1. Introduction
Small interfering RNA (siRNA) is a potent tool for modulating gene expression owing to its high specificity to target mRNA, not easily accessed by traditional drug molecules [
1]. Hence, RNAi therapeutics have demonstrated potential as a more personalized approach in the treatment of many life threatening diseases [
2] caused by faulty protein expression, such as malignancies, fibrosis and amyloidosis [
3,
4,
5]. However, as a naked molecule, siRNA is susceptible to degradation, rapid clearance and a wide bio-distribution due to its small size and high negative charge [
6,
7,
8]. On the other hand, developing carrier systems that can protect and target siRNA to its intended site of action have demonstrated manufacturing and safety challenges [
9,
10].
Whether RNAi therapeutics will make it from bench to bedside will largely depend on the improvement of siRNA molecule’s targetability and pharmacokinetics in terms of plasma stability and circulation time, as well as specific cellular uptake [
11]. In general, systemically administered naked siRNA molecule faces extracellular and intracellular barriers [
12]. Free siRNA molecules are exposed to serum nucleases and phagocyte uptake which markedly reduce their biological half-life. Moreover, once introduced locally the negative charge of the plasma membrane as well as the extracellular matrix (ECM) hinders these molecules from reaching their target and exerting action [
13].
Intracellular barriers are also crucial determinants of the efficiency of a carrier encapsulating siRNA molecules as these are engulfed by endocytosis. The carriers face the challenge of disassembling in a timely manner and escaping the endosome in order to deliver their siRNA cargo to target mRNA, located in the cytoplasm [
9].
Another challenge of siRNA therapeutics is off-target effects or suppression of normal genes after non-targeted systemic administration which can lead to damaging or undesired cell transformation [
14]. Recent reports have also demonstrated that free siRNA can initiate interferon responses via toll-like receptor 7 (TLR-7) leading to cell death in culture [
1]. In addition, phagocytic cells present in bloodstream and extravascular tissues can detect and interact with foreign siRNA molecules causing the activation of further immune responses [
9].
Over the last decade, researchers have been investigating siRNA modification approaches and carrier system development that overcomes its delivery barriers. An ideal vehicle should have the capability to completely encapsulate and protect the siRNA cargo against enzymatic degradation and have an appropriate size to allow extravasation and retention at the target site, while preventing renal clearance. It should also possess appropriate surface properties that prevent serum protein interaction and allow efficient uptake by target cells while evading phagolysosomes. Other desired characteristics that facilitate clinical application are easy functionalization with targeting ligands to enhance tissue specificity, biocompatibility and reduce toxicity [
9,
15].
Although much progress have been accomplished using viral vectors, modified siRNA, and various nanocarriers, their usage as a clinically applicable delivery system is still arguable due to multiple drawbacks related to safety and stability concerns [
16]. On the other hand, synthetic cationic materials have demonstrated some potential as non-viral siRNA delivery vehicles [
15]. Cationic polymers offer several benefits including the ability to facilitate complex formation with negatively charged siRNA molecules through electrostatic interactions, cellular uptake, and proton sponge-mediated endosomal escape [
17]. However, disadvantages of cationic carriers include high toxicity due to cell membrane integrity alteration and high immunogenicity [
18]
.Lipid nanoparticles in general and phospholipids specifically have been recognized as one of the most promising delivery systems for siRNA due to their biocompatibility, relative ease of large scale production and the recent approvals to be used in clinical trials [
19,
20]. Phospholipids are amphiphilic molecules that form spontaneous bilayer structures upon dispersion in water [
21] entrapping the dispersed hydrophilic payload within the aqueous core of the formed structure. Therefore, chemical modification of the head groups of these molecules with cationic arginine molecules should promote entrapment of negatively charged siRNA through electrostatic interactions, making them promising components of siRNA nanocarriers [
22,
23].
