The Use of the Calcitonin Minimal Recognition Module for the Design of DOPA-Containing Fibrillar Assemblies
Abstract
:1. Introduction

2. Results and Discussion
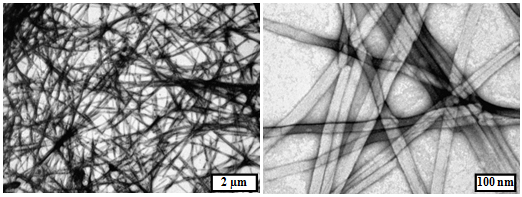
2.1. Morphological Characterization of the Peptide Assemblies

2.2. Secondary Structure Analysis of the Peptide Assemblies




2.3. DOPA-Containing Peptide Assemblies Exhibit Functionality and Reduce Ionic Silver

3. Experimental Section
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- McGaughey, G.B.; Gagné, M.; Rappé, A.K. π-stacking interactions alive and well in proteins. J. Biol. Chem. 1998, 273, 15458–15463. [Google Scholar] [CrossRef]
- Zhang, S. Emerging biological materials through molecular self-assembly. Biotechnol. Adv. 2002, 20, 321–339. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends. Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Harper, J.D.; Lansbury, P.T., Jr. Models of amyloid seeding in Alzheimer’s disease and scrapie: Mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997, 66, 385–407. [Google Scholar] [CrossRef]
- Serpell, L.C. Alzheimer’s amyloid fibrils: Structure and assembly. BBA-Mol. Basis Dis. 2000, 1502, 16–30. [Google Scholar] [CrossRef]
- Geddes, A.J.; Parker, K.D.; Atkins, E.D.T.; Beighton, E. “Cross-β” conformation in proteins. J. Mol. Biol. 1968, 32, 343–358. [Google Scholar] [CrossRef]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.Ø.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-β spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef]
- Gazit, E. Mechanisms of amyloid fibril self-assembly and inhibition. FEBS J. 2005, 272, 5971–5978. [Google Scholar] [CrossRef]
- Balbach, J.J.; Ishii, Y.; Antzutkin, O.N.; Leapman, R.D.; Rizzo, N.W.; Dyda, F.; Reed, J.; Tycko, R. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer’s β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry 2000, 39, 13748–13759. [Google Scholar] [CrossRef]
- Madine, J.; Doig, A.J.; Middleton, D.A. Design of an n-methylated peptide inhibitor of α-synuclein aggregation guided by solid-state NMR. J. Am. Chem. Soc. 2008, 130, 7873–7881. [Google Scholar] [CrossRef]
- Mazor, Y.; Gilead, S.; Benhar, I.; Gazit, E. Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide. J. Mol. Biol. 2002, 322, 1013–1024. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Amyloidogenic hexapeptide fragment of medin: Homology to functional islet amyloid polypeptide fragments. Amyloid 2004, 11, 81–89. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef]
- MacIntyre, I. Calcitonin: A general review. Calcif. Tissue Int. 1967, 1, 173–182. [Google Scholar] [CrossRef]
- Sletten, K.; Westermark, P.; Natvig, J.B. Characterization of amyloid fibril proteins from medullary carcinoma of the thyroid. J. Exp. Med. 1976, 143, 993–998. [Google Scholar] [CrossRef]
- Kedar, I.; Ravid, M.; Sohar, E. In vitro synthesis of “amyloid” fibrils from insulin, calcitonin and parathormone. Isr. J. Med. Sci. 1976, 12, 1137–1140. [Google Scholar]
- Kanaori, K.; Nosaka, A.Y. Study of human calcitonin fibrillation by proton nuclear magnetic resonance spectroscopy. Biochemistry 1995, 34, 12138–12143. [Google Scholar] [CrossRef]
- Kamihira, M.; Naito, A.; Tuzi, S.; Nosaka, A.Y.; Saito, H. Conformational transitions and fibrillation mechanism of human calcitonin as studied by high-resolution solid-state 13C NMR. Protein Sci. 2000, 9, 867–877. [Google Scholar] [CrossRef]
