Biosensing and Delivery of Nucleic Acids Involving Selected Well-Known and Rising Star Functional Nanomaterials
Abstract
1. Introduction
2. AuNPs, AuNWs, Janus and Magnetic Nanoparticles: Synthesis and Functionalization
2.1. AuNPs
2.2. Magnetic Nanomaterials
2.3. Janus Nanoparticles
2.4. AuNWs
3. Biosensing and Delivery of Nucleic Acids: Roles, Opportunities and Cutting-Edge Applications of AuNPs, Magnetic Nanomaterials, Janus Nanoparticles and AuNWs
3.1. Optical Biosensing or Delivery of Nucleic Acids Using Functional Nanomaterials
3.2. Electrochemical Biosensing of Nucleic Acids Using Functional Nanomaterials.
4. Main Conclusions, Challenges and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paleček, E.; Fojta, M. Magnetic beads as versatile tools for electrochemical DNA and protein biosensing. Talanta 2007, 74, 276–290. [Google Scholar] [CrossRef]
- Abi, A.; Mohammadpour, Z.; Zuo, X.; Safavi, A. Nucleic acid-based electrochemical nanobiosensors. Biosens. Bioelectron. 2018, 102, 479–489. [Google Scholar] [CrossRef]
- Esteban-Fernández de Avila, B.; Martín, A.; Soto, F.; López-Ramirez, M.A.; Campuzano, S.; Vásquez-Machado, G.M.; Gao, W.; Zhang, L.; Wang, J. Single cell real-time miRNAs sensing based on nanomotors. ACS Nano 2015, 9, 6756–6764. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Fernández de Ávila, B.; Angell, C.; Soto, F.; Lopez-Ramirez, M.A.; Báez, D.F.; Xie, S.; Wang, J.; Chen, Y. Acoustically propelled nanomotors for intracellular siRNA delivery. ACS Nano 2016, 10, 4997–5005. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, E. Synthesis and electrochemical applications of gold nanoparticles. Anal. Chim. Acta 2007, 598, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-C.; Creran, B.; Rotello, V.M. Gold Nanoparticles: Preparation, Properties, and Applications in Bionanotechnology. Nanoscale 2012, 4, 1871–1880. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Nanoparticles for nucleic-acid-based biosensing: Opportunities, challenges, and prospects. Anal. Bioanal. Chem. 2019, 411, 1791–1806. [Google Scholar] [CrossRef]
- Cai, H.; Xu, C.; He, P.; Fang, Y. Colloid Au-enhanced DNA immobilization for the electrochemical detection of sequence-specific DNA. J. Electroanal. Chem. 2001, 510, 78–85. [Google Scholar] [CrossRef]
- Kang, J.; Li, X.; Wu, G.; Wang, Z.; Lu, X. A new scheme of hybridization based on the Au(nano)-DNA modified glassy carbon electrode. Anal. Biochem. 2007, 364, 165–170. [Google Scholar] [CrossRef]
- Zouari, M.; Campuzano, S.; Pingarrón, J.M.; Raouafi, N. Ultrasensitive determination of microribonucleic acids in cancer cells with nanostructured-disposable electrodes using the viral protein p19 for recognition of ribonucleic acid/microribonucleic acid homoduplexes. Electrochim. Acta 2018, 262, 39–47. [Google Scholar] [CrossRef]
- Zouari, M.; Campuzano, S.; Pingarrón, J.M.; Raouafi, N. Amperometric Biosensing of miRNA-21 in serum and cancer cells at nanostructured platforms using anti-DNA−RNA hybrid antibodies. ACS Omega 2018, 3, 8923–8931. [Google Scholar] [CrossRef] [PubMed]
- de la Escosura-Muñiz, A.; Baptista-Pires, L.; Serrano, L.; Altet, L.; Francino, O.; Sánchez, A.; Merkoçi, A. Magnetic bead/gold nanoparticle double-labeled primers for electrochemical detection of isothermal amplified Leishmania DNA. Small 2016, 12, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Wang, P.; Su, Y.; Wang, L.; Pan, D.; Aldalbahi, A.; Yang, S.; Zuo, X. Nanoprobe-initiated enzymatic polymerization for highly sensitive electrochemical DNA detection. ACS Appl. Mater. Interfaces 2015, 7, 25618–25623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Zhu, C.; Wu, S.; Mandler, D.; Marks, R.S.; Zhang, H. Amplified detection of femtomolar DNA based on a one-to-few recognition reaction between DNA–Au conjugate and target DNA. Nanoscale 2014, 6, 3110–3115. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, F.; Ni, J.; Liao, X.; Zhang, X.; Lin, Z. Facile construction of a highly sensitive DNA biosensor by in-situ assembly of electro-active tags on hairpin-structured probe fragment. Sci. Rep. 2016, 6, 22441. [Google Scholar] [CrossRef]
- Zouari, M.; Campuzano, S.; Pingarrón, J.M.; Raouafi, N. Femtomolar direct voltammetric determination of miRNAs in sera of cancer patients using enzymeless sandwich hybridization biosensors. Anal. Chim. Acta. submitted.
