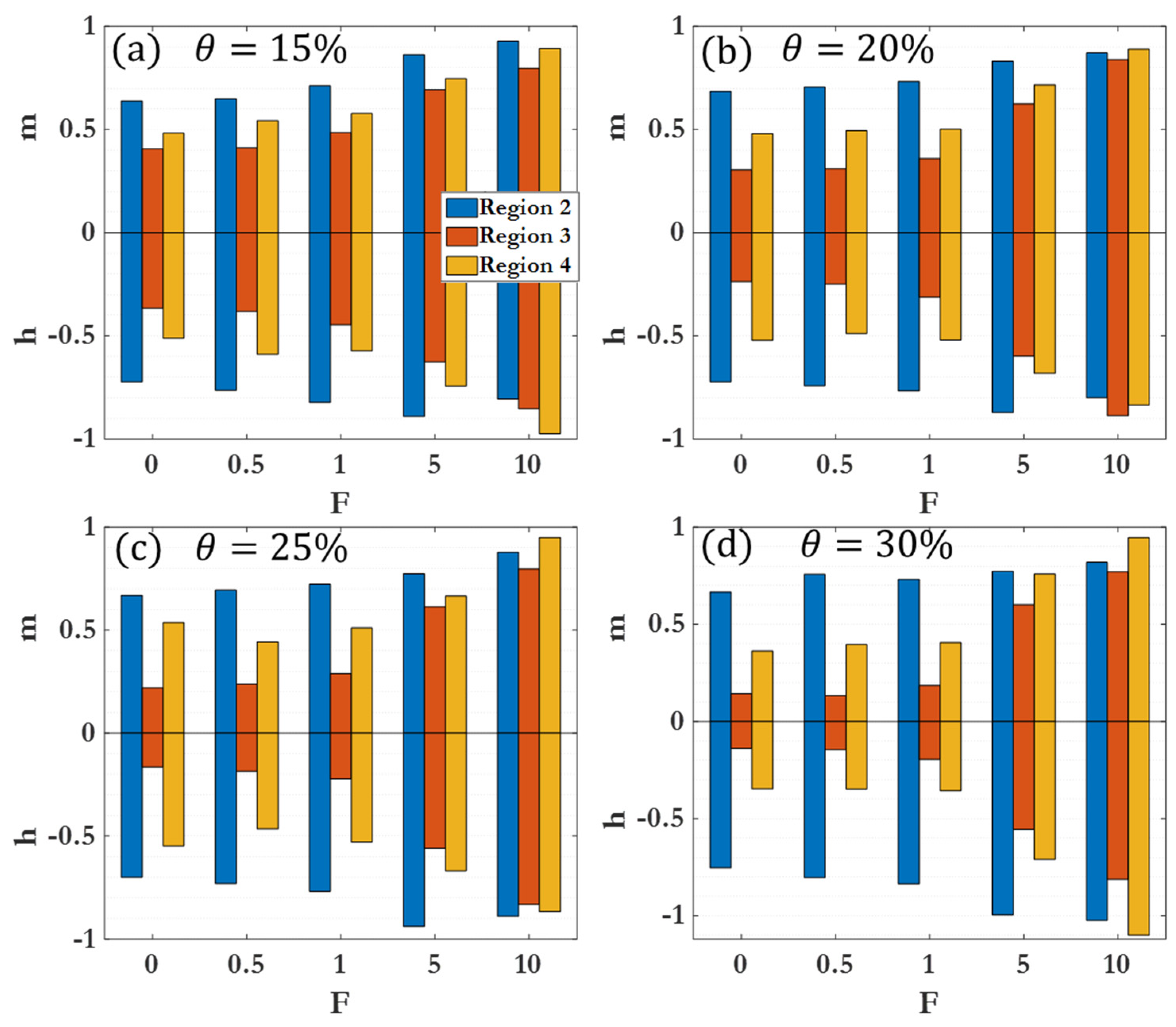
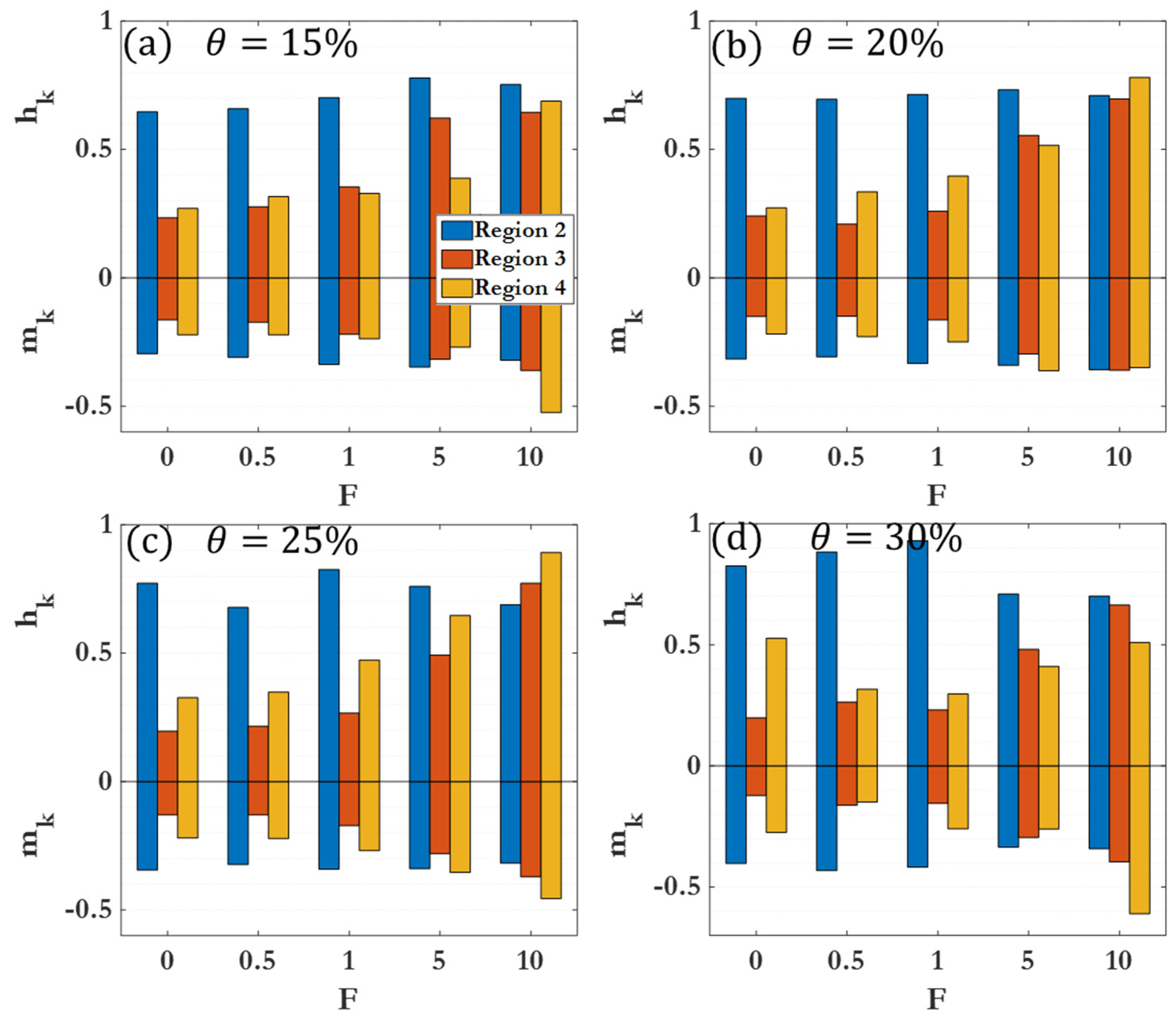
| Region 1 | Effectively no clustering seen | , |
| Region 2 | Initial rapid clustering | |
| Region 3 | Marked slowing of clustering | |
| Region 4 | Long-time clustering behavior |
| 129.4 | 93.82 | 65.05 | 23.88 | 19.52 | |
| 22.05 | 20.71 | 18.42 | 11.83 | 9.78 | |
| 9.49 | 9.17 | 8.72 | 6.44 | 5.21 | |
| 99.93 | 84.58 | 75.60 | 55.16 | 44.21 | |
| 1114 | 1186 | 1207 | 1180 | 1519 | |
| 5.19 | 5.09 | 5.56 | 4.22 | 3.61 | |
| 69.41 | 65.74 | 70.78 | 67.26 | 60.06 | |
| 1094 | 464.3 | 497.2 | 501.7 | 636.7 | |
| 3.66 | 3.15 | 3.11 | 2.88 | 2.25 | |
| 75.41 | 72.80 | 72.29 | 64.64 | 59.54 | |
| 1014 | 901.6 | 803.6 | 687.7 | 383.2 | |
| 2.01 | 1.99 | 1.98 | 1.49 | 1.31 | |
| 102.2 | 91.96 | 88.46 | 80.81 | 73.54 | |
| 1234 | 589.4 | 623.5 | 1813 | 1013 |
| 0.5454 | 0.5837 | 0.5781 | 0.7252 | 0.8177 | |
| 0.4867 | 0.5078 | 0.5587 | 0.6969 | 0.8226 | |
| 0.6388 | 0.6484 | 0.7126 | 0.8629 | 0.9273 | |
| 0.4065 | 0.4115 | 0.4859 | 0.6931 | 0.7962 | |
| 0.4824 | 0.5433 | 0.5779 | 0.7468 | 0.8922 | |
| 0.6840 | 0.7057 | 0.7327 | 0.8302 | 0.8711 | |
| 0.3039 | 0.3100 | 0.3595 | 0.6246 | 0.8389 | |