- Reches, M.; Porat, Y.; Gazit, E. Amyloid fibril formation by pentapeptide and tetrapeptide fragments of human calcitonin. J. Biol. Chem. 2002, 277, 35475–35480. [Google Scholar] [CrossRef]
- Gavin Tsai, H.-H.; Zanuy, D.; Haspel, N.; Gunasekaran, K.; Ma, B.; Tsai, C.J.; Nussinov, R. The stability and dynamics of the human calcitonin amyloid peptide DFNKF. Biophys. J. 2004, 87, 146–158. [Google Scholar] [CrossRef]
- Naito, A.; Kamihira, M.; Inoue, R.; Saitô, H. Structural diversity of amyloid fibril formed in human calcitonin as revealed by site-directed 13C solid-state NMR spectroscopy. Magn. Reson. Chem. 2004, 42, 247–257. [Google Scholar] [CrossRef]
- Haspel, N.; Zanuy, D.; Ma, B.; Wolfson, H.; Nussinov, R. A comparative study of amyloid fibril formation by residues 15–19 of the human calcitonin hormone: A single β-sheet model with a small hydrophobic core. J. Mol. Biol. 2005, 345, 1213–1227. [Google Scholar] [CrossRef]
- Pak, Y.; Shin, J.; Jang, S. Computational study of human calcitonin (hCT) oligomer. Bull. Korean Chem. Soc. 2009, 30, 3006–3010. [Google Scholar] [CrossRef]
- Cherny, I.; Gazit, E. Amyloids: Not only pathological agents but also ordered nanomaterials. Angew. Chem. Int. Ed. 2008, 47, 4062–4069. [Google Scholar] [CrossRef]
- Pilkington, S.M.; Roberts, S.J.; Meade, S.J.; Gerrard, J.A. Amyloid fibrils as a nanoscaffold for enzyme immobilization. Biotechnol. Prog. 2010, 26, 93–100. [Google Scholar]
- Hauser, C.A.E.; Maurer-Stroh, S.; Martins, I.C. Amyloid-based nanosensors and nanodevices. Chem. Soc. Rev. 2014, 43, 5326–5345. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef]
- Scheibel, T.; Parthasarathy, R.; Sawicki, G.; Lin, X.-M.; Jaeger, H.; Lindquist, S.L. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA 2003, 100, 4527–4532. [Google Scholar]
- Orbach, R.; Adler-Abramovich, L.; Zigerson, S.; Mironi-Harpaz, I.; Seliktar, D.; Gazit, E. Self-assembled Fmoc-peptides as a platform for the formation of nanostructures and hydrogels. Biomacromolecules 2009, 10, 2646–2651. [Google Scholar] [CrossRef]
- Scheibel, T. Protein fibers as performance proteins: New technologies and applications. Curr. Opin. Biotechnol. 2005, 16, 427–433. [Google Scholar] [CrossRef]
- Fichman, G.; Gazit, E. Self-assembly of short peptides to from hydrogels: Design of building blocks, physical propertiesand technological applications. Acta Biomater. 2014, 10, 1671–1682. [Google Scholar] [CrossRef]
- Reithofer, M.R.; Chan, K.-H.; Lakshmanan, A.; Lam, D.H.; Mishra, A.; Gopalan, B.; Joshi, M.; Wang, S.; Hauser, C.A.E. Ligation of anti-cancer drugs to self-assembling ultrashort peptides by click chemistry for localized therapy. Chem. Sci. 2014, 5, 625–630. [Google Scholar] [CrossRef]
- Wiegemann, M. Adhesion in blue mussels (mytilus edulis) and barnacles (genus balanus): Mechanisms and technical applications. Aquat. Sci. 2005, 67, 166–176. [Google Scholar] [CrossRef]
- Yu, M.; Hwang, J.; Deming, T.J. Role of L-3,4-dihydroxyphenylalanine in mussel adhesive proteins. J. Am. Chem. Soc. 1999, 121, 5825–5826. [Google Scholar] [CrossRef]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef]
- Xu, C.; Xu, K.; Gu, H.; Zheng, R.; Liu, H.; Zhang, X.; Guo, Z.; Xu, B. Dopamine as a robust anchor to immobilize functional molecules on the iron oxide shell of magnetic nanoparticles. J. Am. Chem. Soc. 2004, 126, 9938–9939. [Google Scholar] [CrossRef]
- Dalsin, J.L.; Hu, B.-H.; Lee, B.P.; Messersmith, P.B. Mussel adhesive protein mimetic polymers for the preparation of nonfouling surfaces. J. Am. Chem. Soc. 2003, 125, 4253–4258. [Google Scholar] [CrossRef]
