CdTe0.5S0.5/ZnS Quantum Dots Embedded in a Molecularly Imprinted Polymer for the Selective Optosensing of Dopamine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of Core/Shell CdTe0.5S0.5/ZnS QDs
2.3. Synthesis of CdTe0.5S0.5/ZnS@MIP and CdTe0.5S0.5/ZnS@NIPs Sensors
2.4. Detection of DA in Aqueous Solution
2.5. Selectivity of DA Detection
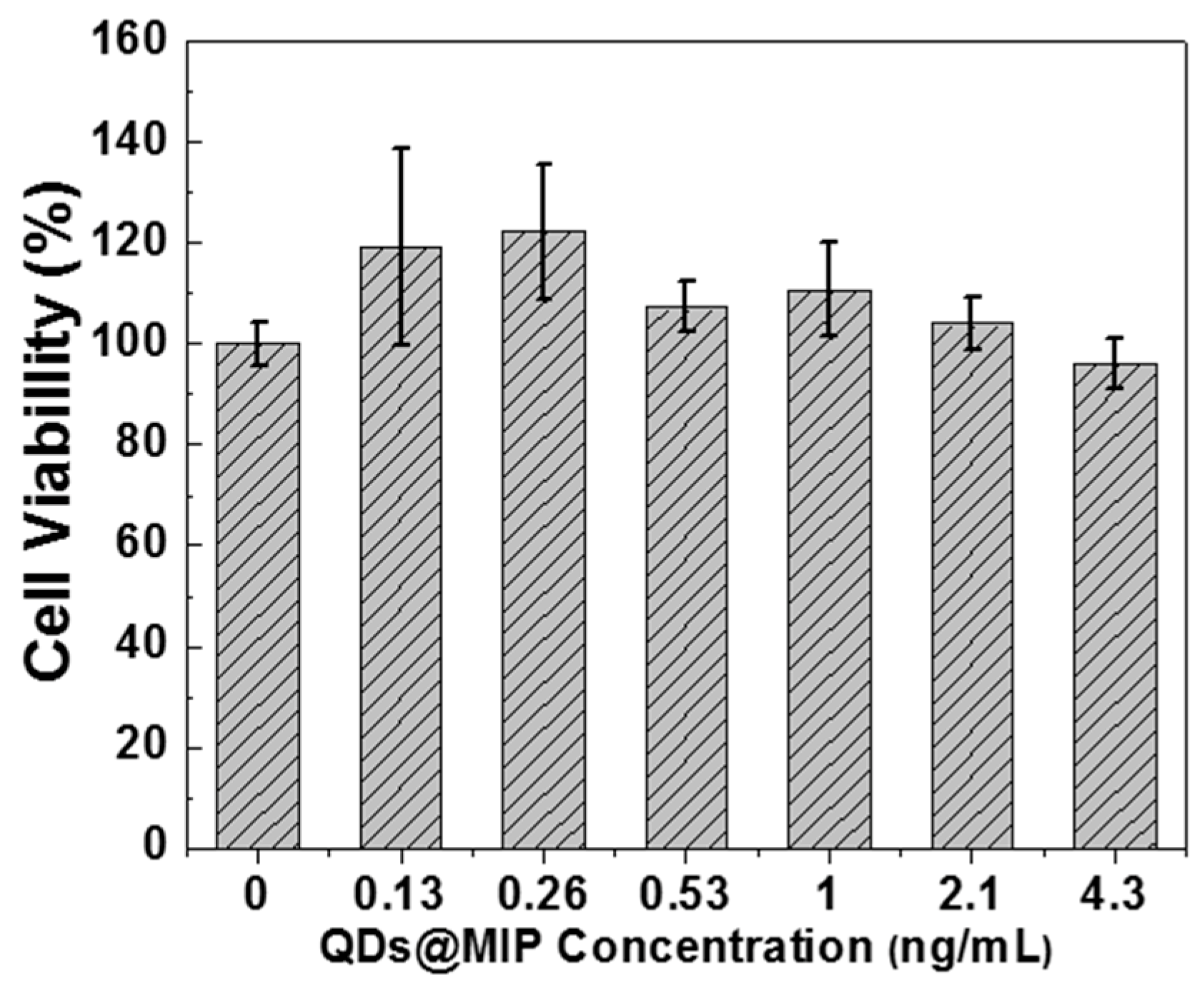
2.6. Biocompatibility
2.7. Characterization
3. Results
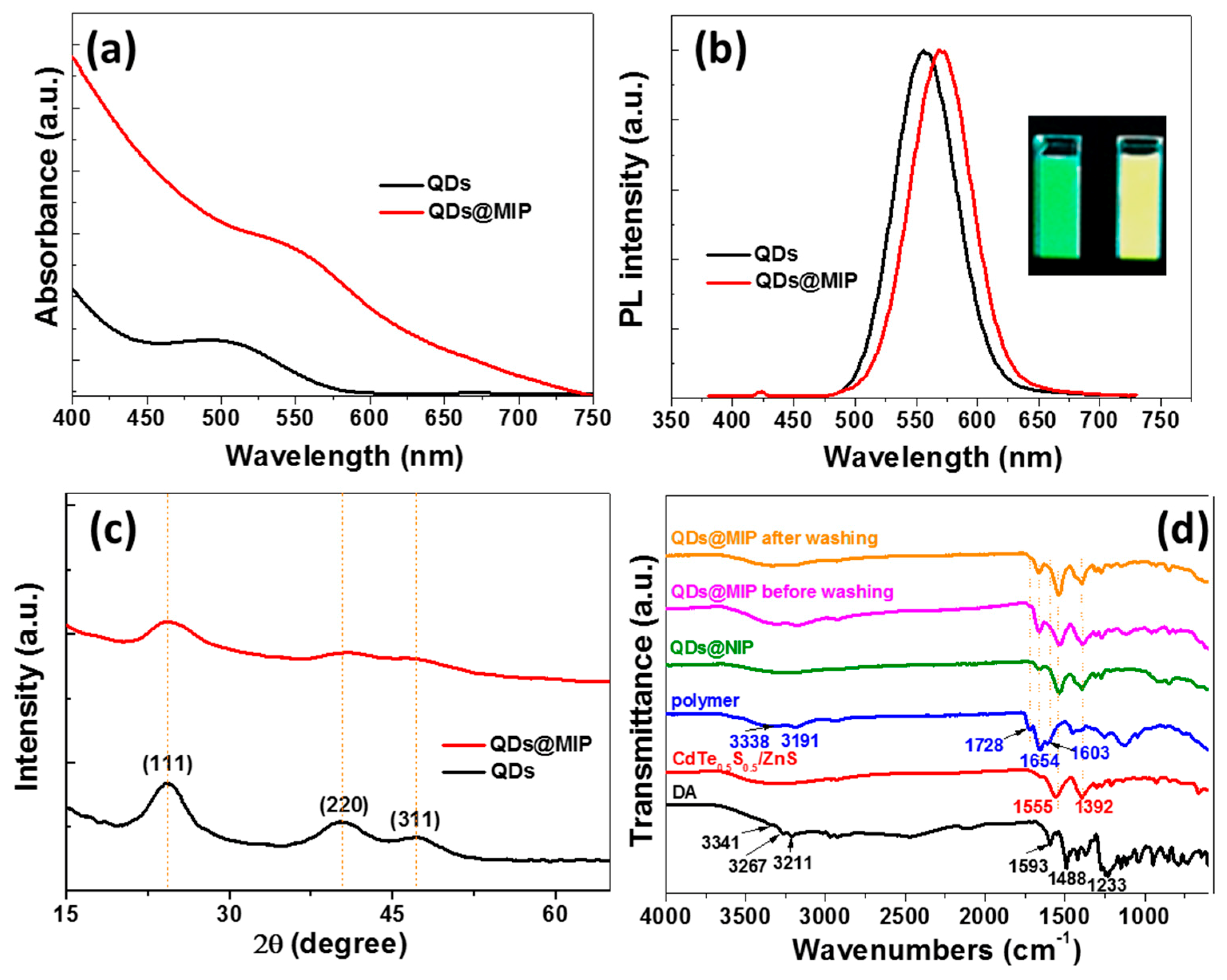
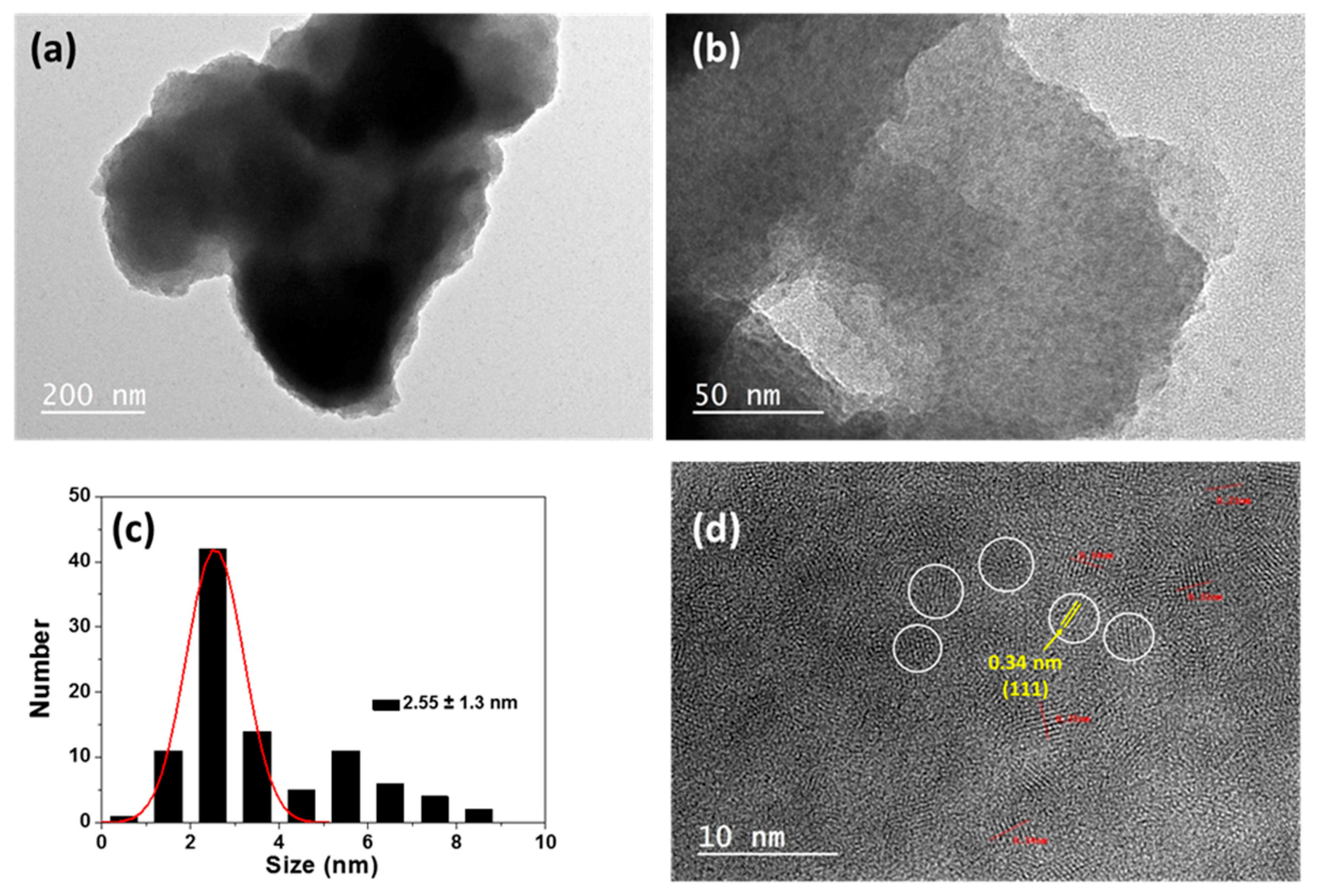
3.1. QDs@MIP Synthesis and Characterization
3.2. Sensitivity of CdTe0.5S0.5/ZnS @MIP Particles for DA Detection and Mechanism
3.3. Selectivity of QDs@MIPs for DA Detection and Toxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, J.M.; Sanyal, S.; Van Tol, H.H.M. Dopamine D2 and D4 receptor ligands: Relation to antipsychotic action. Eur. J. Pharmacol. 1998, 351, 273–286. [Google Scholar] [CrossRef]
- Venton, B.J.; Wightman, R.M. Psychoanalytical electrochemistry: Dopamine and behavior. Correlating neurochemical changes in the brain with behavior marks the beginning of an exciting new interdisciplinary field psychoanalytical chemistry. Anal. Chem. 2003, 75, 414A–421A. [Google Scholar]
- Seema, E.; Niznik, H.B. Dopamine receptors and transporters in Parkinson’s disease and schizophrenia. FASEB J. 1990, 4, 2737–2744. [Google Scholar] [CrossRef]
- Jaber, M.; Robinson, S.W.; Missale, C.; Caron, M.G. Dopamine receptors and brain function. Neuropharmacology 1996, 35, 1503–1519. [Google Scholar] [CrossRef]
- Zhang, A.; Neumeyer, J.L.; Baldessarini, R.J. Recent progress in development of dopamine receptor subtype-selective agents: Potential therapeutics for neurological and psychiatric disorders. Chem. Rev. 2007, 107, 274–302. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, H.; Shen, Y.; Zhang, J.; Zhu, J.-J. Electrogenerated chemiluminescence of Au nanoclusters for the detection of dopamine. Anal. Chem. 2011, 83, 661–665. [Google Scholar] [CrossRef]
