Complementarity of Matrix- and Nanostructure-Assisted Laser Desorption/Ionization Approaches
Abstract
:1. Introduction
2. Matrix-Assisted Laser Desorption/Ionization
3. Protein Identification and Analysis by MALDI-TOF Mass Spectrometry
Problems with Protein Identification and Analysis by MALDI-TOF Mass Spectrometry
4. Non-Protein Analysis by MALDI-TOF Mass Spectrometry
5. Nanostructure-Assisted Laser Desorption/Ionization
Author Contributions
Funding
Conflicts of Interest
References
- Pomastowski, P.; Walczak, J.; Gawin, M.; Bocian, S.; Piekoszewski, W.; Buszewski, B. HPLC separation of casein components on a diol-bonded silica column with MALDI TOF/TOF MS identification. Anal. Methods 2014, 6, 5236–5244. [Google Scholar] [CrossRef]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of microorganisms by modern analytical techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Glish, G.L.; Vachet, R.W. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2003, 2, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Pomastowski, P.; Buszewski, B. Two-dimensional gel electrophoresis in the light of new developments. TrAC Trends Anal. Chem. 2014, 53, 167–177. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Biopolymers. Anal. Chem. 1991, 63, 1193A–1203A. [Google Scholar] [CrossRef]
- Karas, M.; Bachmann, D.; Hillenkamp, F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem. 1985, 57, 2935–2939. [Google Scholar] [CrossRef]
- Meller, K.; Pomastowski, P.; Szumski, M.; Buszewski, B. Preparation of an improved hydrophilic monolith to make trypsin-immobilized microreactors. J. Chromatogr. B 2017, 1043, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Meller, K.; Pomastowski, P.; Grzywiński, D.; Szumski, M.; Buszewski, B. Preparation and evaluation of dual-enzyme microreactor with co-immobilized trypsin and chymotrypsin. J. Chromatogr. A 2016, 1440, 45–54. [Google Scholar] [CrossRef]
- Walczak, J.; Pomastowski, P.; Bocian, S.; Buszewski, B. Determination of phospholipids in milk using a new phosphodiester stationary phase by liquid chromatography-matrix assisted desorption ionization mass spectrometry. J. Chromatogr. A 2016, 1432, 39–48. [Google Scholar] [CrossRef]
- Fukuyama, Y. MALDI Matrix Research for Biopolymers. MALDI MatrIx Res. Biopolym. 2015, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Huwiler, K.G.; Mosher, D.F.; Vestling, M.M. Optimizing the MALDI-TOF-MS observation of peptides containing disulfide bonds. J. Biomol. Tech. 2003, 14, 289–297. [Google Scholar] [PubMed]
- Jackson, S.N.; Barbacci, D.; Egan, T.; Lewis, E.K.; Schultz, J.A.; Woods, A.S. MALDI-Ion Mobility Mass Spectrometry of Lipids in Negative Ion Mode. Anal. Methods 2014, 6, 5001–5007. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.O.; Li, L. Three-Layer Matrix/Sample Preparation Method for MALDI MS Analysis of Low Nanomolar Protein Samples. J. Am. Soc. Mass Spectrom. 2006, 17, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Golding, R.E.; Whittal, R.M. Analysis of Single Mammalian Cell Lysates by Mass Spectrometry. J. Am. Chem. Soc. 1996, 118, 11662–11663. [Google Scholar] [CrossRef]
- Gies, A.P.; Vergne, M.J.; Orndorff, R.L.; Hercules, D.M. MALDI-TOF/TOF CID Study of Polystyrene Fragmentation Reactions. Macromolecules 2007, 40, 7493–7504. [Google Scholar] [CrossRef]
- Trimpin, S.; Keune, S.; Räder, H.J.; Müllen, K. Solvent-Free MALDI-MS: Developmental Improvements in the Reliability and the Potential of MALDI in the Analysis of Synthetic Polymers and Giant Organic Molecules. J. Am. Soc. Mass Spectrom. 2006, 17, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Berry, K.A.Z.; Hankin, J.A.; Barkley, R.M.; Spraggins, J.M.; Caprioli, R.M.; Murphy, R.C. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 2011, 111, 6491–6512. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kim, M.S. A Thermal Mechanism of Ion Formation in MALDI. Annu. Rev. Anal. Chem. 2015, 8, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Molin, L.; Seraglia, R.; Czarnocki, Z.; Maurin, J.K.; Pluciński, F.A.; Traldi, P. On the Primary Ionization Mechanism(s) in Matrix-Assisted Laser Desorption Ionization. J. Anal. Methods Chem. 2012, 2012, 161865. [Google Scholar] [CrossRef]
- Neubert, H.; Halket, J.M.; Fernandez Ocaña, M.; Patel, R.K.P. MALDI post-source decay and LIFT-TOF/TOF investigation of α-cyano-4-hydroxycinnamic acid cluster interferences. J. Am. Soc. Mass Spectrom. 2004, 15, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.O.; Li, L. Discerning matrix-cluster peaks in matrix-assisted laser desorption/ionization time-of-flight mass spectra of dilute peptide mixtures. J. Am. Soc. Mass Spectrom. 2000, 11, 88–93. [Google Scholar] [CrossRef]