- Yang, Y.; Li, C.; Yin, L.; Liu, M.; Wang, Z.; Shu, Y.; Li, G. Enhanced charge transfer by gold nanoparticle at DNA modified electrode and its application to label-free DNA Detection. ACS Appl. Mater. Interfaces 2014, 6, 7579–7584. [Google Scholar] [CrossRef]
- Freitas, M.; Couto, M.S.; Barroso, M.F.; Pereira, C.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J.; Delerue-Matos, C. Highly monodisperse Fe3O4@Au superparamagnetic nanoparticles as reproducible platform for genosensing genetically modified organisms. ACS Sens. 2016, 1, 1044–1053. [Google Scholar] [CrossRef]
- Rocha-Santos, T.A.P. Sensors and biosensors based on magnetic nanoparticles. Trac. Trends Anal. Chem. 2014, 62, 28–36. [Google Scholar] [CrossRef]
- Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Magnetic particles coupled to disposable screen-printed transducers for electrochemical biosensing. Sensors 2016, 16, 1585. [Google Scholar] [CrossRef]
- Reverté, L.; Prieto-Simón, B.; Campàs, M. New advances in electrochemical biosensors for the detection of toxins: Nanomaterials, magnetic beads and microfluidics systems. A review. Anal. Chim. Acta 2016, 908, 8–21. [Google Scholar]
- Serafín, V.; Torrente-Rodríguez, R.M.; Batlle, M.; García de Frutos, P.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Comparative evaluation of the performance of electrochemical immunosensors using magnetic microparticles and nanoparticles. Application to the determination of tyrosine kinase receptor AXL. Microchim. Acta 2017, 184, 4251–4258. [Google Scholar]
- Kudr, J.; Haddad, Y.; Richtera, L.; Heger, Z.; Cernak, M.; Adam, V.; Zitka, O. Magnetic nanoparticles: From design and synthesis to real world applications. Nanomaterials 2017, 7, 243. [Google Scholar] [CrossRef] [PubMed]
- Kudr, J.; Klejdus, B.; Adam, V.; Zitka, O. Magnetic solids in electrochemical analysis. Trac Trends Anal. Chem. 2018, 98, 104–113. [Google Scholar] [CrossRef]
- Pastucha, M.; Farka, Z.; Lacina, K.; Mikušová, Z.; Skládal, P. Magnetic nanoparticles for smart electrochemical immunoassays: A review on recent developments. Microchim. Acta 2019, 186, 312. [Google Scholar] [CrossRef]
- Loaiza, O.A.; Jubete, E.; Ochoteco, E.; Cabanero, G.; Grande, H.; Rodriguez, J. Gold coated ferric oxide nanoparticles based disposable magnetic genosensors for the detection of DNA hybridization processes. Biosens. Bioelectron. 2011, 26, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Lai, Y.J.; Zhang, W.; Jin, L.T. Fe2O3@Au core/shell nanoparticle-based electrochemical DNA biosensor for Escherichia coli detection. Talanta 2011, 84, 607–613. [Google Scholar] [CrossRef]
- Zhao, J.; Lin, F.; Yi, Y.; Huang, Y.; Li, H.; Zhang, Y.; Yao, S. Dual amplification strategy of highly sensitive thrombin amperometric aptasensor based on chitosan–Au nanocomposites. Analyst 2012, 137, 3488–3495. [Google Scholar] [CrossRef]