- Duan, H.; Li, L.; Wang, X.; Wang, Y.; Li, J.; Luo, C. A sensitive and selective chemiluminescence sensor for the determination of dopamine based on silanized magnetic graphene oxide-molecularly imprinted polymer. Spectrochim. Acta A 2015, 139, 374–379. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Tang, L.; Lu, J.; Li, J. Application of graphene-modified electrode for selective detection of dopamine. Electrochem. Commun. 2009, 11, 889–892. [Google Scholar] [CrossRef]
- Liu, L.; Li, S.; Liu, L.L.; Deng, D.H.; Xia, N. Simple, sensitive and selective detection of dopamine using dithiobis(succinimidylpropionate)-modified gold nanoparticles as colorimetric probes. Analyst 2012, 137, 3794–3799. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Xie, K.; Ye, L.; Li, G.; Lu, Z.; He, J. Gold-nanoparticle-based colorimetric array for detection of dopamine in urine and serum. Talanta 2015, 139, 89–95. [Google Scholar] [CrossRef]
- Vuorensola, K.; Siren, H.; Karjalainen, U. Determination of dopamine and methoxycatecholamines in patient urine by liquid chromatography with electrochemical detection and by capillary electrophoresis coupled with spectrophotometry and mass spectrometry. J. Chromatogr. B 2003, 788, 277–289. [Google Scholar] [CrossRef]
- Zhao, D.; Song, H.; Hao, L.; Liu, X.; Zhang, L.; Lv, Y. Luminescent ZnO quantum dots for sensitive and selective detection of dopamine. Talanta 2013, 107, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, X.; Kai, S.; Wang, H.-Y.; Yang, J.; Wu, F.