- Ryan, D.J.; Spraggins, J.M.; Caprioli, R.M. Protein identification strategies in MALDI imaging mass spectrometry: A brief review. Curr. Opin. Chem. Biol. 2019, 48, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Yang, P.-C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [PubMed]
- Edman, P.; Begg, G. A Protein Sequenator. Eur. J. Biochem. 1967, 1, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R. A mass spectrometric journey into protein and proteome research. J. Am. Soc. Mass Spectrom. 2003, 14, 685–695. [Google Scholar] [CrossRef]
- Delom, F.; Chevet, E. Phosphoprotein analysis: From proteins to proteomes. Proteome Sci. 2006, 4, 15. [Google Scholar] [CrossRef]
- Locke, D.; Bian, S.; Li, H.; Harris, A.L. Post-translational modifications of connexin26 revealed by mass spectrometry. Biochem. J. 2009, 424, 385–398. [Google Scholar] [CrossRef]
- Takimori, S.; Shimaoka, H.; Furukawa, J.-I.; Yamashita, T.; Amano, M.; Fujitani, N.; Takegawa, Y.; Hammarström, L.; Kacskovics, I.; Shinohara, Y.; et al. Alteration of the N-glycome of bovine milk glycoproteins during early lactation. FEBS J. 2011, 278, 3769–3781. [Google Scholar] [CrossRef]
- Chen, W.; Lee, P.J.; Stapels, M.; Gebler, J.C. The use of mass spectrometry to determine location and extent of N-glycosylation on folate binding protein from bovine milk. Rapid Commun. Mass Spectrom. 2006, 20, 313–316. [Google Scholar] [CrossRef]
- Slangen, C.J.; Visser, S. Use of mass spectrometry to rapidly characterize the heterogeneity of bovine alpha-lactalbumin. J. Agric. Food Chem. 1999, 47, 4549–4556. [Google Scholar] [CrossRef] [PubMed]
- Ham, J.-S.; Han, G.-S.; Jeong, S.-G.; Seol, K.-H.; Jang, A.-R.; Oh, M.-H.; Kim, D.-H.; Park, Y.W. Determination of molecular weights of caprine milk proteins by matrix-assisted laser desorption/ionization mass spectrometry. J. Dairy Sci. 2012, 95, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zheng, N.; Yang, J.; Bu, D.; Wang, J.; Ma, L.; Sun, P. Animal species milk identification by comparison of two-dimensional gel map profile and mass spectrometry approach. Int. Dairy J. 2014, 35, 15–20. [Google Scholar] [CrossRef]
- Pomastowski, P.; Sprynskyy, M.; Žuvela, P.; Rafińska, K.; Milanowski, M.; Liu, J.J.J.; Yi, M.; Buszewski, B. Silver-Lactoferrin Nanocomplexes as a Potent Antimicrobial Agent. J. Am. Chem. Soc. 2016, 138, 7899–7909. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sun, C.; Wang, D.; Yuan, F.; Gao, Y. Glycosylation improves the functional characteristics of chlorogenic acid–lactoferrin conjugate. RSC Adv. 2015, 5, 78215–78228. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Schoemaker, R.J.W.; Timmer, C.J.A.M.; Kamerling, J.P.; Dijkhuizen, L. N - and O -Glycosylation of a Commercial Bovine Whey Protein Product. J. Agric. Food Chem. 2012, 60, 12553–12564. [Google Scholar] [CrossRef] [PubMed]
- Nwosu, C.C.; Strum, J.S.; An, H.J.; Lebrilla, C.B. Enhanced Detection and Identification of Glycopeptides in Negative Ion Mode Mass Spectrometry. Anal. Chem. 2010, 82, 9654–9662. [Google Scholar] [CrossRef]
- Novotny, M.V.; Alley, W.R. Recent trends in analytical and structural glycobiology. Curr. Opin. Chem. Biol. 2013, 17, 832–840. [Google Scholar] [CrossRef]
- Quaranta, A.; Sroka-Bartnicka, A.; Tengstrand, E.; Thorsén, G. N-Glycan profile analysis of transferrin using a microfluidic compact disc and MALDI-MS. Anal. Bioanal. Chem. 2016, 408, 4765–4776. [Google Scholar] [CrossRef]
- Žuvela, P.; Liu, J.J.; Yi, M.; Pomastowski, P.P.; Sagandykova, G.; Belka, M.; David, J.; Bączek, T.; Szafrański, K.; Żołnowska, B.; et al. Target-based drug discovery through inversion of quantitative structure-drug-property relationships and molecular simulation: CA IX-sulphonamide complexes. J. Enzyme Inhib. Med. Chem. 2018, 33, 1430–1443. [Google Scholar] [CrossRef]
- Papac, D.I.; Shahrokh, Z. Mass Spectrometry Innovations in Drug Discovery and Development. Pharm. Res. 2001, 18, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, H.; Sekine, K.; Fujita, K.; Iigo, M. Cancer prevention by bovine lactoferrin and underlying mechanisms—A review of experimental and clinical studies. Biochem. Cell Biol. 2002, 80, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.P.; Uribe-Luna, S.; Conneely, O.M. Lactoferrin and host defense. Biochem. Cell Biol. 2002, 80, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Orsi, N. The antimicrobial activity of lactoferrin: Current status and perspectives. Biometals 2004, 17, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Poth, A.G.; Deeth, H.C.; Alewood, P.F.; Holland, J.W. Analysis of the Human Casein Phosphoproteome by 2-D Electrophoresis and MALDI-TOF/TOF MS Reveals New Phosphoforms. J. Proteome Res. 2008, 7, 5017–5027. [Google Scholar] [CrossRef] [PubMed]
