Ag+-Mediated Folding of Long Polyguanine Strands to Double and Quadruple Helixes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. DNA Synthesis
2.3. Separation of Poly(dG)-poly(dC) Strands
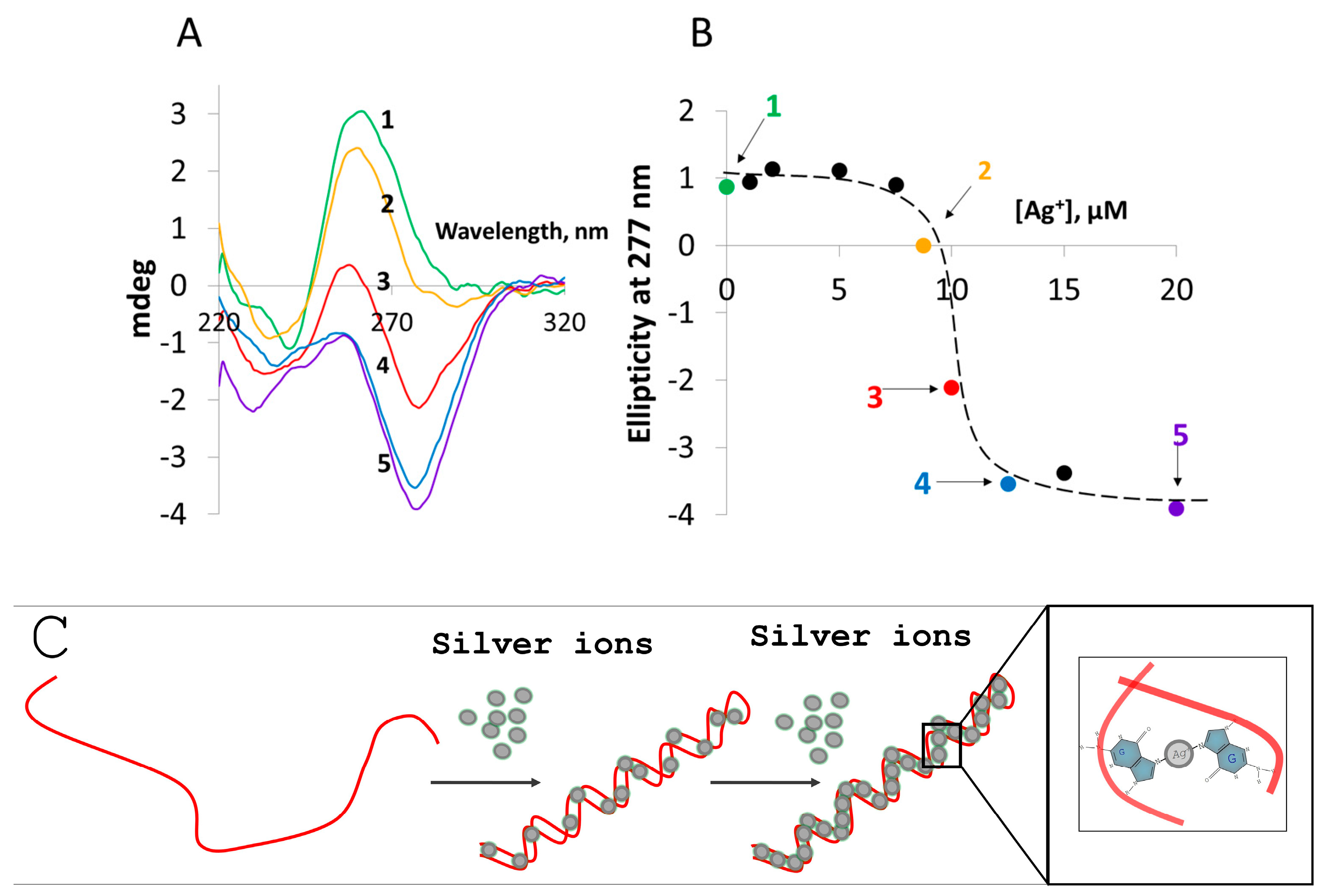
2.4. Preparation of G-Ag+-G Conjugates
2.5. CD Spectroscopy
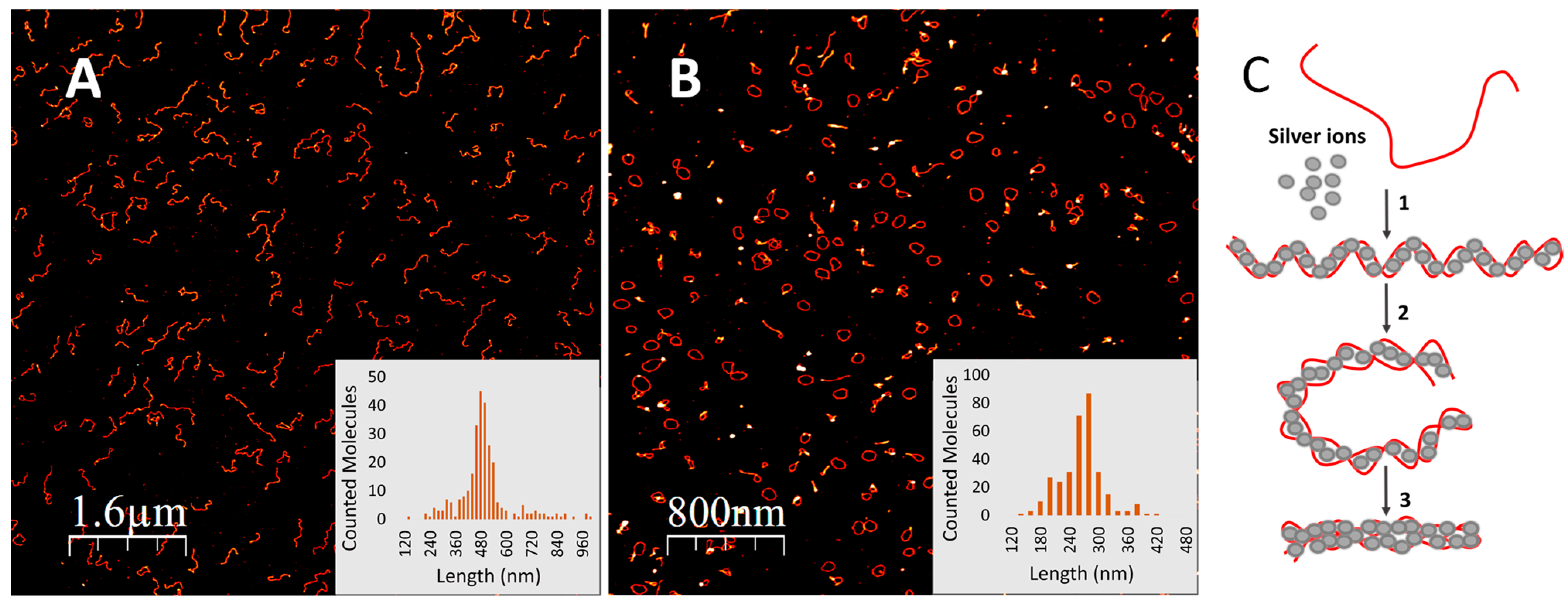
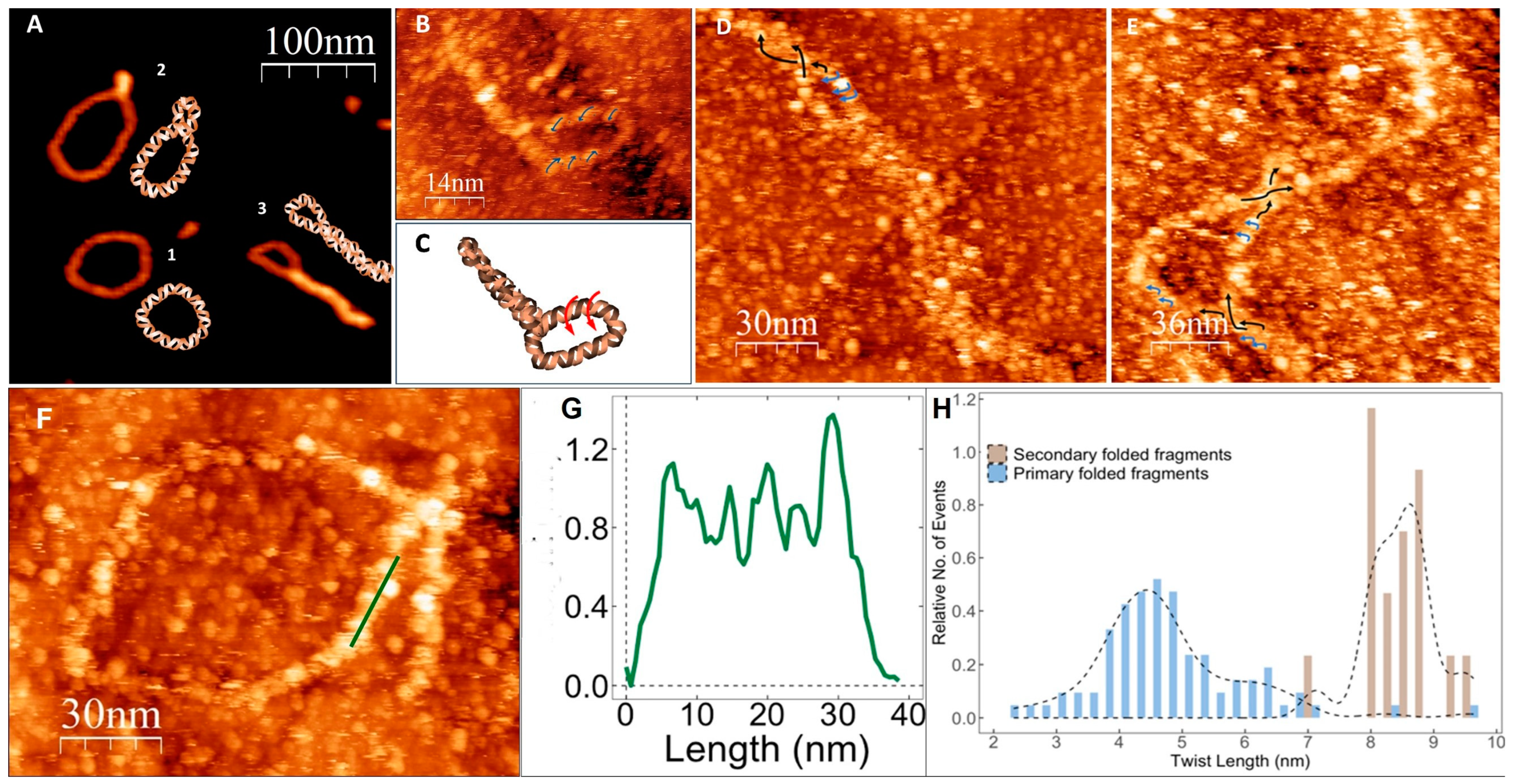
2.6. Atomic Force Microscopy (AFM)
2.7. Scanning Tunneling Microscopy
2.8. Computational Details
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA Secondary Structures: Stability and Function of G-Quadruplex Structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Granzhan, A.; Marquevielle, J.; Cucchiarini, A.; Lacroix, L.; Amrane, S.; Verga, D.; Mergny, J.L. Guidelines for G-Quadruplexes: I. In Vitro Characterization. Biochimie 2023, 214, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, A.B.; Borovok, N.; Molotsky, T.; Cohen, H.; Shapir, E.; Porath, D. Long, Monomolecular Guanine-Based Nanowires. Adv. Mater. 2005, 17, 1901–1905. [Google Scholar] [CrossRef]
- Borovok, N.; Iram, N.; Zikich, D.; Ghabboun, J.; Livshits, G.I.; Porath, D.; Kotlyar, A.B. Assembling of G-Strands into Novel Tetra-Molecular Parallel G4-DNA Nanostructures Using Avidin-Biotin Recognition. Nucleic Acids Res. 2008, 36, 5050–5060. [Google Scholar] [CrossRef] [PubMed]
- Gajarsky, M.; Stadlbauer, P.; Sponer, J.; Cucchiarini, A.; Dobrovolna, M.; Brazda, V.; Mergny, J.L.; Trantirek, L.; Lenarcic Zivkovic, M. DNA Quadruplex Structure with a Unique Cation Dependency. Angew. Chem. Int. Ed. 2024, 63, e202313226. [Google Scholar] [CrossRef] [PubMed]
