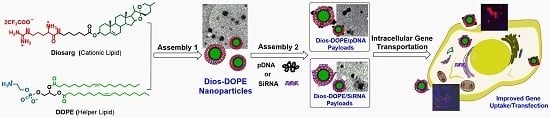
Cationic Nanoparticles Assembled from Natural-Based Steroid Lipid for Improved Intracellular Transport of siRNA and pDNA
Abstract
:1. Introduction
2. Experimental
2.1. Materials
2.2. Analytical Methods
2.2.1. 1H NMR Measurements
2.2.2. Preparation of the Self-Assembled Diosarg-DOPE NPs
2.2.3. Average Particle Sizes and Surface Charges of the Diosarg-DOPE NPs, siRNA and pDNA-Loaded Complexes Measured by the Dynamic Light Scattering Instrument
2.2.4. Morphologies of the Diosarg-DOPE NPs and the siRNA and pDNA-Loaded Complexes Measured by Transmission Electronic Microscopy
2.2.5. MTT Cytotoxicity Assay of the Diosarg Lipid and Diosarg-DOPE NPs
2.2.6. Intracellular Transport of the Cy3-siRNA and Cy3-pDNA-Loaded Complexes of the Diosarg and Diosarg-DOPE NPs Measured by Flow Cytometry and Observed by Fluorescence Microscopy
2.2.7. The Observed Intracellular Transport of the Cy3-siRNA and Cy3-pDNA-Loaded Complexes of the Diosarg and Diosarg-DOPE NPs
2.2.8. Luciferase Gene Transfection for the pLuc DNA-Loaded Complexes of Diosarg and Diosarg-DOPE NPs
2.2.9. Intracellular Co-Localization of the Diosarg/Cy3-pDNA and Diosarg-DOPE NPs/Cy3-pDNA Complexes Observed by Fluorescence Microscopy
3. Results and Discussion
3.1. Preparation of Diosarg-DOPE Nanoparticles and Their Physico-Chemical Properties
3.2. Average Particle Size, Surface Potential and Morphology of the siRNA- and pDNA-Loaded Complexes
3.3. Cytotoxicity of the Diosarg Lipid and Diosarg-DOPE NPs in H1299 Cells by the MTT Assay
3.4. Intracellular Transportation of the Cy3-siRNA- and Cy3-DNA-Loaded Complexes Measured by Flow Cytometry and by Fluorescent Imaging
3.5. Luciferase Gene Transfection Efficiency of the pLuc DNA-Loaded Complexes
3.6. Intracellular Co-Localization of Diosarg/Cy3-pDNA and Diosarg-DOPE NPs/Cy3-pDNA Complexes by Fluorescent Imaging
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2014, 43, 744–764. [Google Scholar] [CrossRef] [PubMed]
- Fortier, C.; Durocher, Y.; De Crescenzo, G. Surface modification of nonviral nanocarriers for enhanced gene delivery. Nanomedicine 2014, 9, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Zhang, S.; Wang, B.; Zhao, Y.; Yang, B.; Yu, S. Transfection efficiency of cationic lipids with different hydrophobic domains in gene delivery. Bioconjugate Chem. 2010, 21, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The headgroup evolution of cationic lipids for gene delivery. Bioconjugate Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Biswas, J. Understanding membranes through the molecular design of lipids. Langmuir 2009, 26, 4642–4654. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ang, C.Y.; Wiradharma, N.; Yong, L.-K.; Liu, S.; Liu, L.; Gao, S.; Yang, Y.Y. Diaminododecane-based cationic bolaamphiphile as a non-viral gene delivery carrier. Biomaterials 2012, 33, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.J.; Zhang, Q.F.; Zhang, J.; Liu, Q.; Ren, L.; Chen, Q.M.; Guo, L.; Yu, X.Q. Cyclen-based lipidic oligomers as potential gene delivery vehicles. Acta Biomater. 2014, 10, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Luo, T.; Zhu, Y.; Li, H.; Sun, J.; Chen, S.; Sun, W.; Cao, A. The intracellular plasmid DNA localization of cationic reducible cholesterol-disulfide lipids. Biomaterials 2011, 32, 3507–3519. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Luo, T.; Li, H.; Sun, J.; Wang, Z.; Cao, A. ‘Click’ synthesized sterol-based cationic lipids as gene carriers, and the effect of skeletons and headgroups on gene delivery. Bioorg. Med. Chem. 2013, 21, 6366–6377. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; Luo, T.; Li, H.; Sun, J.; Wang, Z.; Cao, A. Cholesterol-based cationic lipids for gene delivery: Contribution of molecular structure factors to physico-chemical and biological properties. Colloid. Surf. B 2014, 116, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; McIntosh, T.J.; Grinstaff, M.W. Functional lipids and lipoplexes for improved gene delivery. Biochimie 2012, 94, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, F.; Nishioka, T.; Yamashita, F.; Takakura, Y.; Hashida, M. Effects of erythrocytes and serum proteins on lung accumulation of lipoplexes containing cholesterol or DOPE as a helper lipid in the single-pass rat lung perfusion system. Eur. J. Pharm. Biopharm. 2001, 52, 65–172. [Google Scholar] [CrossRef]
- Wang, X.; Yu, B.; Ren, W.; Mo, X.; Zhou, C.; He, H.; Jia, H.; Wang, L.; Jacob, S.T.; Lee, R.J.; et al. Enhanced hepatic delivery of siRNA and microRNA using oleic acid based lipid nanoparticle formulations. J. Control. Release 2013, 172, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, M.; Alberti, K.A.; Choy, A.; Xu, Q. DOPE facilitates quaternized lipidoids (QLDs) for in vitro DNA delivery. Nanomedicine 2013, 9, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, W.; Szoka, F.C., Jr. Asymmetric 1-alkyl-2-acyl phosphatidylcholine: A helper lipid for enhanced non-viral gene delivery. Int. J. Pharm. 2012, 427, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Scarzello, M.; Chupin, V.; Wagenaar, A.; Stuart, M.C. A.; Engberts, J.B.F.N.; Hulst, R. Polymorphism of pyridinium amphiphiles for gene delivery: Influence of ionic strength, helper lipid content, and plasmid DNA complexation. Biophys. J. 2005, 88, 2104–2113. [Google Scholar] [CrossRef] [PubMed]
- Tabatt, K.; Kneuer, C.; Sameti, M.; Olbrich, C.; Müller, R.H.; Lehr, C.M.; Bakowsky, U. Transfection with different colloidal systems: Comparison of solid lipid nanoparticles and liposomes. J. Control. Release 2004, 97, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Nakamura, T.; Ohno, H.; Fujii, N.; Maitani, Y. siRNA delivery into tumor cells by lipid-based nanoparticles composed of hydroxyethylated cholesteryl triamine. Int. J. Pharm. 2013, 443, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Felnerova, D.; Viret, J.F.; Glück, R.; Moser, C. Liposomes and virosomes as delivery systems for antigens, nucleic acids and drugs. Curr. Opin. Biotech. 2004, 15, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Lonez, C.; Vandenbranden, M.; Ruysschaert, J.M. Cationic liposomal lipids: From gene carriers to cell signaling. Prog. Lipid Res. 2008, 47, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, M.; Khoshhamdam, M.; Dehshahri, A.; Malaekeh-Nikouei, B. The influence of size, lipid composition and bilayer fluidity of cationic liposomes on the transfection efficiency of nanolipoplexes. Colloid. Surf. B 2009, 72, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta 2013, 1828, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Huang, L. Recent advances in nonviral vectors for gene delivery. Acc. Chem. Res. 2011, 45, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Falsini, S.; Ciani, L.; Ristori, S.; Fortunato, A.; Arcangeli, A. Advances in lipid-based platforms for RNAi therapeutics. J. Med. Chem. 2013, 57, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Mével, M.; Kamaly, N.; Carmona, S.; Oliver, M.H.; Jorgensen, M.R.; Crowther, C.; Salazar, F.H.; Marion, P.L.; Fujino, M.; Natori, Y.; et al. DODAG; a versatile new cationic lipid that mediates efficient delivery of pDNA and siRNA. J. Control. Release 2010, 143, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Y.; Xia, K.; Sheng, R.; Jia, L.; Hou, X.; Xu, Y.; Cao, A. Dendritic poly(l-lysine)-b-poly(l-lactide)-b-dendritic poly(l-lysine) amphiphilic gene delivery vectors: Roles of PLL dendritic generation and enhanced transgene efficacies via termini modification. Biomacromolecules 2009, 10, 2284–2293. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liu, X.; Samoshina, N.M.; Samoshin, V.V.; Franz, A.H.; Guo, X. trans-2-Aminocyclohexanol-based amphiphiles as highly efficient helper lipids for gene delivery by lipoplexes. Biochim. Biophys. Acta 2015, 1848, 3113–3125. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wang, J.C.; Zhao, E.Y.; Gao, L.Y.; Feng, Q.; Liu, X.Y.; Zhao, Z.X.; Ma, X.F.; Hou, W.J.; Zhang, L.R.; et al. Self-assembly cationic nanoparticles based on cholesterol-grafted bioreducible poly(amidoamine) for siRNA delivery. Biomaterials 2013, 34, 5303–5316. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Chen, C.K.; Jiang, M.; Fang, L.; Cheng, C.; Pfeifer, B.A. Synthesis of cationic polylactides with tunable charge densities as nanocarriers for effective gene delivery. Mol. Pharm. 2013, 10, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, A.; Sánchez-Hernández, N.; Ticó, J.R.; García-Montoya, E.; Pérez-Lozano, P.; Suñé-Negre, J.M.; Hernández-Munain, C.; Suñé, C.; Miñarro, M. A new optimized formulation of cationic solid lipid nanoparticles intended for gene delivery: Development, characterization and DNA binding efficiency of TCERG1 expression plasmid. Int. J. Pharm. 2014, 473, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.U.; Zuhorn, I.S.; Hoekstra, D. How cationic lipids transfer nucleic acids into cells and across cellular membranes: Recent advances. J. Control. Release 2013, 166, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-F.; Yi, W.-J.; Wang, B.; Zhang, J.; Ren, L.; Chen, Q.-M.; Guo, L.; Yu, X.Q. Linear polycations by ring-opening polymerization as non-viral gene delivery vectors. Biomaterials 2013, 34, 5391–5401. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Chen, Z.; Jin, W.; Cheng, K. Development of cholesteryl peptide micelles for siRNA delivery. J. Control. Release 2013, 172, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhang, S.; Jiang, H.; Zhao, B.; Lv, H. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J. Control. Release 2007, 123, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yang, Y.; Wang, A.; Hu, B.; Luo, X.; Sheng, R.; Dong, Y.; Fan, W. Quantitative and highly selective sensing of sodium houttuyfonate via long-aliphatic chains hydrophobic assembly and aggregation-induced emission. New J. Chem. 2015, 39, 9743–9751. [Google Scholar] [CrossRef]
- Zuhorn, I.S.; Bakowsky, U.; Polushkin, E.; Visser, W.H.; Stuart, M.C.A.; Engberts, J.B.F.N.; Hoekstra, D. Nonbilayer phase of lipoplex-membrane mixture determines endosomal escape of genetic cargo and transfection efficiency. Mol. Therapy 2005, 11, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Sheng, R.; An, F.; Wang, Z.; Li, M.; Cao, A. Assembly of plasmid DNA with pyrene-amines cationic amphiphiles into nanoparticles and their visible lysosome localization. RSC Adv. 2015, 5, 12338–12345. [Google Scholar] [CrossRef]
- Sheng, R.; Xia, K.; Chen, J.; Xu, Y.; Cao, A. Terminal modification on mPEG-dendritic poly-(l)-lysine cationic diblock copolymer for efficient gene delivery. J. Biomater. Sci. Polym. Ed. 2013, 24, 1935–1951. [Google Scholar] [CrossRef] [PubMed]
- Vighi, E.; Montanari, M.; Hanuskova, M.; Iannuccelli, V.; Coppi, G.; Leo, E. Design flexibility influencing the in vitro behavior of cationic SLN as a nonviral gene vector. Int. J. Pharm. 2013, 440, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Swaney, W.; Sorgi, F.; Bahnson, A.; Barranger, J. The effect of cationic liposome pretreatment and centrifugation on retrovirus-mediated gene transfer. Gene Therapy 1997, 4, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Zhang, H.; Shen, Z.; Bi, J.; Dai, S. Developing a chitosan supported imidazole Schiff-base for high-efficiency gene delivery. Polym. Chem. 2013, 4, 840–850. [Google Scholar] [CrossRef]