- Thevis, M.; Loo, R.R.O.; Loo, J.A. Mass spectrometric characterization of transferrins and their fragments derived by reduction of disulfide bonds. J. Am. Soc. Mass Spectrom. 2003, 14, 635–647. [Google Scholar] [CrossRef]
- Gobom, J.; Schuerenberg, M.; Mueller, M.; Theiss, T.; Lehrach, H.; Nordhoff, E. α-Cyano-4-hydroxycinnamic Acid Affinity Sample Preparation. A Protocol for MALDI-MS Peptide Analysis in Proteomics. Anal Chem. 2001, 73, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.K.; Park, Z.-Y.; Russell, D.H. Proteolysis in Mixed Organic−Aqueous Solvent Systems: Applications for Peptide Mass Mapping Using Mass Spectrometry. Anal. Chem. 2001, 73, 2682–2685. [Google Scholar]
- Han, X.; Aslanian, A.; Yates, J.R. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef]
- Zaluzec, E.J.; Gage, D.A.; Watson, J.T. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry: Applications in Peptide and Protein Characterization. Protein Expr. Purif. 1995, 6, 109–123. [Google Scholar] [CrossRef]
- Cohen, S.L.; Chait, B.T. Influence of Matrix Solution Conditions on the MALDI-MS Analysis of Peptides and Proteins. Anal. Chem. 1996, 68, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Bucknall, M.; Fung, K.Y.C.; Duncan, M.W. Practical quantitative biomedical applications of MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2002, 13, 1015–1027. [Google Scholar] [CrossRef]
- Albrethsen, J. Reproducibility in Protein Profiling by MALDI-TOF Mass Spectrometry. Clin. Chem. 2007, 53, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Amado, F.M.L.; Domingues, P.; Graça Santana-Marques, M.; Ferrer-Correia, A.J.; Tomer, K.B. Discrimination effects and sensitivity variations in matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 1997, 11, 1347–1352. [Google Scholar] [CrossRef]
- Kratzer, R.; Eckerskorn, C.; Karas, M.; Lottspeich, F. Suppression effects in enzymatic peptide ladder sequencing using ultraviolet—Matrix assisted laser desorption/ionization—Mass spectrometry. Electrophoresis 1998, 19, 1910–1919. [Google Scholar] [CrossRef]
- Juhasz, P.; Biemann, K. Mass spectrometric molecular-weight determination of highly acidic compounds of biological significance via their complexes with basic polypeptides. Proc. Natl. Acad. Sci. USA 1994, 91, 4333–4337. [Google Scholar] [CrossRef] [PubMed]
- Al-Suod, H.; Pomastowski, P.; Ligor, M.; Railean-Plugaru, V.; Buszewski, B. New approach for fast identification of cyclitols by MALDI-TOF mass spectrometry. Phytochem. Anal. 2018, 29, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Parpinello, G.P.; Palma, A.S.; Teslić, N.; Brilli, C.; Pizzi, A.; Versari, A. Analytical profiling of food-grade extracts from grape (Vitis vinifera sp.) seeds and skins, green tea (Camellia sinensis) leaves and Limousin oak (Quercus robur) heartwood using MALDI-TOF-MS, ICP-MS and spectrophotometric methods. J. Food Compos. Anal. 2017, 59, 95–104. [Google Scholar] [CrossRef]
- Picariello, G.; Sciammaro, L.; Siano, F.; Volpe, M.G.; Puppo, M.C.; Mamone, G. Comparative analysis of C -glycosidic flavonoids from Prosopis spp. and Ceratonia siliqua seed germ flour. Food Res. Int. 2017, 99, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Hoong, Y.B.; Pizzi, A.; Tahir, P.M.; Pasch, H. Characterization of Acacia mangium polyflavonoid tannins by MALDI-TOF mass spectrometry and CP-MAS 13C NMR. Eur. Polym. J. 2010, 46, 1268–1277. [Google Scholar] [CrossRef]
- Sastre, F.; Ferreira, F.; Pedreschi, F. MALDI-TOF mass spectrometry and reversed-phase HPLC-ELSD chromatography for structural and quantitative studies of major steroid saponins in commercial extracts of Yucca schidigera Roezl. J. Pharm. Biomed. Anal. 2016, 120, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Szultka-Mlynska, M.; Pomastowski, P.; Buszewski, B. Application of solid phase microextraction followed by liquid chromatography-mass spectrometry in the determination of antibiotic drugs and their metabolites in human whole blood and tissue samples. J. Chromatogr. B 2018, 1086, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Amelin, V.G.; Krasnova, T.A. Identification and determination of antibiotics from various classes in food- and feedstuffs by matrix-or surface-assisted laser desorption/ionization mass spectrometry. J. Anal. Chem. 2015, 70, 850–859. [Google Scholar] [CrossRef]