-G.; Chen, Z. Highly sensitive and selective detection of dopamine using one-pot synthesized highly photoluminescent silicon nanoparticles. Anal. Chem. 2015, 87, 3360–3365. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, G.; Song, Q.; Tang, T.; Wang, X.; Wei, F.; Hu, Q. Highly sensitive determination of dopamine by a turn-on fluorescent biosensor based on aptamer labeled carbon dots and nano-graphite. Sens. Actuator. B: Chem. 2016, 231, 506–512. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, L.; Lan, C.; Zhao, S. Graphene quantum dots as effective probes for label-free fluorescence detection of dopamine. Sens. Actuators B Chem. 2016, 223, 246–251. [Google Scholar] [CrossRef]
- Ren, X.; Ge, J.; Mang, X.; Qiu, X.; Ren, J.; Tang, F. Sensitive detection of dopamine and quinone drugs based on the quenching of the fluorescence of carbon dots. Sci. Bull. 2016, 61, 1615–1623. [Google Scholar] [CrossRef]
- Haupt, K.; Mosbach, K. Molecularly imprinted polymers and their use in biomimetic sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–28. [Google Scholar] [CrossRef]
- Spivak, D.A. Optimization, evaluation, and characterization of molecularly imprinted polymers. Adv. Drug. Delivery Rev. 2005, 57, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Wackerling, J.; Lieberzeit, P.A. Molecularly imprinted polymer nanoparticles in chemical sensing—Synthesis, characterisation and application. Sens. Actuators B Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Uzun, L.; Turner, A.P.F. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Karimi, M. Novel developments and trends of analytical methods for drug analysis in biological and environmental samples by molecularly imprinted polymers. Trends Anal. Chem. 2017, 89, 146–162. [Google Scholar] [CrossRef]