- Perez, A.P.; Cosaka, M.L.; Romero, E.L.; Morilla, M.J. Uptake and intracellular traffic of siRNA dendriplexes in glioblastoma cells and macrophages. Int. J. Nanomed. 2011, 6, 2715–2728. [Google Scholar]
- Li, H.; Luo, T.; Sheng, R.; Sun, J.; Wang, Z.; Cao, A. Endoplasmic reticulum localization of poly(ω-aminohexyl methacrylamide)s conjugated with (l-)-arginines in plasmid DNA delivery. Biomaterials 2013, 34, 7923–7938. [Google Scholar] [CrossRef] [PubMed]
- Mével, M.; Haudebourg, T.; Colombani, T.; Peuziat, P.; Dallet, L.; Chatin, B.; Lambert, O.; Berchel, M.; Montier, T.; Jaffrès, P.A.; et al. Important role of phosphoramido linkage in imidazole-based dioleyl helper lipids for liposome stability and primary cell transfection. J. Gene Med. 2015, 18. [Google Scholar] [CrossRef] [PubMed]
- Koynova, R.; Tenchov, B.; MacDonald, R.C. Nonlamellar phases in cationic phospholipids, relevance to drug and gene delivery. ACS Biomater. Sci. Eng. 2015, 1, 130–138. [Google Scholar] [CrossRef]
- Khalil, I.A.; Kogure, K.; Akita, H.; Harashima, H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharm. Rev. 2006, 58, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Kou, L.; Sun, J.; Zhai, Y.; He, Z. The endocytosis and intracellular fate of nanomedicines: Implication for rational design. Asia J. Pharm. Sci. 2013, 8, 1–10. [Google Scholar] [CrossRef]
- Chu, Z.; Miu, K.; Lung, P.; Zhang, S.; Zhao, S.; Chang, H.-C.; Lin, G.; Li, Q. Rapid endosomal escape of prickly nanodiamonds: Implications for gene delivery. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Caratti, G.; Ince, L.M.; Poolman, T.M.; Trebble, P.J.; Holt, C.M.; Ray, D.W.; Matthews, L.C. Serum cholesterol selectively regulates glucocorticoid sensitivity through activation of JNK. J. Endocrinol. 2014, 223, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Morgenstern, R.; Nyström, A.M. Nanoparticle-directed sub-cellular localization of doxorubicin and the sensitization breast cancer cells by circumventing GST-Mediated drug resistance. Biomaterials 2014, 35, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ho, W.; Zhang, X.; Bertrand, N.; Farokhzad, O. Cancer nanomedicine: From targeted delivery to combination therapy. Trend. Mol. Med. 2015, 21, 223–232. [Google Scholar] [CrossRef] [PubMed]









| Cationic Carrier Samples | Hydrodynamic Mean Particle Size (nm) a | PDI | Zeta Potential (mv) |
|---|---|---|---|
| Diosarg | / | / | / |
| Diosarg-DOPE (2:1) NPs | 116 ± 2 | 0.275 | 49.1 ± 5.1 |
| Diosarg-DOPE (1:1) NPs | 117 ± 3 | 0.225 | 46.8 ± 7.2 |
| Diosarg-DOPE (1:2) NPs | 105 ± 2 | 0.274 | 49.5 ± 5.6 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, R.; Zhuang, X.; Wang, Z.; Cao, A.; Lin, K.; Zhu, J.X.X. Cationic Nanoparticles Assembled from Natural-Based Steroid Lipid for Improved Intracellular Transport of siRNA and pDNA. Nanomaterials 2016, 6, 69. https://doi.org/10.3390/nano6040069
Sheng R, Zhuang X, Wang Z, Cao A, Lin K, Zhu JXX. Cationic Nanoparticles Assembled from Natural-Based Steroid Lipid for Improved Intracellular Transport of siRNA and pDNA. Nanomaterials. 2016; 6(4):69. https://doi.org/10.3390/nano6040069
Chicago/Turabian StyleSheng, Ruilong, Xiaoqing Zhuang, Zhao Wang, Amin Cao, Kaili Lin, and Julian X. X. Zhu. 2016. "Cationic Nanoparticles Assembled from Natural-Based Steroid Lipid for Improved Intracellular Transport of siRNA and pDNA" Nanomaterials 6, no. 4: 69. https://doi.org/10.3390/nano6040069
APA StyleSheng, R., Zhuang, X., Wang, Z., Cao, A., Lin, K., & Zhu, J. X. X. (2016). Cationic Nanoparticles Assembled from Natural-Based Steroid Lipid for Improved Intracellular Transport of siRNA and pDNA. Nanomaterials, 6(4), 69. https://doi.org/10.3390/nano6040069