- Plácido, A.; Pereira, C.; Guedes, A.; Barroso, M.F.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Delerue-Matos, C. Electrochemical genoassays on gold-coated magnetic nanoparticles to quantify genetically modified organisms (GMOs) in food and feed as GMO percentage. Biosens. Bioelectron. 2018, 110, 147–154. [Google Scholar] [CrossRef]
- Plácido, A.; Pereira, C.; Barroso, M.F.; de-los-Santos-Álvarez, N.; Delerue-Matos, C. Chronoamperometric magnetogenosensing for simultaneous detection of two Roundup Ready™ soybean lines: GTS 40-3-2 and MON89788. Sens. Actuators B Chem. 2019, 283, 262–268. [Google Scholar]
- Pal, S.; Alocilja, E.C. Electrically active magnetic nanoparticles as novel concentrator and electrochemical redox transducer in Bacillus anthracis DNA detection. Biosens. Bioelectron. 2010, 26, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Situ, B.; Cao, N.; Li, B.; Liu, Q.; Lin, L.; Dai, Z.; Zou, X.; Cai, Z.; Wang, Q.; Yan, X.; et al. Sensitive electrochemical analysis of BRAFV600E mutation based on an amplification-refractory mutation system coupled with multienzyme functionalized Fe3O4/Au nanoparticles. Biosens. Bioelectron. 2013, 43, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.H.; Arias, J.L.; Nicolas, J.; Couvreur, P. Magnetic nanoparticles: Design and characterization, toxicity and biocompatibility, pharmaceutical and biomedical applications. Chem. Rev. 2012, 112, 5818–5878. [Google Scholar] [CrossRef] [PubMed]
- Urbanova, V.; Magro, M.; Gedanken, A.; Baratella, D.; Vianello, F.; Zboril, R. Nanocrystalline iron oxides, composites, and related materials as a platform for electrochemical, magnetic, and chemical biosensors. Chem. Mater. 2014, 26, 6653–6673. [Google Scholar] [CrossRef]
- Güzel, R.; Üstündağ, Z.; Ekşi, H.; Keskin, S.; Taner, B.; Durgun, Z.G.; Turan, A.A.İ.; Solak, A.O. Effect of Au and Au@Ag core–shell nanoparticles on the SERS of bridging organic molecules. J. Colloid Interface Sci. 2010, 351, 35–42. [Google Scholar]
- Yola, M.L.; Eren, T.; Atar, N. A novel and sensitive electrochemical DNA biosensor based on Fe@Au nanoparticles decorated grapheme oxide. Electrochim. Acta 2014, 125, 38–47. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, E. Electrochemical biosensors based on magnetic micro/nano particles. Electrochim. Acta 2012, 84, 62–73. [Google Scholar] [CrossRef]
- Pan, J.; Yang, Q. Antibody-functionalized magnetic nanoparticles for the detection of carcinoembryonic antigen using a flow-injection electrochemical device. Anal. Bioanal. Chem. 2007, 388, 279–286. [Google Scholar] [CrossRef]
- Tran, L.D.; Nguyen, B.H.; Hieu, N.V.; Tran, H.V.; Nguyen, H.L.; Nguyen, P.X. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen-printed electrodes. Mater. Sci. Eng. C 2011, 31, 477–485. [Google Scholar] [CrossRef]
- Yi, Y.; Sanchez, L.; Gao, Y.; Yu, Y. Janus particles for biological imaging and sensing. Analyst 2016, 141, 3526–3539. [Google Scholar] [CrossRef]
- Shao, Z.; Cao, H.; Yang, Y.; Chen, X. Intelligent Janus nanoparticles for intracellular real-time monitoring of dual drug release. Nanoscale 2016, 8, 6754–6760. [Google Scholar]