- Wackerlig, J.; Schirhagl, R. Applications of molecularly imprinted polymer nanoparticles and their advances toward industrial use: A Review. Anal. Chem. 2016, 88, 250–261. [Google Scholar] [CrossRef]
- Pan, J.; Chen, W.; Mab, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587. [Google Scholar] [CrossRef] [PubMed]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Michalet, X.; Pinaud, F.F.; Bentolila, L.A.; Tsay, J.M.; Doose, S.; Li, J.J.; Sundaresan, G.; Wu, A.M.; Gambhir, S.S.; Weiss, S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 2005, 307, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Bao, Y.; Han, D.; Li, F.; Niu, L. Efficient one-pot synthesis of molecularly imprinted silica nanospheres embedded carbon dots for fluorescent dopamine optosensing. Biosens. Bioelectron. 2012, 38, 55–60. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, A.; Yu, C.; Wu, S.; Shen, J. Facile synthesis of molecularly imprinted graphene quantum dots for the determination of dopamine with affinity-adjustable. ACS Appl. Mater. Interfaces 2015, 7, 11741–11747. [Google Scholar] [CrossRef]
- Zhou, X.; Gao, X.; Song, F.; Wang, C.; Chu, F.; Wu, S. A sensing approach for dopamine determination by boronic acid-functionalized molecularly imprinted graphene quantum dots composite. Appl. Surf. Sci. 2017, 423, 810–816. [Google Scholar] [CrossRef]
- Kunstman, P.; Coulon, J.; Kolmykov, O.; Moussa, H.; Balan, L.; Medjahdi, G.; Lulek, J.; Schneider, R. One step synthesis of bright luminescent core/shell CdTexS1-x/ZnS quantum dots emitting from the visible to the near infrared. J. Lumin. 2018, 194, 760–767. [Google Scholar] [CrossRef]
- Berridge, M.V.; Tan, A.S.; McCoy, K.D. The biochemical and cellular basis of cell proliferation assays that use tetrazolium salts. Biochemica 1996, 4, 14–19. [Google Scholar]