- Williamson, J.R.; Raghuraman, M.K.; Cech, T.R. Monovalent Cation-Induced Structure of Telomeric DNA: The G-Quartet Model. Cell 1989, 59, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.M.; Payet, L.; Huppert, J.L. Function and Targeting of G-Quadruplexes. Curr. Opin. Mol. Ther. 2009, 11, 146–155. [Google Scholar]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-Quartet Formation by UV-Spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Chaires, J.B.; Gray, R.D.; Trent, J.O. Stability and Kinetics of G-Quadruplex Structures. Nucleic Acids Res. 2008, 36, 5482–5515. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Brown, T.; Fox, K.R. Sequence Effects of Single Base Loops in Intramolecular Quadruplex DNA. FEBS Lett. 2007, 581, 1657–1660. [Google Scholar] [CrossRef]
- Bugaut, A.; Balasubramanian, S. A Sequence-Independent Study of the Influence of Short Loop Lengths on the Stability and Topology of Intramolecular DNA G-Quadraplexes. Biochemistry 2008, 47, 689–697. [Google Scholar] [CrossRef]
- Liu, S.P.; Weisbrod, S.H.; Tang, Z.; Marx, A.; Scheer, E.; Erbe, A. Direct Measurement of Electrical Transport through G-Quadruplex DNA with Mechanically Controllable Break Junction Electrodes. Angew. Chem. Int. Ed. 2010, 49, 3313–3316. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.I.; Stern, A.; Rotem, D.; Borovok, N.; Eidelshtein, G.; Migliore, A.; Penzo, E.; Wind, S.J.; Di Felice, R.; Skourtis, S.S.; et al. Long-Range Charge Transport in Single G-Quadruplex DNA Molecules. Nat. Nanotechnol. 2014, 9, 1040–1046. [Google Scholar] [CrossRef]
- Zhuravel, R.; Stern, A.; Fardian-Melamed, N.; Eidelshtein, G.; Katrivas, L.; Rotem, D.; Kotlyar, A.B.; Porath, D. Advances in Synthesis and Measurement of Charge Transport in DNA-Based Derivatives. Adv. Mater. 2018, 30, e1706984. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Eidelshtein, G.; Zhuravel, R.; Livshits, G.I.; Rotem, D.; Kotlyar, A.; Porath, D. Highly Conductive Thin Uniform Gold-Coated DNA Nanowires. Adv. Mater. 2018, 30, e1800433. [Google Scholar] [CrossRef]