- Schrittwieser, S.; Pelaz, B.; Parak, W.J.; Lentijo-Mozo, S.; Soulantica, K.; Dieckho, J.; Ludwig, F.; Guenther, A.; Tschöpe, A.; Schotter, J. Homogeneous biosensing based on magnetic particle labels. Sensors 2016, 16, 828. [Google Scholar] [CrossRef] [PubMed]
- Yánez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Janus particles for (bio)sensing. Appl. Mat. Today 2017, 9, 276–288. [Google Scholar] [CrossRef]
- Campuzano, S.; Gamella, M.; Serafín, V.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. Magnetic Janus particles for static and dynamic (bio)sensing. Magnetochemistry 2019, 5, 47. [Google Scholar] [CrossRef]
- Jung, C.W.; Jalani, G.; Ko, J.; Choo, J.; Lim, D.W. Synthesis, characterization, and directional binding of anisotropic biohybrid microparticles for multiplexed biosensing. Macromol. Rapid Commun. 2014, 35, 56–65. [Google Scholar] [CrossRef]
- Li, B.; Wang, M.; Chen, K.; Cheng, Z.; Chen, G.; Zhang, Z. Synthesis of biofunctional Janus particles. Macromol. Rapid Commun. 2015, 36, 1200–1204. [Google Scholar] [CrossRef]
- Sánchez, A.; Díez, P.; Martínez-Ruíz, P.; Villalonga, R.; Pingarrón, J.M. JanusAu-mesoporous silica nanoparticles as electrochemical biorecognition-signaling system. Electrochem. Commun. 2013, 30, 51–54. [Google Scholar] [CrossRef]
- Boujakhrout, A.; Sánchez, E.; Díez, P.; Sánchez, A.; Martínez-Ruiz, P.; Parrado, C.; Pingarrón, J.M.; Villalonga, R. Single-walled carbon nanotubes/Au–mesoporoussilica Janus nanoparticles as building blocks for the preparation of a bienzyme biosensor. ChemElectroChem 2015, 2, 1735–1741. [Google Scholar] [CrossRef]
- Paniagua, G.; Villalonga, A.; Eguílaz, M.; Vegas, B.; Parrado, C.; Rivas, G.; Díez, P.; Villalonga, R. Amperometric aptasensor for carcinoembryonic antigen based on the use of bifunctionalized Janus nanoparticles as biorecognition-signaling element. Anal. Chim. Acta 2019, 1061, 84–91. [Google Scholar] [CrossRef]
- Jimenez-Falcao, S.; Parra-Nieto, J.; Pérez-Cuadrado, H.; Martínez-Máñez, R.; Martínez-Ruiz, P.; Villalonga, R. Avidin-gated mesoporous silica nanoparticles for signal amplification in electrochemical biosensor. Electrochem. Commun. 2019, 108, 109556. [Google Scholar] [CrossRef]
- Campuzano, S.; Kagan, D.; Orozco, J.; Wang, J. Motion-driven sensing and biosensing using electrochemically propelled nanomotors. Analyst 2011, 136, 4621–4630. [Google Scholar] [CrossRef] [PubMed]
- Kagan, D.; Calvo-Marzal, P.; Balasubramanian, S.; Sattayasamitsathit, S.; Manesh, K.M.; Flechsig, G.-U.; Wang, J. Chemical sensing based on catalytic nanomotors: Motion-based detection of trace silver. J. Am. Chem. Soc. 2009, 131, 12082–12083. [Google Scholar] [CrossRef] [PubMed]
- Kline, T.R.; Paxton, W.F.; Mallouk, T.E.; Sen, A. Catalytic nanomotors: Remote-controlled autonomous movement of striped metallic nanorods. Angew. Chem. Int. Ed. 2005, 44, 744–746. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Sánchez, B. Nanoscale biosensors based on self-propelled objects. Biosensors 2018, 8, 59. [Google Scholar] [CrossRef]