| 0.4793 | 0.4939 | 0.5018 | 0.7161 | 0.8896 | |
| 0.6682 | 0.6946 | 0.7227 | 0.7740 | 0.8778 | |
| 0.2197 | 0.2379 | 0.2886 | 0.6132 | 0.7971 | |
| 0.5359 | 0.4422 | 0.5107 | 0.6647 | 0.9489 | |
| 0.6660 | 0.7582 | 0.7302 | 0.7729 | 0.8195 | |
| 0.1432 | 0.1323 | 0.1848 | 0.6006 | 0.7693 | |
| 0.3621 | 0.3955 | 0.4053 | 0.7583 | 0.9453 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becton, M.; Hou, J.; Zhao, Y.; Wang, X. Dynamic Clustering and Scaling Behavior of Active Particles under Confinement. Nanomaterials 2024, 14, 144. https://doi.org/10.3390/nano14020144
Becton M, Hou J, Zhao Y, Wang X. Dynamic Clustering and Scaling Behavior of Active Particles under Confinement. Nanomaterials. 2024; 14(2):144. https://doi.org/10.3390/nano14020144
Chicago/Turabian StyleBecton, Matthew, Jixin Hou, Yiping Zhao, and Xianqiao Wang. 2024. "Dynamic Clustering and Scaling Behavior of Active Particles under Confinement" Nanomaterials 14, no. 2: 144. https://doi.org/10.3390/nano14020144
APA StyleBecton, M., Hou, J., Zhao, Y., & Wang, X. (2024). Dynamic Clustering and Scaling Behavior of Active Particles under Confinement. Nanomaterials, 14(2), 144. https://doi.org/10.3390/nano14020144