- Wuister, S.F.; de Mello Donega, C.; Meijerink, A. Influence of thiol capping on the exciton luminescence and decay kinetics of CdTe and CdSe quantum dots. J. Phys. Chem. B 2004, 108, 17393–17397. [Google Scholar] [CrossRef]
- Aldeek, F.; Balan, L.; Medjahdi, G.; Roques-Carmes, T.; Malval, J.-P.; Mustin, C.; Ghanbaja, J.; Schneider, R. Enhanced optical properties of core/shell/shell CdTe/CdS/ZnO quantum dots prepared in aqueous solution. J. Phys. Chem. C 2009, 113, 19458–19467. [Google Scholar] [CrossRef]
- Haro-Gonzalez, P.; Martinez-Maestro, L.; Martin, I.R.; Garcia-Solé, J.; Jaque, D. High-sensitivity fluorescence lifetime thermal sensing based on CdTe quantum dots. Small 2012, 8, 2652–2658. [Google Scholar] [CrossRef]
- Yang, P.; Murase, N. Preparation-condition dependence of hybrid SiO2-coated CdTe nanocrystals with intense and tunable photoluminescence. Adv. Funct. Mater. 2010, 20, 1258–1265. [Google Scholar] [CrossRef]
- Byrne, S.J.; Corr, S.A.; Rakovich, T.Y.; Gun’ko, Y.K.; Rakovich, Y.P.; Donegan, J.F.; Mitchell, S.; Volkov, Y. Optimisation of the synthesis and modification of CdTe quantum dots for enhanced live cell imaging. J. Mater. Chem. 2006, 16, 2896–2902. [Google Scholar] [CrossRef]
- Xia, H.; He, G.; Peng, J.; Li, W.; Facy, Y. Preparation and fluorescent sensing applications of novel CdSe–chitosan hybrid films. Appl. Surf. Sci. 2010, 256, 7270–7275. [Google Scholar] [CrossRef]
- He, Q.; Zhang, J.; Shi, J.; Zhu, Z.; Zhang, L.; Bu, W.; Guo, L.; Chen, Y. The effect of PEGylation of mesoporous silica nanoparticles on nonspecific binding of serum proteins and cellular responses. Biomaterials 2010, 31, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Bosshart, H.; Heinzelmann, M. THP-1 cells as a model for human monocytes. Ann. Transl. Med. 2016, 4, 438. [Google Scholar] [CrossRef]