- Wang, W.; Li, S.; Mair, L.; Ahmed, S.; Huang, T.J.; Mallouk, T.E. Acoustic propulsion of nanorod motors inside living cells. Angew. Chem. Int. Ed. 2014, 53, 3201–3204. [Google Scholar] [CrossRef]
- Wu, J.; Balasubramanian, S.; Kagan, D.; Manesh, K.M.; Campuzano, S.; Wang, J. Motion-based DNA detection using catalytic nanomotors. Nat. Commun. 2010, 1, 36. [Google Scholar] [CrossRef]
- Wang, J. Self-propelled affinity biosensors: Moving the receptor around the sample. Biosens. Bioelectron. 2016, 76, 234–242. [Google Scholar] [CrossRef]
- Qualliotine, J.R.; Bolat, G.; Beltrán-Gastélum, M.; Esteban-Fernández de Ávila, B.; Wang, J.; Califano, J.A. Acoustic nanomotors for detection of human papilloma virus–associated head and neck cancer. Otolaryngol. Head Neck Surg. 2019, 161, 814–822. [Google Scholar] [CrossRef]
- Hansen-Bruhn, M.; Esteban-Fernández de Ávila, B.; Beltrán-Gastélum, M.; Zhao, J.; Ramírez-Herrera, D.E.; Angsantikul, P.; Vesterager Gothelf, K.; Zhang, L.; Wang, J. Active intracellular delivery of a Cas9/sgRNA complex using ultrasound-propelled nanomotors. Angew. Chem. Int. Ed. 2018, 57, 2657–2661. [Google Scholar] [CrossRef]
- Altay, C.; Senay, R.H.; Eksina, E.; Congur, G.; Erdem, A.; Akgol, S. Development of amino functionalized carbon coated magnetic nanoparticles and their application to electrochemical detection of hybridization of nucleic acids. Talanta 2017, 164, 175–182. [Google Scholar] [CrossRef]
- Sousa, J.B.; Ramos-Jesus, J.; Silva, L.C.; Pereira, C.; de-los-Santos-Alvarez, N.; Fonseca, R.A.S.; Miranda-Castro, R.; Delerue-Matos, C.; Ribeiro Santos Junior, J.; Barroso, M.F. Fe3O4@Au nanoparticles-based magnetoplatform for the HMGA maize endogenous gene electrochemical genosensing. Talanta 2020, 206, 120220. [Google Scholar] [CrossRef] [PubMed]





| Objective | Functional Nanomaterial | Rationale Behind the Strategy | Detection Technique | Analyte Detected/Delivered | LOD | Sample | Assay Time, min | Ref. |
|---|---|---|---|---|---|---|---|---|
| Biosensing | Magnetic Janus Au–Ni–PtNWs | Increased speed of Au–Ni–PtNWs in the presence of Ag+ enriched H2O2 solution generated by performing sandwich hybridization assays at photolithography-prepared 16×AuEs array involving an AgNPs-labeled detector probe (AgNPs-Dp) | Optical | DNA and E. coli 16S mRNA | 40 amol synthetic DNA and 2000 cfu·mL−1 of E. coli | Raw bacterial lysate | ~60 min starting from Cp-16×AuEs array (preparation of Au–Ni–Pt NWs: ~2 h; AgNPs-Dp: ~62 h; Cp-16×AuEs array: ~13 h) | [56] |
| Biosensing (in vitro and intracellular) | GO-AuNWs | “Off–On” fluorescence switching due to the displacement of the dye-ssDNA probe from the ssDNA@GO-AuNWs surface after cell internalization in the presence of the target miRNA | Fluorescent | miRNA-21 | Single cell | Intact cancer cells (MCF-7 and HeLa) | 5–10 min starting from ssDNA@GO-AuNWs (AuNWs: ~2 h; ssDNA@GO-AuNWs ~14 h 15 min) | [3] |