Connective tissue growth factor (CTGF) is considered the master switch in chronic fibrotic diseases [
24,
25] and provides a unique strategy for siRNA targeted therapeutics [
26]. Following chronic organ injury, CTGF is over expressed as a part of the wound healing response exerting its own pro-fibrotic effect as well as facilitating production of profibrotic cytokine transforming growth factor β1 (TGF-β1). These two factors work synergistically to activate endothelial cells to exert a phenotype of proliferative myofibroblasts, in turn, causing accumulation of collagen and other proteins in the surrounding ECM, thus affecting the organ morphology and function [
25,
27]. Downregulation of CTGF expression has shown to be an effective strategy for the reversal of endothelial cell activation and accumulation of fibrotic ECM [
28,
29]. Recently it has been demonstrated that targeting CTGF by an siRNA based cationic solid lipid nanoparticle in liver fibrosis successfully reversed symptoms of fibrosis as well as reduced content of key downstream mediators regulating this disease [
30]. Furthermore, CTGF has been targeted via another delivery vehicle for reducing cardiac fibrosis indicating the important role of the protein during the pathogenesis of disease [
31].
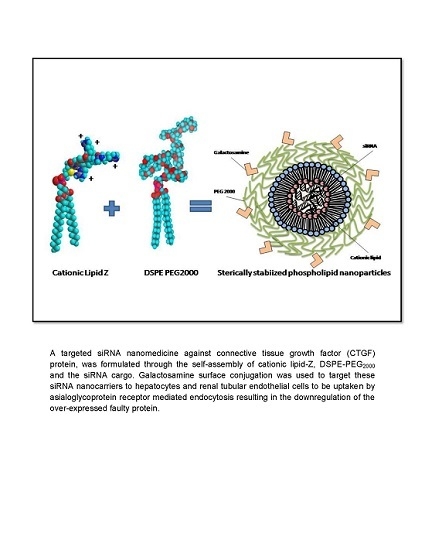
In this study, we formulated a biocompatible and relatively inexpensive sterically stabilized phospholipid nanoparticles (SSLNPs) composed of naturally occuring phospholipids and amino acid components in addition to the US Food and Drug Administration (FDA) approved DSPE-PEG
2000 monomers [
32]. This nanocarrier is designed to effectively load and deliver sufficient amounts of siRNA against CTGF to hepatocytes or renal tubular epithelial cells through passive and active targeting mechanisms established by the nanosize of the particle and surface conjugation with galactosamine (GalN). GalN is known to target asialoglycoprotein receptors, expressed on the surface of hepatocytes [
33,
34]. Scientific evidence suggests that receptor mediated endocytosis results in the internalization of the siRNA and sequence-specific degradation of CTGF mRNA causing the down-modulation of CTGF activity. This effect, in turn shifts the TGF-β1/Bone morphogenic protein 7 (BMP-7-a natural antagonist of TGF-β1) balance in the direction of anti-fibrosis,
i.e., inhibiting ECM synthesis and increasing its degradation [
25].
In the present study, we first performed physicochemical characterization and stability studies of siRNA encapsulated in SSLNPs. Then, we demonstrated the low cytotoxicity of SSLNP on different cell lines as well as their significant uptake in cell culture. Efficacy of developped siRNA-SSLNP nanomedicine against CTGF and the reversal of endothelial cell activation was demonstrated through the downregulation of key protein players of fibrosis in vitro.
Finally, bio-distribution (BD) and pharmacokinetic (PK) studies in mice were performed to confirm the potential of using these nanocarriers for targeted delivery to liver and kidney tissues. To achieve steric stability and improve BD/PK properties [
35] of our carrier, we used a polyethylene glycol conjugated (PEGylated) lipid, which is approved for human use by FDA in another pharmaceutical product. We believe that the entire particle surface is covered by PEG so that possible immune reaction and activation of the complement system [
36,
37] is minimal.