- Eidelshtein, G.; Fardian-Melamed, N.; Gutkin, V.; Basmanov, D.; Klinov, D.; Rotem, D.; Levi-Kalisman, Y.; Porath, D.; Kotlyar, A. Synthesis and Properties of Novel Silver-Containing DNA Molecules. Adv. Mater. 2016, 28, 4839–4844. [Google Scholar] [CrossRef]
- Vecchioni, S.; Lu, B.; Livernois, W.; Ohayon, Y.P.; Yoder, J.B.; Yang, C.F.; Woloszyn, K.; Bernfeld, W.; Anantram, M.P.; Canary, J.W.; et al. Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction. Adv. Mater. 2023, 35, e2210938. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, E.G.; O’Neill, P.; Guerrero, A.J.; Bouwmeester, D.; Fygenson, D.K. Sequence-Dependent Fluorescence of DNA-Hosted Silver Nanoclusters. Adv. Mater. 2008, 20, 279–283. [Google Scholar] [CrossRef]
- Izatt, R.M.; Christensen, J.J.; Rytting, J.H. Sites and Thermodynamic Quantities Associated with Proton and Metal Ion Interaction with Ribonucleic Acid, Deoxyribonucleic Acid, and Their Constituent Bases, Nucleosides, and Nucleotides. Chem. Rev. 1971, 71, 439–481. [Google Scholar] [CrossRef]
- Swasey, S.M.; Rosu, F.; Copp, S.M.; Gabelica, V.; Gwinn, E.G. Parallel Guanine Duplex and Cytosine Duplex DNA with Uninterrupted Spines of AgI-Mediated Base Pairs. J. Phys. Chem. Lett. 2018, 9, 6605–6610. [Google Scholar] [CrossRef]
- Megger, D.A.; Müller, J. Silver(I)-Mediated Cytosine Self-Pairing is Preferred over Hoogsteen-Type Base Pairs with the Artificial Nucleobase 1,3-Dideaza-6-Nitropurine. Nucleosides Nucleotides Nucleic Acids 2010, 29, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Cao, S.; Togashi, H.; Tashiro, M.; Fujimoto, T.; Machinami, T.; Oda, S.; Miyake, Y.; Okamoto, I.; Tanaka, Y. Specific Interactions between Silver(i) Ions and Cytosine–Cytosine Pairs in DNA Duplexes. Chem. Commun. 