| Biosensing (in vitro and intracellular) | GO-AuNWs | “OFF–ON” fluorescence switching due to the displacement of the dye-ssDNA probe from the ssDNA@GO-AuNWs surface after cell internalization in the presence of the target mRNA | Fluorescent | HPV16 E6 mRNA transcripts | Single cell | Total RNA extracted from HPV-positive OPC cells and intact cells (HPV-positive or HPV-negative human OPC cells) | 15 min starting from ssDNA@GO-AuNWs (AuNWs: ~2 h; ssDNA@GO-AuNWs ~14 h 15 min) | [58] |
| Delivery | siRNA/RCA- AuNWs | Intracellular delivery of GFP/RCA to knockout GFP gene | Fluorescent | siRNA/RCA | Single cell | Intact HEK-293 and MCF-7 cells | ~5 min starting from siRNA/RCA- AuNWs (AuNWs: ~2 h; RCA: ~19 h 45 min; siRNA/RCA- AuNWs: ~13 h) | [4] |
| Delivery | Cas9–sgRNA–AuNWs | Intracellular delivery of Cas9–sgRNA complex to silence the GFP response | Fluorescent | Cas9–sgRNA complex | Single cell | Intact B16F10 cells | ~5 min starting from Cas9–sgRNA–AuNWs (AuNWs: ~2 h; Cas9–sgRNA complex: ~10 min; Cas9–sgRNA–AuNWs: ~16 h) | [59] |
| Electrode | Functional Nanomaterial (Role) | Method | Detection Technique | Target Analyte | Linear Range | LOD | Sample | Assay Time, Min | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| AuNPs–SPCE | AuNPs (electrode modifier) | Direct hybridization approach at SH-RNA-Cp/MCH-AuNPs–SPCE and selective recognition of the RNA/miRNA hybrid with the p19-MBP fusion protein further conjugated with and HRP anti-MBP antibody | Amperometry (H2O2/HQ) | miRNA-21 | 0.5–50 pmol·L−1 | 142 fmol·L−1 | RNAt extracted from healthy and cancerous breast cells | ~60 min starting from SH-RNA-Cp/MCH-AuNPs–SPCE (SPCE modification: ~9 h 5 min) | [10] |
| SPCE | AuNPs (catalytic label) | Isothermal amplification of Leishmania DNA using primers labeled with MBs and AuNPs and magnetic capture of the MB–amplified DNA–AuNP complexes on SPCEs | Chronoamperometry | Leishmania DNA | 500–0.5 parasite mL−1 blood | 0.8 parasite mL−1 blood | Dog’s blood | ~10 min starting from primers conjugated with MBs and AuNPs (primers conjugation: ~65 h 55 min) | [12] |
| AuE | AuNPs (nanocarriers of redox-labeled DNA probes) | Sandwich hybridization assay developed at an Au electrode modified with thiolated Cps; use of AuNPs modified with two different probes labeled with methylene blue (just one complementary to the target DNA) | DPV (methylene blue) | Target DNA | 10−13–10−8 M | 50 fM | — | ~2 h starting from SH-Cp/MCH-AuE (modified AuE: ~1 h and DNA-AuNPs conjugates: ~5 h 30 min) | [14] |
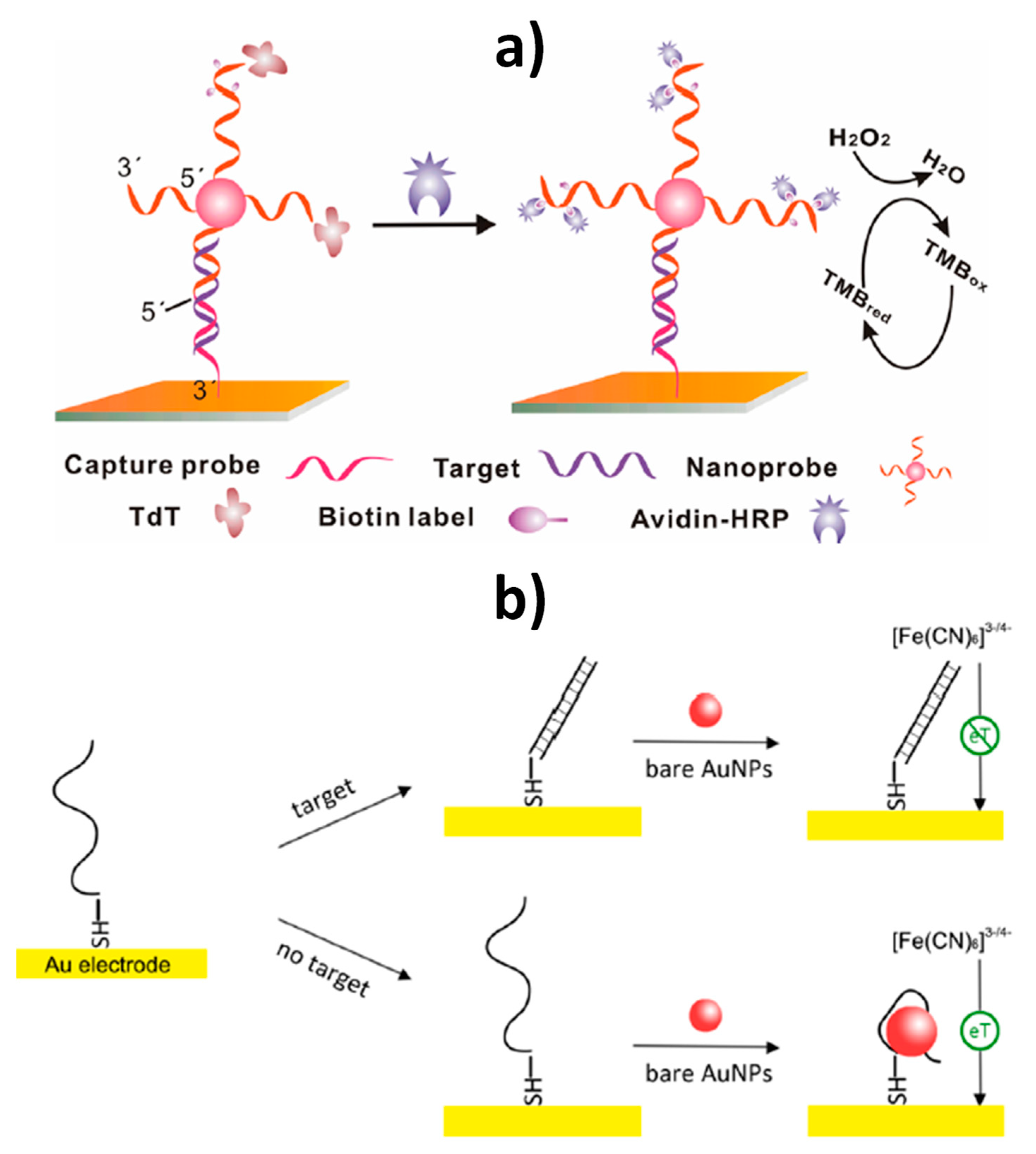
| AuE | AuNPs (nanocarriers of reporter probes and enzymes) | Sandwich hybridization between SH-Cp/SH-OEG-AuE and reporter probe-linked AuNPs, and terminal deoxynucleotidyl transferase (TdT)-catalyzed elongation of the free 3´-terminal of DNA on the nanoprobe to incorporate multiple biotin moieties further conjugated with avidin-modified HRP molecules | Amperometry (TMB/H2O2) | Target DNA | 10 fM–10 pM | 10 fM | — | ~2 h 45 min starting from SH-Cp/SH-OEG-AuE (modified AuE: ~16 h and DNA-AuNPs conjugates: ~56 h 15 min) | [13] |