3. Experimental Section
3.1. Materials
1,2-Distearoyl-sn-glycero-3-phosphatidylethanolamine-N-[methoxy(polyethyleneglycol)-2000] sodium salt (DSPE-PEG2000) was purchased from Lipoid GmbH (Ludwigshafen, Germany). 1,2-Dipalmitoyl-sn-Glycero-3-Phosphothioethanol Sodium Salt (Ptd Thioethanol) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carboxy(polyethyleneglycol)-2000] ammonium salt (DSPE-PEG2000 Carboxylic Acid) were from Avanti polar lipids, Inc. (Alabaster, AL, USA). d-galactosamine hydrochloride and N-(3-dimethylaminopropyl)-Nʹ-ethylcarbodiimide (EDC) were from Thermo Fisher Scientific (Pittsburgh, PA, USA). N-hydroxysuccinimide (NHS), negative siRNA control and Cy5-labeled siRNA were purchased from Sigma-Aldrich (St. Louis, MO, USA). siRNA against CTGF was obtained from Santa Cruz biotechnology (Dallas, TX, USA). RNase One Ribonuclease, CellTiter-96 AQ-one Solution Cell proliferation Assay and CytoTox-one homogeneous membrane integrity assay were purchased from Promega, Inc. (Madison, WI, USA). FAM-labeled siRNA, lipofectamine, SYBR Green-II and Alexa-fluoro 488 donkey-anti-rabbit IgG secondary antibody were from Invitrogen™ Life Technologies (Grand Island, NY, USA). Trypsin-EDTA (0.25% with 0.53 mM EDTA), Minimum essential media (MEM), fetal bovine serum (FBS), non-essential amino acids, antibiotic solution (penicillin 10,000 units/mL with streptomycin 10 mg/mL) and sodium pyruvate were all purchased from Mediatech-Cellgro (Manassas, VA, USA). Keratinocyte serum free medium and supplements (K-SFM) were purchased from Invitrogen™ Life Technologies (Grand Island, NY, USA). Human hepatocellular carcinoma cells (Hep-G2) and immortalized proximal renal tubular epithelial cells (HK-2) were obtained from the American Type Culture Collection (Manassas, VA, USA). CTGF-ELISA kit was from Antigenix, Inc. (Huntington Station, NY, USA). Human primary hepatic stellate cells (HSC) as well as corresponding stellate cell media (SteCM) and supplements were from Sciencell (Carlsbad, CA, USA). Sirius red/fast green kit was from Chondrex, Inc. (Redmond, WA, USA). Primary anti-collagen I, anti-collagen-III and anti-α-SMA antibodies were from Abcam (Cambridge, MA, USA). Six-week old male Balb/c mice were obtained from Harlan Laboratories (Indianapolis, IN, USA). Other materials, if not specified, were purchased from Thermo Fisher Scientific (Pittsburgh, PA, USA) or Sigma-Aldrich (St. Louis, MO, USA).
3.2. Cationic Lipid-Z Synthesis
Four arginine (4R) peptide synthesis was performed by solid phase peptide synthesis method using Fmoc-AA-Wang resin (50 µmole) and Symphony® Peptide Synthesizer (Protein Technologies Inc, Tucson, AZ, USA). Peptide was synthesized using cycles that started with the removal of Fmoc group, using 20% piperidine in N,N-Dimethylformamide (DMF) (2 × 5 min) followed by washing the resin with DMF (6 × 30 s). The first amino acid (Fmoc protected, 2 equivalent) was added in the presence of 0.4 M O-Benzotriazole-N,N,Nʹ,Nʹ-tetramethyl-uronium-hexafluoro-phosphate (HBTU, 1.9 equivalent), and 0.8 M 4-methylmorpholine (NMM, 4 equivalent) in DMF (3 × 30 min), amino acids were added in cycles. Excess reagents were washed (6 × 30 s) with DMF. The synthesis took place from C-terminal to N-terminal; amino acids side groups were protected during the synthesis.
For the coupling of 4R peptide to phospholipid, resin was washed with 0.5%
N,
N-Di isopropylethylamine (DIEA) in DMF (5 × 1 mL).
m-maleimidobenzoyl-
N-hydroxysuccinimide ester (MBS, 1.1 equivalent) and DIEA (1.1 equivalent) in 1 mL DMF were added to the resin and stirred for 2 h at room temperature. Second coupling was done with the same amounts of reagents, stirred at 4 °C, overnight. Resin was then washed with DMF (5 × 1 mL). Ptd Thioethanol Lipid (1.1 equivalent) was dissolved in chloroform and was added to the resin along with 1.1 equiv of DIEA. The reaction was run for several hours at room temperature. A second coupling was done with the lipid to ensure the reaction has gone to completion. The resin was then washed with DMF (5 × 1 mL), methylene chloride (5 × 1 mL) and dried. The conjugated peptide was cleaved from resin with 100% Trifluoroacetic acid (TFA) for 1.5 h and product was purified by reversed phase high performance liquid chromatography (RP-HPLC) on a Vydac™ protein and peptide C18 column. The final product was then identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (
Figure S1).