- Grant, G.A.; Frison, S.L.; Sporns, P. A Sensitive Method for Detection of Sulfamethazine and N4-Acetylsulfamethazine Residues in Environmental Samples Using Solid Phase Immunoextraction Coupled with MALDI-TOF MS. J. Agric. Food Chem. 2003, 51, 5367–5375. [Google Scholar] [CrossRef] [PubMed]
- Jerome Jeyakumar, J.M.; Zhang, M.; Thiruvengadam, M. Determination of mycotoxins by HPLC, LC-ESI-MS/MS, and MALDI-TOF MS in Fusarium species-infected sugarcane. Microb. Pathog. 2018, 123, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.; Zschörnig, O.; Petković, M.; Müller, M.; Arnhold, J.; Arnold, K. Lipid analysis of human HDL and LDL by MALDI-TOF mass spectrometry and (31)P-NMR. J. Lipid Res. 2001, 42, 1501–1508. [Google Scholar] [PubMed]
- Serna, J.; García-Seisdedos, D.; Alcázar, A.; Lasunción, M.Á.; Busto, R.; Pastor, Ó. Quantitative lipidomic analysis of plasma and plasma lipoproteins using MALDI-TOF mass spectrometry. Chem. Phys. Lipids 2015, 189, 7–18. [Google Scholar] [CrossRef]
- Hirata, K.; Uchida, T.; Nakajima, Y.; Maekawa, T.; Mizuki, T. Chemical synthesis and cytotoxicity of neo-glycolipids; rare sugar-glycerol-lipid compounds. Heliyon 2018, 4, e00861. [Google Scholar] [CrossRef]
- Adeuya, A.; Price, N.P.J. Enumeration of hydroxyl groups of sugars and sugar alcohols by aqueous-based acetylation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2012, 26, 1372–1376. [Google Scholar] [CrossRef]
- Abe, T.; Kikuchi, H.; Aritsuka, T.; Takata, Y.; Fukushi, E.; Fukushi, Y.; Kawabata, J.; Ueno, K.; Onodera, S.; Shiomi, N. Structural confirmation of novel oligosaccharides isolated from sugar beet molasses. Food Chem. 2016, 202, 284–290. [Google Scholar] [CrossRef]
- Longuespée, R.; Wefers, A.K.; De Vita, E.; Miller, A.K.; Reuss, D.E.; Wick, W.; Herold-Mende, C.; Kriegsmann, M.; Schirmacher, P.; von Deimling, A.; et al. Rapid detection of 2-hydroxyglutarate in frozen sections of IDH mutant tumors by MALDI-TOF mass spectrometry. Acta Neuropathol. Commun. 2018, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, T.; Liu, B.; Du, Y.; Li, H.; Zhao, S.; Lu, Y. Matrix selection for polymer guanidine analysis by MALDI–TOF MS. Int. J. Mass Spectrom. 2013, 356, 1–6. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, C.; Ren, H.; Xi, Y.; Du, Y.; Peng, Y. Solvent effect in polymer analysis by MALDI-TOF mass spectrometry. Int. J. Polym. Anal. Charact. 2017, 22, 160–168. [Google Scholar] [CrossRef]
- Li, Y.; Hoskins, J.N.; Sreerama, S.G.; Grayson, S.M. MALDI−TOF Mass Spectral Characterization of Polymers Containing an Azide Group: Evidence of Metastable Ions. Macromolecules 2010, 43, 6225–6228. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.J.; Guttman, C.M.; Flynn, K.M.; Filliben, J.J. Significant parameters in the optimization of MALDI-TOF-MS for synthetic polymers. J. Am. Soc. Mass Spectrom. 2006, 17, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Railean-Plugaru, V.; Pomastowski, P.; Meller, K.; Złoch, M.; Rafinska, K.; Buszewski, B. Lactococcus lactis as a safe and inexpensive source of bioactive silver composites. Appl. Microbiol. Biotechnol. 2017, 101, 7141–7153. [Google Scholar]
- Railean-Plugaru, V.; Pomastowski, P.; Kowalkowski, T.; Sprynskyy, M.; Buszewski, B. Physicochemical study of natural fractionated biocolloid by asymmetric flow field-flow fractionation in tandem with various complementary techniques using biologically synthesized silver nanocomposites. Anal. Bioanal. Chem. 2018, 410. [Google Scholar] [CrossRef]
- Arendowski, A.; Nizioł, J.; Ossoliński, K.; Ossolińska, A.; Ossoliński, T.; Dobrowolski, Z.; Ruman, T. Laser desorption/ionization MS imaging of cancer kidney tissue on silver nanoparticle-enhanced target. Bioanalysis 2018, 10, 83–94. [Google Scholar] [CrossRef]
- Wei, J.; Buriak, J.M.; Siuzdak, G. Desorption–ionization mass spectrometry on porous silicon. Nature 1999, 399, 243–246. [Google Scholar] [CrossRef]
- Lowe, R.D.; Guild, G.E.; Harpas, P.; Kirkbride, P.; Hoffmann, P.; Voelcker, N.H.; Kobus, H. Rapid drug detection in oral samples by porous silicon assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2009, 23, 3543–3548. [Google Scholar] [CrossRef]
- Kraj, A.; Dylag, T.; Gorecka-Drzazga, A.; Bargiel, S.; Dziubanand, J.; Silberring, J. Desorption/ionization on silicon for small molecules: A promising alternative to MALDI TOF. Acta Biochim. Pol. 2003, 50, 783–787. [Google Scholar] [PubMed]
- Gulbakan, B.; Park, D.; Kang, M.; Kececi, K.; Martin, C.R.; Powell, D.H.; Tan, W. Laser desorption ionization mass spectrometry on silicon nanowell arrays. Anal. Chem. 2010, 82, 7566–7575. [Google Scholar] [CrossRef] [PubMed]