| AuE | AuNPs (nanocarriers of melamine–Cu2+ complexes) | Hybridization-induced structural variation of electrode-immobilized SH-hCp with attached Cu2+-Mel-AuNPs | DPV (Cu2+/Cu+) | Target DNA | 1.0 × 10−18 M–1.0 × 10−12 M | 1.2 × 10−19 M | 10% spiked human serum | ~40 min starting from Cu2+-Mel-AuNPs/SH-hCp/MCH/AuE (AuE modification: ~77 h 20 min) | [15] |
| AuNPs/rGO/SPCEs | AuNPs (nanocarriers of Strep and Fc) | Sandwich hybridization approach at a MCH/HS-DNACp-AuNPs/rGO/SPCEs using a biotinylated Dp conjugated with Fc-AuNPs-Strep conjugates | DPV (Fc) | miRNA-21 | 10 fM–2 pM | 5 fM | RNAt, extracted from breast adenocarcinoma cells and serum from cancer patients | ~1 h 45 min starting from Fc-AuNPs-Strep (AuNPs modification: ~24 h and HS-DNACp-AuNPs/rGO/SPCE: ~9 h 30 min) | [16] |
| AuE | AuNPs (electron transfer regulator) | Enhancement of the interfacial electron transfer process between the electrode and the redox couple ([Fe(CN)6]3−/4−) in the absence of target DNA due to AuNPs–DNA binding | EIS ([Fe(CN)6]3−/4−) | Target DNA (BRCA1 gene) | 1 pM–500 nM | 1 pM | — | ~2 h starting from AuNPs (AuNPs preparation: ~30 min and HS-DNACp-AuE: ~3 h) | [17] |
| PGE | NH2-CC-MNPs | Direct DNA hybridization at DNA Cp immobilized onto NH2-CC-MNPs | DPV (guanine oxidation) | HBV target DNA | 5–25 μg mL−1 | 1.15 μg mL−1 | — | ~35 min starting from Cp-NH2-CC-MNPs (synthesis: ~23 h 30 min + Cp immobilization: ~1 h 20 min) | [60] |
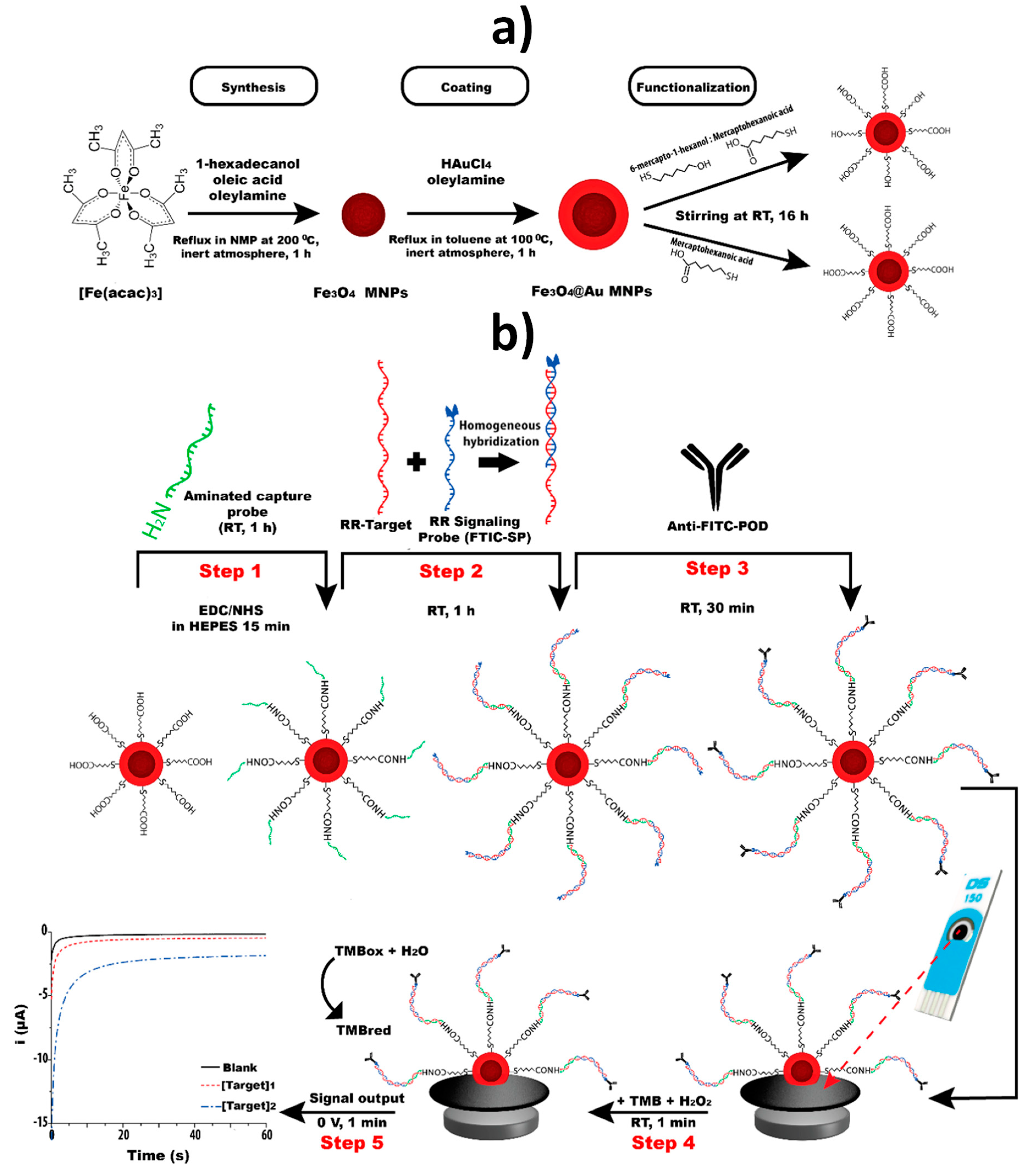
| SPCE | Fe3O4@Au MNPs | Sandwich hybridization approach involving covalent immobilization of an NH2-DNA Cp onto Fe3O4@Au MNPs modified with a TOA/MCH SAM and a FITC signaling probe further conjugated with anti-FITC-HRP Fab fragment | Chronoamperometry (TMB/H2O2) | GMO (a specific fragment of the transgenic construct from maize MON810 maize) | 0.25–2.5 nM | 0.15 nM | PCR amplicons obtained from CRMs of maize MON810 | ~2 h starting from Cp-Fe3O4@Au MNPs (MNPs synthesis: ~20 h;TOA/MCH SAM: ~24 h; Cp immobilization: ~2 h) | [18] |
| SPCE | Fe3O4@Au MNPs | Sandwich hybridization approach involving covalent immobilization of an NH2-DNA Cp onto Fe3O4@Au MNPs modified with a MHA/MCH SAM and a FITC signaling probe further conjugated with anti-FITC-HRP Fab fragment | Chronoamperometry (TMB/H2O2) | DNA fragments from the insertion point of the transgenic construct of RR GTS 40-3-2 soybean, an event-specific sequence, and of the taxon-specific soybean gene, lectin | 0.1–10.0 nM (event specific) 0.1–5.0 nM (taxon-specific) | 0.02 nM (event specific) 0.05 nM (taxon-specific) | PCR amplicons obtained from soybean seeds and cat feed | ~1 h 40 min starting from Cp- Fe3O4@Au MNPs (synthesis: ~21 h; MHA/MCH SAM: ~16 h; Cp immobilization: ~1 h 40 min) | [29] |