3.3. Galactosamine-DSPE-PEG2000 Coupling
DSPE-PEG
2000-COOH (1 equiv) was activated by the reaction with EDC (10 equivalent) in 2 mL dimethyl sulfoxide (DMSO) for 2 h at room temperature. NHS (10 equivalent) was then added to the mixture and stirred overnight at room temperature.
d-galactosamine HCl (2 equiv) was reacted with triethylamine (2 equivalent) overnight at room temperature to produce the free base.
d-galactosamine base was then added to the activated DSPE-PEG
2000-COOH and the reaction was run with continuous stirring in the dark at room temperature for 48 h. The resulting solution was then dialyzed using pre-treated regenerated cellulose (RC) Spectra/Por 7 dialysis membrane with molecular weight cut off size (MwCO) of 1000 Da (Spectrum Laboratories, Inc., Rancho Dominguez, CA, USA) against phosphate buffered saline (PBS) for 24 h, then against distilled water for another 24 h (to remove unreacted galactosamine, coupling reagents and DMSO). The resulting solution was lyophilized using the LabconcoFreeZone
® 6 L FreezeDry System (Labconco, Kansas, MO, USA). The obtained powder was evaluated for successful conjugation using MALDI-TOF MS (
Figure S2) and nuclear magnetic resonance (NMR) spectroscopy (
Figure S3).
3.4. Preparation of siRNA-SSLNP Complexes
siRNA-SSLNP complexes were prepared by film rehydration method [
45] with different nitrogen to phosphate (N/P) ratios (30, 20 and 10). Briefly, Lipid-Z and DSPE-PEG
2000 were dissolved separately in methanol then mixed in round bottom flasks at appropriate ratios. The solvent was subsequently removed using a vacuum rotary evaporator (BUCHI Labortechnik AG; Flawil, Switzerland) under a stream of argon and vacuum (600 mm Hg pressure) at 50 °C and 150 rpm for 30 min. The residual solvent from the resulting film was removed under vacuum overnight in dark. Thereafter, the dried film was rehydrated with 5 nmol of siRNA in nuclease free water. The resulting dispersion was vortexed until the film was dissolved, followed by bath sonication for 5 min. Flasks were then flushed with argon, sealed, and allowed to equilibrate in the dark for 2 h at 37 °C with continuous stirring to produce siRNA-SSLNP. Samples were then extruded through Nylon membranes with pore sizes of 200, 100 and 50 nm to ensure uniformity and particle size of <100 nm. Previously, we showed with an aid of isothermal titration calorimetry that insertion of ligand-conjugated lipid to pre-formed particles via self-association has a surface saturation point of approximately 5.6% without significant changes in particle properties [
46]. Based on this data we aimed for 10% galactosamine conjugation taking into account for the smaller size of GalN as compared to the ligand used in the previous study. Accordingly, appropriate amounts of DSPE-PEG
2000-GalN was incubated with the preformed particles and allowed to self-associate for 2 h. Empty SSMM were prepared using equal ratios of Lipid-Z and DSPE-PEG
2000 following the same procedure described above and reconstituted with siRNA-free nuclease-free water.