- Owega, S.; Lai, E.P.C.; Bawagan, A.D.O. Surface Plasmon Resonance-Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry. Anal. Chem. 1998, 70, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N. Nanoparticle assisted laser desorption/ionization mass spectrometry for small molecule analytes. Microchim. Acta 2018, 185, 200. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-W.; Unnikrishnan, B.; Anand, A.; Mao, J.-Y.; Huang, C.-C. Nanoparticle-based laser desorption/ionization mass spectrometric analysis of drugs and metabolites. J. Food Drug Anal. 2018, 26, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Tornatore, P.; Weinberger, S.R. Current developments in SELDI affinity technology. Mass Spectrom. Rev. 2004, 23, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Skytt, Å.; Thysell, E.; Stattin, P.; Stenman, U.-H.; Antti, H.; Wikström, P. SELDI-TOF MS versus prostate specific antigen analysis of prospective plasma samples in a nested case–control study of prostate cancer. Int. J. Cancer 2007, 121, 615–620. [Google Scholar] [CrossRef]
- Qiong-ying, H.; Wang, K.; Ding, Y.; Zheng, L.; Liang, S.; Lei, Z.; Fu, W.; Yan, L. Application of SELDI-TOF-MS Coupled with an Artificial Neural Network Model to the Diagnosis of Pancreatic Cancer. Lab. Med. 2010, 41, 676–681. [Google Scholar] [CrossRef]
- Wegdam, W.; Moerland, P.D.; Meijer, D.; de Jong, S.M.; Hoefsloot, H.C.J.; Kenter, G.G.; Buist, M.R.; Aerts, J.M.G. A critical assessment of SELDI-TOF-MS for biomarker discovery in serum and tissue of patients with an ovarian mass. Proteome Sci. 2012, 10, 45. [Google Scholar] [CrossRef]
- Sabel, M.S.; Liu, Y.; Lubman, D.M. Proteomics in melanoma biomarker discovery: Great potential, many obstacles. Int. J. Proteom. 2011, 2011, 181890. [Google Scholar] [CrossRef]
- Al-Tarawneh, S.K.; Bencharit, S. Applications of Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight (SELDI-TOF) Mass Spectrometry in Defining Salivary Proteomic Profiles. Open Dent. J. 2009, 3, 74–79. [Google Scholar] [CrossRef]
- Zhou, Z.-Y.; Ji, T.; Luo, H.-S. Surface-enhanced laser desorption ionization time-of-flight mass spectrometry used to screen serum diagnostic markers of colon cancer recurrence in situ following surgery. Oncol. Lett. 2015, 9, 2313–2316. [Google Scholar] [CrossRef] [PubMed]
- Angel, M.; Rodrigo, M.; Zitka, O.; Krizkova, S.; Moulick, A.; Adam, V.; Kizek, R. MALDI-TOF MS as evolving cancer diagnostic tool: A review. J. Pharm. Biomed. Anal. 2014, 95, 245–255. [Google Scholar]
- Guerreiro, N.; Gomez-Mancilla, B.; Charmont, S. Optimization and evaluation of surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry for protein profiling of cerebrospinal fluid. Proteome Sci. 2006, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cozzi, P.; Zhao, Z.; Giannakis, E.; Jingjing, Y.; Russell, P.; Walsh, B.; Willcox, M. SELDI-TOF-MS Analysis of Urine and Tear Samples to Discover Novel Biomarkers for Diagnosis and Prognosis of Prostate Cancer; Waverly Press: Baltimore, MD, USA, 2008; Volume 68. [Google Scholar]
- Frankfort, S.V.; van Campen, J.P.C.M.; Tulner, L.R.; Beijnen, J.H. Serum amyloid beta peptides in patients with dementia and age-matched non-demented controls as detected by surface-enhanced laser desorption ionisation-time of flight mass spectrometry (SELDI-TOF MS). Curr. Clin. Pharmacol. 2008, 3, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Singh, A.N. Exploring Biomarkers for Alzheimer’s Disease. J. Clin. Diagn. Res. 2016, 10, KE01-6. [Google Scholar] [CrossRef] [PubMed]
- Humpel, C. Identifying and validating biomarkers for Alzheimer’s disease. Trends Biotechnol. 2011, 29, 26–32. [Google Scholar] [CrossRef]
- Poon, T.C. Opportunities and limitations of SELDI-TOF-MS in biomedical research: Practical advices. Expert Rev. Proteom. 2007, 4, 51–65. [Google Scholar] [CrossRef]
- Lu, M.; Yang, X.; Yang, Y.; Qin, P.; Wu, X.; Cai, Z. Nanomaterials as Assisted Matrix of Laser Desorption/Ionization Time-of-Flight Mass Spectrometry for the Analysis of Small Molecules. Nanomaterials 2017, 7, 87. [Google Scholar] [CrossRef]
- Sekuła, J.; Nizioł, J.; Misiorek, M.; Dec, P.; Wrona, A.; Arendowski, A.; Ruman, T. Gold nanoparticle-enhanced target for MS analysis and imaging of harmful compounds in plant, animal tissue and on fingerprint. Anal. Chim. Acta 2015, 895, 45–53. [Google Scholar] [CrossRef]
- Tata, A.; Fernandes, A.M.A.P.; Santos, V.G.; Alberici, R.M.; Araldi, D.; Parada, C.A.; Braguini, W.; Veronez, L.; Silva Bisson, G.; Reis, F.H.Z.; et al. Nanoassisted laser desorption-ionization-MS imaging of tumors. Anal. Chem. 2012, 84, 6341–6345. [Google Scholar] [CrossRef] [PubMed]