| SPdCE | Fe3O4@Au MNPs | Sandwich hybridization approaches involving covalent immobilization of NH2-DNA capture probes onto Fe3O4@Au MNPs modified with a MHA SAM and FITC or DIG signaling probes further conjugated with anti-FITC-HRP or anti-DIG-HRP Fab fragments | Chronoamperometry (TMB/H2O2) | GMO (fragments of the transgenic construct from GTS 40-3-2 and MON89788 soybean lines) | 0.1–2.5 nM (GTS 40-3-2) 0.1–1.0 nM (MON89788) | 0.1 nM (both events) | — | ~2 h 5 min starting from Cp- Fe3O4@Au MNPs (synthesis: ~21 h; MHA SAM: ~16 h; Cp immobilization: ~1 h 35 min) | [30] |
| Homemade AuE | Fe3O4@Au MNPs | Sandwich hybridization approach involving covalent immobilization of an NH2-DNA Cp onto Fe3O4@Au MNPs modified with a MHA/MCH SAM and a FITC signaling probe further conjugated with anti-FITC-HRP Fab fragment | Chronoamperometry (TMB–H2O2) | Maize taxon-specific (HMGA gene) | 0.5–5 nM | 90 pM | PCR amplicons obtained from maize flour | ~2 h starting from Cp-Fe3O4@Au MNPs (synthesis: ~20 h; MHA/MCH SAM: ~24 h; Cp immobilization: ~2 h) | [61] |
| SPCE | Au-MSN JNPs | Au-MS JNPs functionalized with HRP and a dual biotin thiol-functionalized anti-CEA DNA hairpin aptamer in connection with avidin-modified Fe3O4@SiO2 NanoCaptors | Amperometry (H2O2/HQ) | CEA | 5.5 pM–28 nM | 1.2 pM | Spiked lyophilized human serum samples | ~1 h starting from bifunctionalized Au-MSN JNPs (synthesis of Au-MSN JNPs: ~38 h 30 min; bifunctionalization: ~4 h 5 min) | [49] |
| AuNPs–SPCE | MSNs | MSNs loaded with methylene blue molecules and capped with an avidin/imminobiotin stimulus-responsive gate-like ensemble in connection with an AuNPs–SPCE modified with a biotin and thiol-functionalized anti-CEA DNA hairpin (Apt-AuNPs–SPCE) | DPV (methylene blue) | CEA | 1.0 pg mL−1–160 ng mL−1 | 280 fg mL−1 | 5-fold diluted human serum samples | ~45 min starting from bifunctionalized MSNs and Apt-AuNPs–SPCE (MSNs: ~2 h 5 min; bifunctionalization: ~72 h; Apt-AuNPs–SPCE: ~1 h 5 min) | [50] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campuzano, S.; Gamella, M.; Serafín, V.; Pedrero, M.; Yáñez-Sedeño, P.; Pingarrón, J.M. Biosensing and Delivery of Nucleic Acids Involving Selected Well-Known and Rising Star Functional Nanomaterials. Nanomaterials 2019, 9, 1614. https://doi.org/10.3390/nano9111614
Campuzano S, Gamella M, Serafín V, Pedrero M, Yáñez-Sedeño P, Pingarrón JM. Biosensing and Delivery of Nucleic Acids Involving Selected Well-Known and Rising Star Functional Nanomaterials. Nanomaterials. 2019; 9(11):1614. https://doi.org/10.3390/nano9111614
Chicago/Turabian StyleCampuzano, Susana, Maria Gamella, Verónica Serafín, María Pedrero, Paloma Yáñez-Sedeño, and José Manuel Pingarrón. 2019. "Biosensing and Delivery of Nucleic Acids Involving Selected Well-Known and Rising Star Functional Nanomaterials" Nanomaterials 9, no. 11: 1614. https://doi.org/10.3390/nano9111614
APA StyleCampuzano, S., Gamella, M., Serafín, V., Pedrero, M., Yáñez-Sedeño, P., & Pingarrón, J. M. (2019). Biosensing and Delivery of Nucleic Acids Involving Selected Well-Known and Rising Star Functional Nanomaterials. Nanomaterials, 9(11), 1614. https://doi.org/10.3390/nano9111614