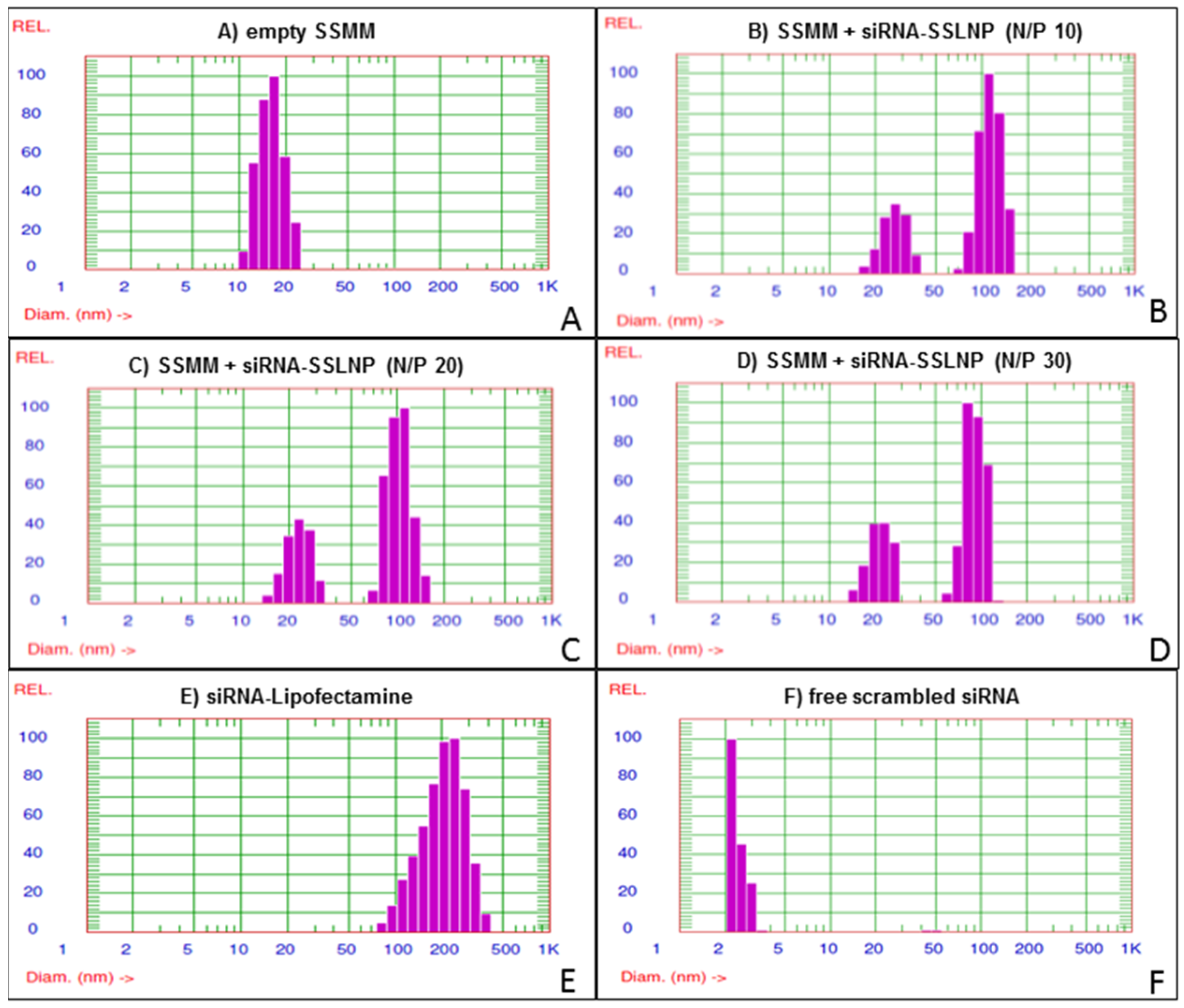
3.5. Physicochemical Characterization
Particle size distribution and zeta potential of the prepared samples were measured using dynamic light scattering (DLS) and electrophoretic light scattering (ELS) respectively by the particle sizer (Agilent 7030 NICOMP DLS/ZLS, Santa Clara, CA, USA) equipped with a 100 mW He-Ne laser (632.8 nm excitation wavelength) and set up at a fixed scattering angle of 90°. Solvent viscosity and refractive index of water were used with values of 0.933 cP, and 1.333 respectively. Samples were measured at room temperature and 1 atm pressure. The mean hydrodynamic particle diameters (dh) of particles in the aqueous dispersion, were calcaulated according to the Stokes-Einstein equation using the measured diffusion of particles in solution, while zeta potential (ζ) was determined using the Smoluchowski approximation. The reported experimental results were the average of at least three values obtained from analysis of the autocorrelation function accumulated for at least 15 min.
Transmission electron microscopy (TEM) images of the prepared siRNA-SSLNP and siRNA-SSLNP-GalN were acquired using a JOEL manufactured JEM-1220 transmission electron microscope (JEOL USA, Inc, Peabody, MA, USA) fitted with a tungsten electron source. Briefly, freshly prepared siRNA-SSLNP complexes (5 μL) were spotted onto 300-mesh format carbon-coated grids (Electron Microscopy Sciences, Hatfield, PA) and incubated for 5 min at room temperature. Negative staining was performed with 0.5% uranyl acetate (40 μL). Samples were air-dried. All TEM images were acquired at an accelerating voltage of 80 kV by GatanEs1000W 11MP CCD camera. Digital Micrograph software was used to analyze the resulting images.