- Ronzani, C.; Safar, R.; Diab, R.; Chevrier, J.; Paoli, J.; Abdel-Wahhab, M.A.; Le Faou, A.; Rihn, B.H.; Joubert, O. Viability and gene expression responses to polymeric nanoparticles in human and rat cells. Cell Biol. Toxicol. 2014, 30, 137–146. [Google Scholar] [CrossRef]




| [DA] (µM) | τ1 (µs) | τ2 (µs) |
|---|---|---|
| 0 | 0.65 | 9.4 |
| 5 | 0.64 | 8.5 |
| 10 | 0.80 | 8.0 |
| 15 | 0.97 | 8.5 |
| 20 | 0.82 | 6.8 |
| 25 | 0.50 | 3.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadem-Abbassi, K.; Rinnert, H.; Balan, L.; Doumandji, Z.; Joubert, O.; Masteri-Farahani, M.; Schneider, R. CdTe0.5S0.5/ZnS Quantum Dots Embedded in a Molecularly Imprinted Polymer for the Selective Optosensing of Dopamine. Nanomaterials 2019, 9, 693. https://doi.org/10.3390/nano9050693
Khadem-Abbassi K, Rinnert H, Balan L, Doumandji Z, Joubert O, Masteri-Farahani M, Schneider R. CdTe0.5S0.5/ZnS Quantum Dots Embedded in a Molecularly Imprinted Polymer for the Selective Optosensing of Dopamine. Nanomaterials. 2019; 9(5):693. https://doi.org/10.3390/nano9050693
Chicago/Turabian StyleKhadem-Abbassi, Kiana, Hervé Rinnert, Lavinia Balan, Zahra Doumandji, Olivier Joubert, Majid Masteri-Farahani, and Raphaël Schneider. 2019. "CdTe0.5S0.5/ZnS Quantum Dots Embedded in a Molecularly Imprinted Polymer for the Selective Optosensing of Dopamine" Nanomaterials 9, no. 5: 693. https://doi.org/10.3390/nano9050693
APA StyleKhadem-Abbassi, K., Rinnert, H., Balan, L., Doumandji, Z., Joubert, O., Masteri-Farahani, M., & Schneider, R. (2019). CdTe0.5S0.5/ZnS Quantum Dots Embedded in a Molecularly Imprinted Polymer for the Selective Optosensing of Dopamine. Nanomaterials, 9(5), 693. https://doi.org/10.3390/nano9050693