- Black, K.C.L.; Liu, Z.; Messersmith, P.B. Catechol redox induced formation of metal core-polymer shell nanoparticles. Chem. Mater. 2011, 23, 1130–1135. [Google Scholar] [CrossRef]
- Charlot, A.; Sciannaméa, V.; Lenoir, S.; Faure, E.; Jérôme, R.; Jérôme, C.; van de Weerdt, C.; Martial, J.; Archambeau, C.; Willet, N.; et al. All-in-one strategy for the fabrication of antimicrobial biomimetic films on stainless steel. J. Mater. Chem. 2009, 19, 4117–4125. [Google Scholar]
- Fullenkamp, D.E.; Rivera, J.G.; Gong, Y.-K.; Lau, K.H.A.; He, L.; Varshney, R.; Messersmith, P.B. Mussel-inspired silver-releasing antibacterial hydrogels. Biomaterials 2012, 33, 3783–3791. [Google Scholar] [CrossRef]
- Saha, A.; Bolisetty, S.; Handschin, S.; Mezzenga, R. Self-assembly and fibrillization of a Fmoc-functionalized polyphenolic amino acid. Soft Matter 2013, 9, 10239–10242. [Google Scholar] [CrossRef]
- Ceylan, H.; Urel, M.; Erkal, T.S.; Tekinay, A.B.; Dana, A.; Guler, M.O. Mussel inspired dynamic cross-linking of self-healing peptide nanofiber network. Adv. Funct. Mater. 2013, 23, 2081–2090. [Google Scholar] [CrossRef] [Green Version]
- Fichman, G.; Adler-Abramovich, L.; Manohar, S.; Mironi-Harpaz, I.; Guterman, T.; Seliktar, D.; Messersmith, P.B.; Gazit, E. Seamless metallic coating and surface adhesion of self-assembled bio-inspired nanostructures based on di-(3,4-dihydroxy-L-phenylalanine) peptide motif. ACS Nano 2014, 8, 7220–7228. [Google Scholar] [CrossRef]
- Howie, A.J.; Brewer, D.B. Optical properties of amyloid stained by Congo red: History and mechanisms. Micron 2009, 40, 285–301. [Google Scholar] [CrossRef]
- Liang, G.; Xu, K.; Li, L.; Wang, L.; Kuang, Y.; Yang, Z.; Xu, B. Using Congo red to report intracellular hydrogelation resulted from self-assembly of small molecules. Chem. Commun. 2007, 40, 4096–4098. [Google Scholar]
- Hamill, A.C.; Wang, S.-C.; Lee, C.T., Jr. Solution structure of an amyloid-forming protein during photoinitiated hexamer-dodecamer transitions revealed through small-angle neutron scattering. Biochemistry 2007, 46, 7694–7705. [Google Scholar] [CrossRef]
- Naiki, H.; Higuchi, K.; Hosokawa, M.; Takeda, T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavine T. Anal. Biochem. 1989, 177, 244–249. [Google Scholar] [CrossRef]
- Levine, H. Thioflavine T teraction with synthetic Alzheimer’s disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar] [CrossRef]
- Shtainfeld, A.; Sheynis, T.; Jelinek, R. Specific mutations alter fibrillation kinetics, fiber morphologies, and membrane interactions of pentapeptides derived from human calcitonin. Biochemistry 2010, 49, 5299–5307. [Google Scholar] [CrossRef]
- Byler, D.M.; Susi, H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 1986, 25, 469–487. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophy. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Surewicz, W.K.; Mantsch, H.H. New insight into protein secondary structure from resolution-enhanced infrared spectra. Biochim. Biophys. Acta 1988, 952, 115–130. [Google Scholar] [CrossRef]
- Gilmanshin, R.; Williams, S.; Callender, R.H.; Woodruff, W.H.; Dyer, R.B. Fast events in protein folding: Relaxation dynamics and structure of the I form of apomyoglobin. Biochemistry 1997, 36, 15006–15012. [Google Scholar] [CrossRef]