For gel retardation studies, samples containing 200 ng of siRNA, with varying N/P ratios in nuclease free water, were electrophoresed through 15% Novex TBE-urea gel (Invitrogen™-life technologies, Grand Island, NY, USA) in TBE running buffer. Gels were run at a voltage of 180V for 60 min, then stained with 1:5000 SYBR Green-II in TBE with mild agitation for 30 min, after which they were photographed under UV light using BioRad Gel-Doc imaging system (Life Science Research, Hercules, CA, USA).
SYBR Green-II exclusion assay was performed to quantify the encapsulation of siRNA within SSLNP using the fluorescence quenching method. These experiments were carried out by measuring the fluorescence intensity of siRNA-SSLNP complexes, prepared with different N/P ratios, as a result of the intercalation between siRNA and SYBR Green-II. Fluorescence was measured using 96-well plate reader BioTek Synergy 4, manufactured by BioTek (Winooski, VT, USA) at excitation and emission wave lengths of 497 nm and at 520 nm respectively. Percentage of encapsulated siRNA was determined from the relative fluorescence obtained with each sample to that of SYBR Green-II and siRNA in the absence of lipids.
Nuclease resistance of SSLNP incorporated siRNA was determined after the treatment of samples with 1U of RNase I ribonuclease/µg siRNA for 30 min at 37 °C. 0.1% Triton-X 100 was used to terminate RNase activity and Heparin sodium 50 U/µg siRNA was used to disassemble SSLNPs nanoparticles. Gel retardation technique and SYBR Green-II exclusion assay were repeated to determine the integrity of the preserved SSLNP siRNA compared to free siRNA.
3.6. Cytotoxicity and Cell Uptake Studies
Hepatic Hep-G2 cells were seeded at a density of 6000 cells/well in 96-well plates and allowed to attach for 24 h at 37 °C and 5% CO2. Hep-G2 cells were cultured in 100 µL/well MEM medium, containing 10% FBS, 1 mM sodium pyruvate, 1% nonessential aminoacids, 100 U/mL penicillin, and 100 µg/mL streptomycin. After cell attachment, the medium was replaced with 100 µL/well fresh complete media, containing serial dilutions of vectors with a molar siRNA concentration ranging from 1 to 1000 nM, and incubated for 72 h. At the end of the incubation period, Hep-G2 were treated with CellTiter-96 AQ-one solution cell proliferation (MTS) assay according to manufacturer’s instructions. Cell viability in each well was determined by absorbance of the formazan product recorded at 490 nm by a BioTek Synergy 4 plate reader (Winooski, VT, USA) normalized to untreated control.
Primary HSC were seeded in 96-well plates at a density of 6000 cells/well and allowed to attach for 24 h at 37 °C and 5% CO2. HSC were cultured in 100 µL/well Stellate Cell Medium (SteSM), supplemented with 10% FBS, stellate cell growth supplements, and 100 U/mL penicillin, and 100 µg/mL streptomycin. After cell attachment, the medium was replaced with FBS free medium containing serial dilutions of vectors with a molar siRNA concentration from 1 to 1000 nM, and incubated overnight (16 h). At the end of the incubation period cells were subjected to CytoTox-one homogeneous membrane integrity (LDH) assay according to manufacturer’s protocols. Cell integrity was assessed by fluorometry at an excitation wavelength of 560 nm and an emission of 590 nm using BioTek Synergy 4 plate reader, manufactured by BioTek (Winooski, VT, USA). Data were normalized to untreated controls.
To assess the effect of SSLNPs on the proliferation of renal HK-2 cells over an incubation period of 72 h, cells were seeded at a density of 5000 cells/well in 96-well plates and allowed to attach for 24 h at 37 °C and 5% CO2. Cells were cultured in Keratinocyte Serum Free Medium supplemented with 0.05 mg/mL bovine pituitary extract (BPE), 5 ng/mL human recombinant epidermal growth factor (EGF) and antibiotic solution (penicillin 10,000 units/mL with streptomycin 10 mg/mL). After overnight attachment, medium was replaced with 100 μL/well fresh media containing serial dilutions of the following formulations, with a molar siRNA concentration ranging from 1 to 1000 nM: free scrambled-siRNA, scrambled -siRNA in SSLNP (with N/P ratios of 30), scrambled-siRNA in SSLNP-GalN or scrambled-siRNA in lipofectamine (LF). In addition, HK-2 cells were also incubated for 24, 48 or 72 h at the same incubation conditions with various siRNA formulations at a set siRNA concentration of 250 nM. At the end of incubation periods MTS solution was added to wells and plates were further incubated in the dark for 3 h after which the absorbance of formazan was measured at 490 nm using a BioTek Synergy 4 plate reader (Winooski, VT, USA). The results were normalized to untreated control and percentage of cell viability was calculated per treatment.