- Nizioł, J.; Ossoliński, K.; Ossoliński, T.; Ossolińska, A.; Bonifay, V.; Sekuła, J.; Dobrowolski, Z.; Sunner, J.; Beech, I.; Ruman, T. Surface-Transfer Mass Spectrometry Imaging of Renal Tissue on Gold Nanoparticle Enhanced Target. Anal. Chem. 2016, 88, 7365–7371. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, Z.; Yan, X.; Cai, Z. Zeolitic imidazolate framework nanocrystals for enrichment and direct detection of environmental pollutants by negative ion surface-assisted laser desorption/ionization time-of-flight mass spectrometry. RSC Adv. 2016, 6, 23790–23793. [Google Scholar] [CrossRef]
- Ma, Y.R.; Zhang, X.L.; Zeng, T.; Cao, D.; Zhou, Z.; Li, W.H.; Niu, H.; Cai, Y.Q. Polydopamine-coated magnetic nanoparticles for enrichment and direct detection of small molecule pollutants coupled with MALDI-TOF-MS. ACS Appl. Mater. Interfaces 2013, 5, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.O.; Madkour, M.; Al-Hetlani, E. Metal oxide nanoparticles for latent fingerprint visualization and analysis of small drug molecules using surface-assisted laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 2018, 410, 4815–4827. [Google Scholar] [CrossRef] [PubMed]
- Sekuła, J.; Nizioł, J.; Rode, W.; Ruman, T. Gold nanoparticle-enhanced target (AuNPET) as universal solution for laser desorption/ionization mass spectrometry analysis and imaging of low molecular weight compounds. Anal. Chim. Acta 2015, 875, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Nizioł, J.; Rode, W.; Laskowska, B.; Ruman, T. Novel monoisotopic 109AgNPET for laser desorption/ionization mass spectrometry. Anal. Chem. 2013, 85, 1926–1931. [Google Scholar] [CrossRef]
- Arendowski, A.; Nizioł, J.; Ruman, T. Silver-109-based laser desorption/ionization mass spectrometry method for detection and quantification of amino acids. J. Mass Spectrom. 2018, 53, 369–378. [Google Scholar] [CrossRef]
- Kawasaki, H.; Ozawa, T.; Hisatomi, H.; Arakawa, R. Platinum vapor deposition surface-assisted laser desorption/ionization for imaging mass spectrometry of small molecules. Rapid Commun. Mass Spectrom. 2012, 26, 1849–1858. [Google Scholar] [CrossRef]
- Tang, H.H.H.; Lu, W.; Che, C.C.; Ng, K.K. Gold Nanoparticles and Imaging Mass Spectrometry: Double Imaging of Latent Fingerprints. Anal. Chem. 2010, 82, 1589–1593. [Google Scholar] [CrossRef]
- Nayak, R.; Knapp, D.R. Matrix-free LDI mass spectrometry platform using patterned nanostructured gold thin film. Anal. Chem. 2010, 82, 7772–7778. [Google Scholar] [CrossRef]
- Cha, S.; Song, Z.; Nikolau, B.J.; Yeung, E.S. Direct profiling and imaging of epicuticular waxes on Arabidopsis thaliana by LDI mass spectrometry using silver colloid as a matrix. Anal. Chem. 2009, 81, 2991–3000. [Google Scholar]
- Sacks, C.D.; Stumpo, K.A. Gold nanoparticles for enhanced ionization and fragmentation of biomolecules using LDI-MS. J. Mass Spectrom. 2018, 53, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.P.; Yu, C.J.; Lin, C.Y.; Lin, Y.H.; Tseng, W.L. Gold Nanoparticles as Assisted Matrices for the Detection of Biomolecules in a High-Salt Solution through Laser Desorption/Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009, 20, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kawasaki, H.; Yonezawa, T.; Arakawa, R. Surface-assisted laser desorption/ionization mass spectrometry (SALDI-MS) of low molecular weight organic compounds and synthetic polymers using zinc oxide (ZnO) nanoparticles. J. Mass Spectrom. 2008, 43, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xu, N.; Huang, W.Y.; Han, H.M.; Xiao, S.J. Electroless plating of silver nanoparticles on porous silicon for laser desorption/ionization mass spectrometry. Int. J. Mass Spectrom. 2009, 281, 1–7. [Google Scholar] [CrossRef]
- Dufresne, M.; Thomas, A.; Breault-Turcot, J.; Masson, J.F.; Chaurand, P. Silver-assisted laser desorption ionization for high spatial resolution imaging mass spectrometry of olefins from thin tissue sections. Anal. Chem. 2013, 85, 3318–3324. [Google Scholar] [CrossRef] [PubMed]
- Gamez, R.C.; Castellana, E.T.; Russell, D.H. Sol-Gel-derived silver-nanoparticle-embedded thin film for mass spectrometry-based biosensing. Langmuir 2013, 29, 6502–6507. [Google Scholar] [CrossRef]
- Du, J.; Zhu, Q.; Teng, F.; Wang, Y.; Lu, N. Ag nanoparticles/ZnO nanorods for highly sensitive detection of small molecules with laser desorption/ionization mass spectrometry. Talanta 2019, 192, 79–85. [Google Scholar] [CrossRef]
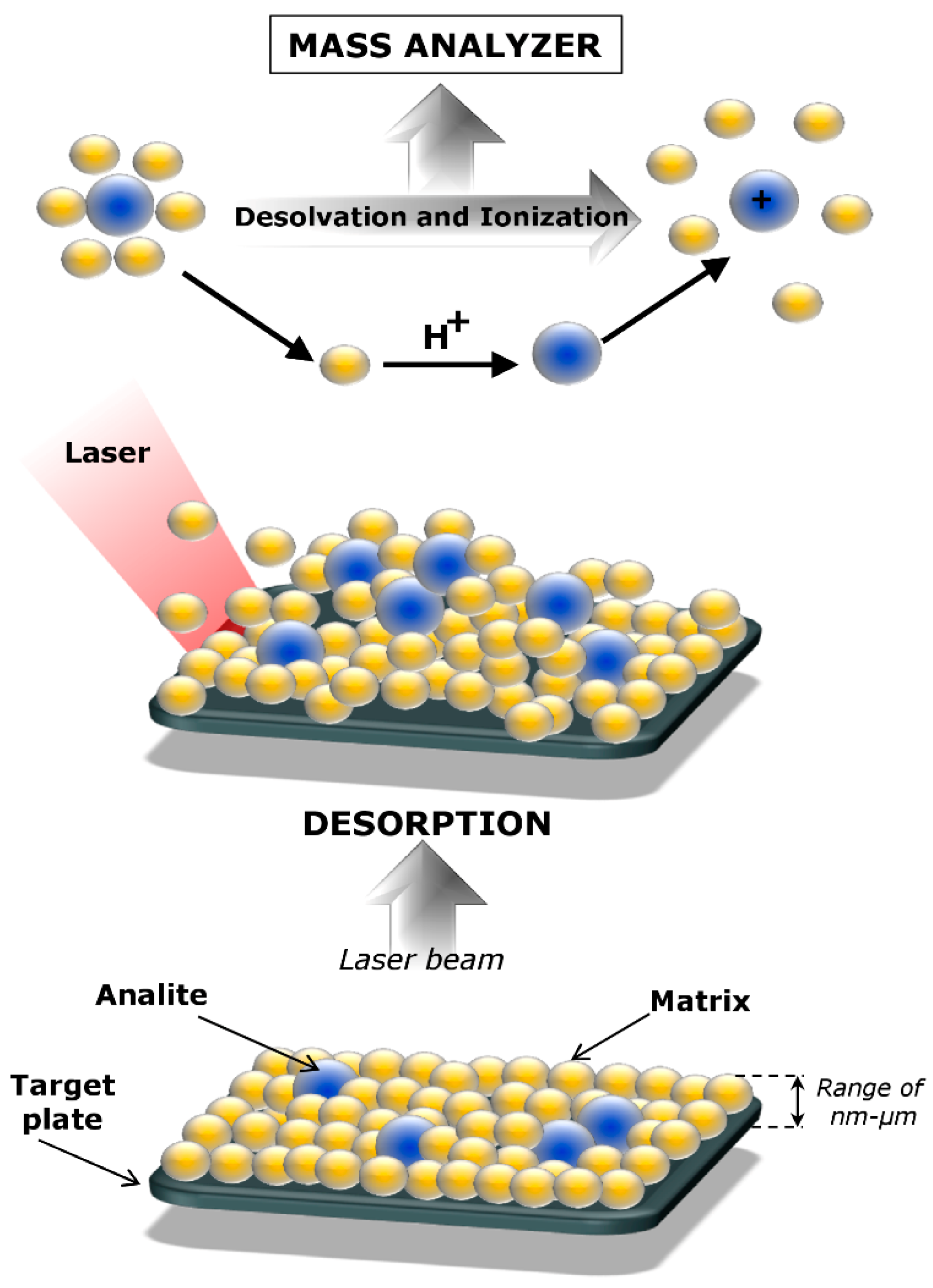

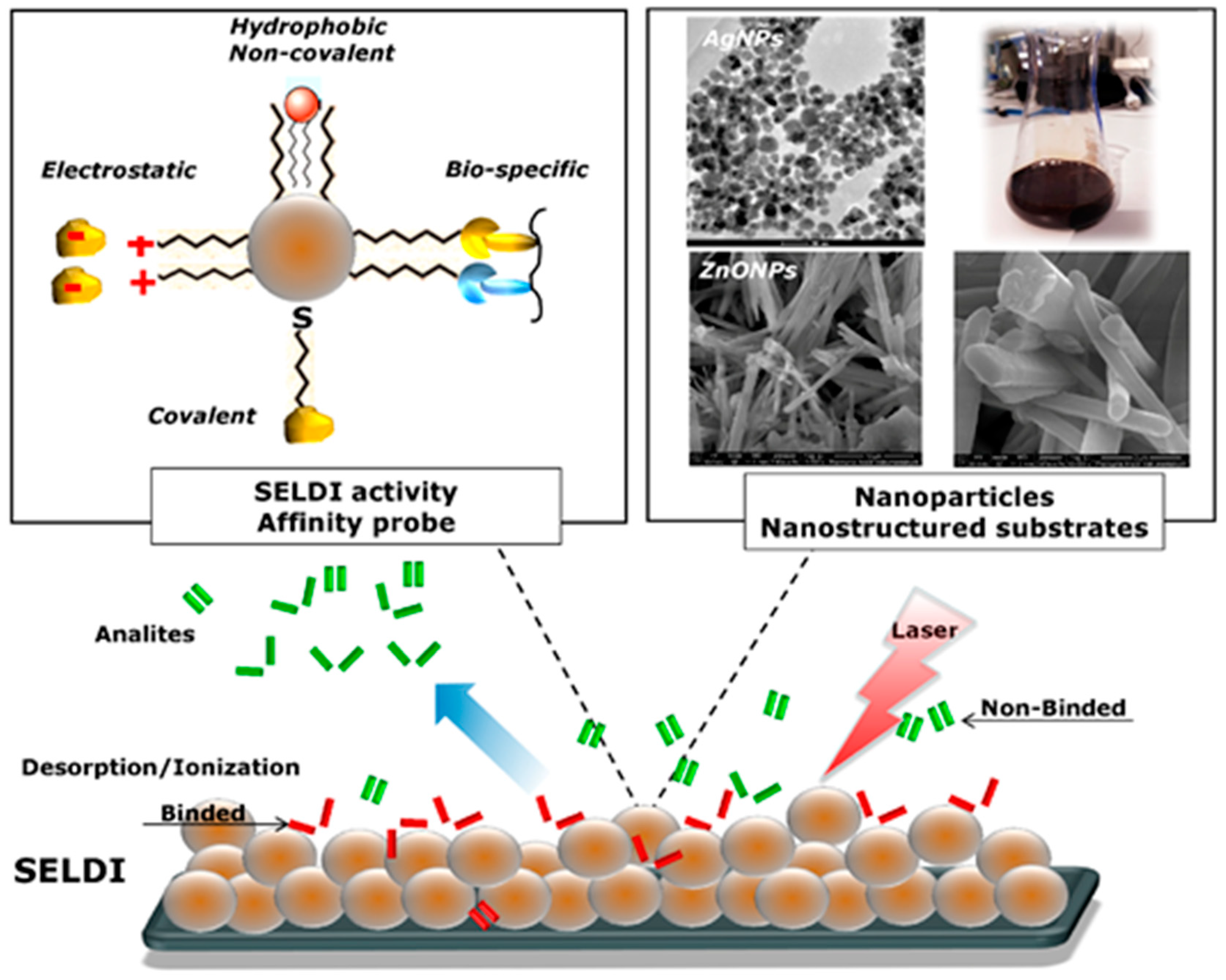
| Protein | Molecular Mass [kDa] | Sequence Coverage (SC) | Post-Translation Modifications | Reference |
|---|---|---|---|---|
| Connexin26 | - | 71.3% | acetylation, hydroxylation, γ-carboxyglutamation, methylation, phosphorylation | [28] |