2008, 4825–4827. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, C.M.; Johnsen, K.R.; Kiser, J.R.; Antoku, Y.; Dickson, R.M.; Petty, J.T. Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ono, A.; Torigoe, H.; Tanaka, Y.; Okamoto, I. Binding of Metal Ions by Pyrimidine Base Pairs in DNA Duplexes. Chem. Soc. Rev. 2011, 40, 5855–5866. [Google Scholar] [CrossRef] [PubMed]
- Swasey, S.M.; Leal, L.E.; Lopez-Acevedo, O.; Pavlovich, J.; Gwinn, E.G. Silver (I) as DNA Glue: Ag+-Mediated Guanine Pairing Revealed by Removing Watson-Crick Constraints. Sci. Rep. 2015, 5, 10163. [Google Scholar] [CrossRef] [PubMed]
- Vecchioni, S.; Capece, M.C.; Toomey, E.; Nguyen, L.; Ray, A.; Greenberg, A.; Fujishima, K.; Urbina, J.; Paulino-Lima, I.G.; Pinheiro, V.; et al. Construction and Characterization of Metal Ion-Containing DNA Nanowires for Synthetic Biology and Nanotechnology. Sci. Rep. 2019, 9, 6942. [Google Scholar] [CrossRef] [PubMed]
- Katrivas, L.; Fardian-Melamed, N.; Rotem, D.; Porath, D.; Kotlyar, A. Formation of Novel Octuplex DNA Molecules from Guanine Quadruplexes. Adv. Mater. 2021, 33, 2006932. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, A.B.; Borovok, N.; Molotsky, T.; Fadeev, L.; Gozin, M. In Vitro Synthesis of Uniform Poly(DG)-Poly(DC) by Klenow Exo- Fragment of Polymerase I. Nucleic Acids Res. 2005, 33, 525–535. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. Density Functionals with Broad Applicability in Chemistry. Acc. Chem. Res. 2008, 41, 157–167. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum Mechanical Continuum Solvation Models. Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, F.J.A.; Cerezo, J.; Stendardo, E.; Improta, R.; Santoro, F. Insights for an Accurate Comparison of Computational Data to Experimental Absorption and Emission Spectra: Beyond the Vertical Transition Approximation. J. Chem. Theory Comput. 2013, 9, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Kasumov, A.Y.; Klinov, D.V.; Roche, P.-E.; Guéron, S.; Bouchiat, H. Thickness and Low-Temperature Conductivity of DNA Molecules. Appl. Phys. Lett. 2004, 84, 1007–1009. [Google Scholar] [CrossRef]
- Asha, H.; Green, J.A.; Esposito, L.; Martinez-Fernandez, L.; Santoro, F.; Improta, R. Effect of the Thermal Fluctuations of the Photophysics of GC and CG DNA Steps: A Computational Dynamical Study. J. Phys. Chem. B 2022, 126, 10608–10621. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katrivas, L.; Makarovsky, A.; Kempinski, B.; Randazzo, A.; Improta, R.; Rotem, D.; Porath, D.; Kotlyar, A.B. Ag+-Mediated Folding of Long Polyguanine Strands to Double and Quadruple Helixes. Nanomaterials 2024, 14, 663. https://doi.org/10.3390/nano14080663
Katrivas L, Makarovsky A, Kempinski B, Randazzo A, Improta R, Rotem D, Porath D, Kotlyar AB. Ag+-Mediated Folding of Long Polyguanine Strands to Double and Quadruple Helixes. Nanomaterials. 2024; 14(8):663. https://doi.org/10.3390/nano14080663
Chicago/Turabian StyleKatrivas, Liat, Anna Makarovsky, Benjamin Kempinski, Antonio Randazzo, Roberto Improta, Dvir Rotem, Danny Porath, and Alexander B. Kotlyar. 2024. "Ag+-Mediated Folding of Long Polyguanine Strands to Double and Quadruple Helixes" Nanomaterials 14, no. 8: 663. https://doi.org/10.3390/nano14080663