To assess the ability of SSLNP and SSLNP-GalN to transfect siRNA into cells in comparison to lipofectamine (LF), carboxyfluorescein (FAM)-labeled siRNA was formulated in SSLNPs at N/P ratio of 30 as described above. Hep-G2 cells were seeded in 6-well plates at a density of 200,000 cells/well as described above and incubated for 24 h prior to transfection. Cells were treated with either free FAM-siRNA, siRNA-SSLNP, siRNA-SSLNP-GalN at siRNA concentration of 200 nM or siRNA-LF at 50 nM concentration according to manufacture recommendations. Treated cells were incubated overnight then washed with PBS and trypsinized. The uptake of FAM-siRNA mediated with different vectors was detected using Beckman Coulter Cyan ADP flow cytometry and analyzer, manufactured by Beckman (Indianapolis, IN, USA).
3.7. In Vitro Protein Downregulation
To evaluate CTGF downregulation, hepatic Hep-G2 cells and renal HK-2 cells were seeded in 24-well plates at a density of 50,000 cells/well at conditions described above and incubated for 24 h prior to treatment. CTGF-siRNA complexes with SSLNP and SSLNP-GalN were prepared at N/P ratio of 30 as previously described. Cells were treated with either free CTGF-siRNA, CTGF-siRNA in SSLNP, or CTGF-siRNA in SSLNP-GalN at siRNA concentrations of 50, 100 and 200 nM, while positive control cells were treated with CTGF-siRNA in LF at 50 nM concentration according to manufacture recommendations. Treated cells were incubated overnight then analyzed for CTGF expression 24 h post-transfection using CTGF-ELISA kit (Antigenix, Inc., Huntington Station, NY, USA) according to the manufacture’s protocol. Results were normalized to total protein in samples measured by Bradford protein assay.
For the evaluation of collagen type I and III as well as α-SMA expression immunocytochemistry technique was used. Primary HSC were seeded at a density of 5000 cells/well on German glass slide with cover in 0.25 mL SteCM/well as described above. 24 h after incubation, cells were treated with either free CTGF-siRNA or CTGF-siRNA in SSLNP at siRNA concentration of 200 nM, while positive control cells were treated with CTGF-siRNA in LF at 50 nM siRNA concentration and incubated overnight. Cells were then washed three times with (37 °C) PBS with Ca2+ and Mg2+ and fixed in ice-cold methanol for 10 min, washed three times with PBST (0.1% tween in PBS), and incubated in PBST containing 1% bovine serum albumin (BSA) for 30 min. All primary antibodies (anti-collagen I, anti-collagen-III and anti-α-SMA) incubations were performed overnight at 4 °C in 1% BSA in PBST. Following three PBST washes, cells were incubated with Alexa-Fluor 488 conjugated secondary donkey-anti-rabbit antibody in 1% BSA in PBST for 1 h at room temperature and followed by three washes with PBST. Cell nuclei were stained with DAPI (4,6-diamidino-2-phenylindole) included in Vectashield mounting media (Vector Laboratories, Burlingame, CA, USA). Images were acquired using an Olympus IX70 inverted fluorescence microscope, manufactured by Olympus microscopy, Pennsylvania, USA. coupled with a QImaging RETIGA 1300 cooled-CCD digital camera; and processed using QCapture Pro™ 6 software.