| Carbonic anhydrase IX (CA IX) | 44.48 | - | - | [40] |
| αs1-CN | 23.61 | 75% | phosphorylation | [1] |
| 25.0 | 54% | [45] | ||
| β-CN | 23.99 | 37–49% | [1] | |
| 29.0 | 51% | [45] | ||
| κ-CN | 19.0 | 33–35% | phosphorylation | [1] |
| - | - | O-glycosylation | [37] | |
| α-lactalbumin | 14.199 | - | - | [31] |
| - | - | N-glycosylation | [29] | |
| β-lactoglobulin | 18.397 | - | - | [32] |
| - | - | N-glycosylation | [29] | |
| Lactoferrin | 77.167-81.189 | 19–25% | Glycosylation | [34] |
| 84.011 | - | [35] | ||
| Bovine serum albumin (BSA) | 62.0 | 35.7–43.9% | - | [8] |
| Transferrin | - | 41.8–56.4% | - | [9] |
| - | - | N-glycosylation | [39] | |
| 79.562 | - | O- and N-glycosylation Acetylation | [46] |
| Matrix | Analytes, Sample | Synthesis | Analytes Deposition | LOD (Limit of Detection) | Reference |
|---|---|---|---|---|---|
| Gold nanoparticles, target plate (AuNPET) | Pentedrone, diphenylamine, metronidazole and endogenous compound (saccharides, ionic and non-ionic glycerides, amino acids, fatty acids, sulfides, sulfoxides, phenols) in human FP, onion bulb, chicken liver | In situ on target plate (84 h reaction) | A volume of 0.5 µL of extract of liver, 1 µL of onion extract, was placed directly on AuNPET, air dried, and measured within 60–1000 m/z range | n/d (non-detected) | [101] |
| AuNPET | Nucleosides, saccharides, amino acids, glycosides, nucleic bases | In situ | Stock solution (1 mg/mL) of each analyte was prepared, diluted, and 0.3 µL of the final solution was applied to the AuNPET and air- dried | n/d | [107] |
| Gold nanoparticles (AuNPs) (2 and 5 nm) | Carbohydrates, steroids, bile acids | Ready to use from manufacturer | Analytes were dissolved into 3:1 acetonitrile: water with 0.1% formic acid (FA) at a 2.0 × 10−2 M and diluted to make matrix-to-analyte ratios of 1 × 104:1, 1 × 105:1, and 1 × 106, 3 μL of each solution was spotted and dried on separate wells of a stainless steel plate | n/d | [114] |
| AgNPET | Amino acids (AAs) from blood samples | In situ from silver-109 trifluoroacetate dissolved in tetrahydrofuran (THF) | Volumes of 0.5 μL of amino acid solutions diluted 10 times were placed directly on target plate and air dried, target was inserted into MS apparatus; 1 µL of plasma was dissolved in 999 μL of ultrapure water and used in this diluted form; plasma was spiked with 515 ng of Ser or 38 ng of Phe | LOD for AAs (pg/mm2): Arg—0.9; His—0.13; Ile—0.06; Met—0.13; Ser—0.16; Phe—0.1; Tyr—2.6; Ala—64 (fg/mm2); Asp—12; Cys—41; Lys—2.3 pg/spot | [109] |
| AgNPET (monoisotopic 109Ag) | AA: Trp, His, Ser, Met, Arg, Pro, Cys, Gln, Glu, Asp, Ala, Tyr, Leu, and Phe | Silver trifluoroacetate (200 mg) was dissolved in anhydrous, inhibitor-free THF (250 mL) and the solution poured into a large beaker containing a target plate. Solid 2,5-dihydroxy acid benzoic (DHB) (400 mg) was added and, following stirring, the solution was left for 24 h | Samples (0.5 μL) of the final solutions (dissolved in water) were applied to the sample plate and air-dried | Signal-to-noise (S/N) ratio: Gln, Ala, Phe, Leu, Glu, Tyr, Cys, Ser higher than 200; Arg—10. | [110] |
| Silver nanoparticles (AgNPs) | Different lipid classes from mouse and rat tissues, including brain, kidney, liver, and testis | Silver layers were deposited on top of the tissue sections using a sputter coater | All tissues were sectioned at 14 μm thickness using a cryostat and thaw-mounted on ITO coated slides; tissues were dried in desiccators prior to the silver deposition | n/d | [118] |
| AgNPs | triglycerides (TAG, C8−C16) and phosphatidylcholines (PC): 1,2-dimyristoyl-sn-glycero-3-PC (DMPC), 1,2-dipalmitoyl-sn-glycero-3-PC (DPPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-PC (POPC) | Porous AgNPs-impregnated thin films were prepared by the sol-gel method | A mixture of analytes was spotted directly to the surface and analyzed | n/d | [119] |
| AgNPs | Tetrapyridinporphyrin (TPyP), oligomers of polyethylene glycol, peptide of oxytocin | Electroless plating of nanoparticles on porous silicon for desorption–ionization on porous silicon (DIOS) | PEG was dissolved in acetonitrile (ACN), TPyP in dichloromethane (DCM), and oxytocin in MILLIQ water, diluted and 1 µL was dropped onto the silver film of the substrate, air-dried, and chip was introduced immediately into the mass spectrometer | LODs, fmol: TPyP—1.3 Oxytocin—1.5 | [117] |