- Manas, E.S.; Getahun, Z.; Wright, W.W.; DeGrado, W.F.; Vanderkooi, J.M. Infrared spectra of amide groups in α-helical proteins: Evidence for hydrogen bonding between helices and water. J. Am. Chem. Soc. 2000, 122, 9883–9890. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hayakawa, T. Conformational studies of sequential polypeptides containing L-β-(3,4-dihydroxyphenyl)-α-alanine (dopa) and L-lysine. Macromolecules 1983, 16, 1058–1063. [Google Scholar] [CrossRef]
- Dutt, A.; Dutta, A.; Mondal, R.; Spencer, E.C.; Howard, J.A.K.; Pramanik, A. Studies of β-turn opening with model peptides containing non-coded α-amino isobutyric acid. Tetrahedron 2007, 63, 10282–10289. [Google Scholar] [CrossRef]
- Kar, S.; Dutta, A.; Drew, M.G.B.; Koley, P.; Pramanik, A. Design of supramolecular β-sheet forming β-turns containing aromatic rings and non-coded α-aminoisobutyric acid and the formation of flat fibrillar structures through self-assembly. Supramol. Chem. 2009, 21, 681–690. [Google Scholar] [CrossRef]
- Maji, S.K.; Haldar, D.; Drew, M.G.B.; Banerjee, A.; Das, A.K.; Banerjee, A. Self-assembly of β-turn forming synthetic tripeptides into supramolecular β-sheets and amyloid-like fibrils in the solid state. Tetrahedron 2004, 60, 3251–3259. [Google Scholar] [CrossRef]
- Lakshmanan, A.; Cheong, D.W.; Accardo, A.; di Fabrizio, E.; Riekel, C.; Hauser, C.A.E. Aliphatic peptides show similar self-assembly to amyloid core sequences, challenging the importance of aromatic interactions in amyloidosis. Proc. Natl. Acad. Sci. USA 2013, 110, 519–524. [Google Scholar]
- Tifany, M.L.; Krimm, S. Effect of temperature on the circular dichroism spectra of polypeptides in the extended state. Biopolymers 1972, 11, 2309–2316. [Google Scholar] [CrossRef]
- McFarland, A.D.; van Duyne, R.P. Single silver nanoparticles as real-time optical sensors with zeptomole sensitivity. Nano Lett. 2003, 3, 1057–1062. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Ong, B.S. Facile synthesis of silver nanoparticles useful for fabrication of high-conductivity elements for printed electronics. J. Am. Chem. Soc. 2005, 127, 3266–3267. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, Y.; Feng, X.; Wang, W.; Xu, F.; Zhang, L. Antibacterial surfaces through dopamine functionalization and silver nanoparticle immobilization. Mater. Chem. Phys. 2010, 121, 534–540. [Google Scholar]
- Xu, H.; Shi, X.; Ma, H.; Lv, Y.; Zhang, L.; Mao, Z. The preparation and antibacterial effects of DOPA-cotton/AgNPs. Appl. Surf. Sci. 2011, 257, 6799–6803. [Google Scholar]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2007, 1, 2876–2890. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fichman, G.; Guterman, T.; Adler-Abramovich, L.; Gazit, E. The Use of the Calcitonin Minimal Recognition Module for the Design of DOPA-Containing Fibrillar Assemblies. Nanomaterials 2014, 4, 726-740. https://doi.org/10.3390/nano4030726
Fichman G, Guterman T, Adler-Abramovich L, Gazit E. The Use of the Calcitonin Minimal Recognition Module for the Design of DOPA-Containing Fibrillar Assemblies. Nanomaterials. 2014; 4(3):726-740. https://doi.org/10.3390/nano4030726
Chicago/Turabian StyleFichman, Galit, Tom Guterman, Lihi Adler-Abramovich, and Ehud Gazit. 2014. "The Use of the Calcitonin Minimal Recognition Module for the Design of DOPA-Containing Fibrillar Assemblies" Nanomaterials 4, no. 3: 726-740. https://doi.org/10.3390/nano4030726
APA StyleFichman, G., Guterman, T., Adler-Abramovich, L., & Gazit, E. (2014). The Use of the Calcitonin Minimal Recognition Module for the Design of DOPA-Containing Fibrillar Assemblies. Nanomaterials, 4(3), 726-740. https://doi.org/10.3390/nano4030726