To measure the amount of collagen deposit in the extracellular matrix of HSC, cells were seeded in 24-well plate at a density of 50,000 cells/well 24 h prior to treatment as described above. Cells were treated with either free CTGF-siRNA, CTGF-siRNA in SSLNP, or CTGF-siRNA in SSLNP-GalN at siRNA concentrations of 50, 100 and 200 nM, while positive control cells were treated with CTGF-siRNA in LF at 50 nM concentration and incubated overnight. Cells were then washed three times with (37 °C) PBS with Ca
2+ and Mg
2+ and fixed in ice-cold ethanol for 10 min, then dyed with Sirius red/fast green (Chondrex, Inc., Redmond, WA, USA) according to the supplier’s protocol. Briefly, dye solution was added and plates incubated for 30 min at room temperature. Dye solution was then removed by washing for multiple times until fluid appeared colorless. Dye extraction solution, provided with kit, was added to each well and mixed gently until color eluted from cells and ECM. Absorbance was measured using a BioTek Synergy 4 plate reader at 540 nm and 605 nm from which total collagen was calculated and normalized to total non-collagenous protein in well using the following formulas according to supplier’s protocol:
Renal HK-2 cells were seeded in 12-well plate at a density of 100,000 cells/well and allowed to attach and activate into myofibroblasts with 3 ng/mL TGF-β1 for 48 h at 37 °C and 5% CO
2 [
47]. The cells were incubated with 1 mL/well keratinocyte serum free media and supplements described earlier.
After verification of fibroblast phenotype by light microscopy, cells were treated with free CTGF-siRNA, CTGF-siRNA in SSLNP, or CTGF-siRNA in SSLNP-GalN at siRNA concentrations of 50, 100 and 200 nM. 50 nM CTGF-siRNA in LF was used as positive control, while 200 nM scrambled siRNA in SSLNP-GalN was used as a negative control.
Treated cells were then washed and re-incubated with fresh media for 48 h to allow the downregulation of CTGF and degradation of collagenous matrix. At the end of incubation period, cells were washed three times with (37 °C) PBS with Ca2+ and Mg2+ and fixed with ice-cold ethanol for 10 min, then dyed with Sirius red/fast green (Chondrex, Inc., Redmond, WA, USA) according to the supplier’s protocol. Data were expressed as percentage of collagen normalized to non-collagenous proteins, determined as described above for HSC cells.
3.8. In Vivo Biodistribution and Pharmacokinetic Studies
Biodistribution studies were performed on healthy 6 weeks old Balb/c male mice. Mice were randomized into four groups of 4 animals/group and treated with one of the following formulations: free Cy5 in 5% dextrose (D5W) (60 µg/animal corresponding to 76 nmol/kg), free Cy5-labeled siRNA, Cy5-labeled siRNA in SSLNP or Cy5-labeled siRNA in SSLNP-GalN with the later three formulations administered at 1mg siRNA/animal corresponding to 76 nmol/kg dose. Formulations were injected via tail vein at 0.1 mL and mice were anesthetized with intraperitoneal (IP) injection using ketamine/xylazine (90 mg/kg/3 mg/kg) then sacrificed by exsanguination at predetermined time points of 15 min, 1 h, 3 h, 6 h, 9 h and 24 h. Organs (heart, spleen, lungs, kidneys, and liver) as well as blood and urine were collected from each animal and photographed using Xenogen (Caliper Life Sciences, MA, USA) IVIS Spectrum 100 imaging system at excitation and emission wavelengths of 640 nm and 680 nm respectively. Fluorescence signals were quantified using Living Image 4.0 acquisition and analysis software, by Caliper Life Sceinces, MA, USA. Blood, collected by cardiac puncture into EDTA coated BD™ microtainer tubes, was centrifuged at 3000 rpm for 10 min to separate plasma. Cy5-siRNA concentration in plasma was quantified using a 96-well plate reader BioTek Synergy 4 (Winooski, VT, USA) at excitation and emission wavelengths of 640 nm and 680 nm respectively. siRNA concentration at tested time points was used to plot plasma concentration vs. time curve and calculate PK parameters. Pharmacokinetic parameters were calculated according to two-compartmental modeling using Pharsight Phoenix WinNonlin 6.3 PK/PD modeling and simulation software, by Certara, NJ, USA.
3.9. Data and Statistical Analysis
All results are expressed as the mean ± standard deviation (SD) of at least three experiments. For statistical analysis, student’s t-test or one-way analysis of variance (ANOVA) followed by Fisher least significant difference post-hoc test were used. p values less than 0.05 (p < 0.05) were considered to be statistically significant.