| Silver colloids | Cuticular wax metabolites from Arabidopsis thaliana leaves and flowers | Commercial colloidal silver was sprayed to the target; the parameters for spraying were optimized; the sample was air-dried after each application | Collected flower and leaf samples from plants were stored in the ice before attaching them to sample plates; they were attached onto a stainless steel target plate of similar dimensions as a microscope glass slide using a conductive double-sided tape | n/d | [113] |
| AgNPs/zinc oxide nanorods (ZnO NRs) | Amino acids, polyethylene glycol (PEG) (MW 2000), Rhodamine 6G (R6G) | ZnO NRs were fabricated through seed layer-assisted hydrothermal method; then ZnO NRs were modified with TPFS and decorated with evaporated Ag NPs; seed layers was formed on silicon wafers | A volume of 3 µL of analyte solution was added on substrate, then dried in air at room temperature; the substrate was mounted on a target plate using double-sided carbon tape | Arg—1.0 × 10–15 M; PEG—2000–1.0 × 10–10 M; R6G—1.0 × 10–15 M | [120] |
| Zinc oxide nanoparticles (ZnO NPs) | Small drug molecules (nortriptyline, amitriptyline, imipramine, promazine) in latent fingerprint | Nanoparticles were synthesized by microemulsions and dried at 110 °C overnight | The thumb was wiped across the forehead for 10 s and then pressed against the target plate or a glass slide for 10 s, leaving an impression on the surface; following deposition, NPs or the DHB organic matrix were applied to the LFP by dusting using a brush; fingerprint surface was spiked with 3 protocols | n/d; (relative standard deviations (RSDs) for [M-H]+ in %: nortriptyline—0.094; amitriptyline—0.202; imipramine—0.036; promazine—0.199 | [108] |
| AuNPs | Peptide fragments from standard protein digests of bovine serum albumin, bovine catalase, and bovine lactoperoxidase | Gold thin film was deposited on indium tin oxide (ITO) -conductive glass | Protein digests were dissolved in 80% ACN and 20% citrate buffer solution (3:1 50 mM ammonium citrate/100 mM citric acid) and 0.2 µL of each digest were spotted on the hydrophilic etched gold spots | angiotensin I peptide—8 fmol | [112] |
| AuNPs | Testosterone, progesterone, cortisol, ribose, glucose, maltose, 5- 5-hydroxyindolacetic acid (HIAA), tryptophan, gangliozyd (GM1), angiotensin I from urine samples | A solution of the AuNPs was prepared by the chemical reduction of metal salt precursor in a liquid solution | Urine samples were directly deposited onto the sample plate and allowed to dry in air; then, an equal volume of 13 nM Au NPs or 20 mg/mL DHB was deposited onto the first layer and allowed to dry in air before MALDI—TOF-MS measurements | LOD, nm: Testosterone—188; Progesterone—389.8; Cortisol—641; Ribose—1395; Glucose—393.4; Maltose—785.3; 5-HIAA—46.5; Trp—141.5; GM1—1648.4; Angiotensin I—5115.7 | [115] |
| ZnO NPs | Polyethylene glycol, polystyrene and polymethylmethacrylate, oligosaccharides, lipids | ZnO was suspended to achieve 10 wt % in methanol; the suspended solution was irradiated by ultrasonic agitation for 2 h | Each 0.6 µL of the ZnO dispersed solution and analyte solutions were placed on a stainless-steel sample target (384 wells) and dried at room temperature; NaI (0.1 mM) was added to all sample solutions as cationizing agent, except for verapamil hydrochloride | Β-cyclodextrin and hexa-N-acetylchitohexaose—1 pmol | [116] |
| Platinum nanoparticles (PtNPs) | Saccharides, pigments, and drugs | Vapor deposition Pt deposition on the target imaging sample was performed by commercially available magnetron sputtering device | Pt-deposited sample was mounted onto a holder plate and fixed using electrically conductive carbon tape | n/d | [110] |
| AuNPs | Endogenous chemicals in latent fingerprints (LFPs) | Vapor deposition by sputtering | To prepare sebum-rich LFPs, the donor wiped his thumb on his forehead for about 10 s, and then pressed his thumb on the desired substrates gently for about 10 s | n/d | [111] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomastowski, P.; Buszewski, B. Complementarity of Matrix- and Nanostructure-Assisted Laser Desorption/Ionization Approaches. Nanomaterials 2019, 9, 260. https://doi.org/10.3390/nano9020260
Pomastowski P, Buszewski B. Complementarity of Matrix- and Nanostructure-Assisted Laser Desorption/Ionization Approaches. Nanomaterials. 2019; 9(2):260. https://doi.org/10.3390/nano9020260
Chicago/Turabian StylePomastowski, Pawel, and Boguslaw Buszewski. 2019. "Complementarity of Matrix- and Nanostructure-Assisted Laser Desorption/Ionization Approaches" Nanomaterials 9, no. 2: 260. https://doi.org/10.3390/nano9020260
APA StylePomastowski, P., & Buszewski, B. (2019). Complementarity of Matrix- and Nanostructure-Assisted Laser Desorption/Ionization Approaches. Nanomaterials, 9(2), 260. https://doi.org/10.3390/nano9020260






