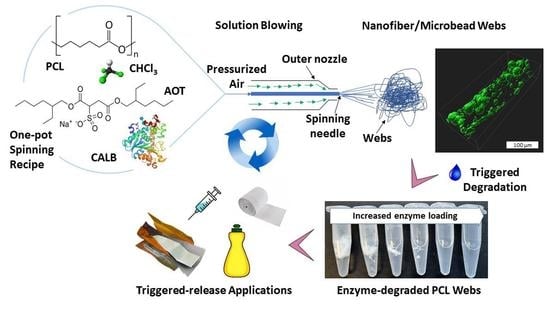
Controllable Water-Triggered Degradation of PCL Solution-Blown Nanofibrous Webs Made Possible by Lipase Enzyme Entrapment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. SBN-PCL and EFSBN-PCL Web Production
2.3. Protein Content and Immobilization Yield of EFSBN-PCL
2.4. Lipase Assay of Free and Immobilized CALB
2.5. FTIR Spectroscopy Analysis of SBN-PCL and EFSBN-PCL
2.6. Enzyme Distribution in EFSBN-PCL by Confocal Microscopy
2.7. Surface Analysis Using ToF-SIMS
2.8. EFSBN-PCL Degradation Analysis
2.9. Morphological Analysis of SBN-PCL, EFSBN-PCL, and Degraded EFSBN-PCL Using SEM
2.10. Storage Stability of Free and Immobilized CALB-EFSBN-PCL
3. Results and Discussion
3.1. SBN-PCL and EFSBN-PCL Preparation by Solution Spinning Process
3.2. Enzyme Loading and Immobilization Yield of EFSBN-PCL
3.3. FTIR, ToF-SIMS and Laser Confocal Microscopy Analysis
3.4. Enzymatic Degradation of EFSBN-PCL Webs
3.5. Morphology Analysis of EFSBN-PCL and Partially Degraded EFSBN-PCL
3.6. Biocatalytic Activity of Free and Immobilized CALB
3.7. Storage Stability of Free CALB and CALB-EFSBN-PCL
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milovanovic, S.; Hollermann, G.; Errenst, C.; Pajnik, J.; Frerich, S.; Kroll, S.; Rezwan, K.; Ivanovic, J. Supercritical CO2 Impregnation of PLA/PCL Films with Natural Substances for Bacterial Growth Control in Food Packaging. Food Res. Int. 2018, 107, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Tawakkal, I.S.M.A.; Cran, M.J.; Miltz, J.; Bigger, S.W. A Review of Poly(Lactic Acid)-Based Materials for Antimicrobial Packaging. J. Food Sci. 2014, 79, R1477–R1490. [Google Scholar] [CrossRef] [PubMed]
- Naser, A.Z.; Deiab, I.; Darras, B.M. Poly(Lactic Acid) (PLA) and Polyhydroxyalkanoates (PHAs), Green Alternatives to Petroleum-Based Plastics: A Review. RSC Adv. 2021, 11, 17151–17196. [Google Scholar] [CrossRef] [PubMed]
- Egan, J.; Salmon, S. Strategies and Progress in Synthetic Textile Fiber Biodegradability. SN Appl. Sci. 2022, 4, 22. [Google Scholar] [CrossRef]
- Hadj-Hamou, A.S.; Metref, F.; Yahiaoui, F. Thermal Stability and Decomposition Kinetic Studies of Antimicrobial PCL/Nanoclay Packaging Films. Polym. Bull. 2017, 74, 3833–3853. [Google Scholar] [CrossRef]
- Khan, R.A.; Beck, S.; Dussault, D.; Salmieri, S.; Bouchard, J.; Lacroix, M. Mechanical and Barrier Properties of Nanocrystalline Cellulose Reinforced Poly (Caprolactone) Composites: Effect of Gamma Radiation. J. Appl. Polym. Sci. 2013, 110, 3038–3046. [Google Scholar] [CrossRef]
- Englert, C.; Brendel, J.C.; Majdanski, T.C.; Yildirim, T.; Schubert, S.; Gottschaldt, M.; Windhab, N.; Schubert, U.S. Pharmapolymers in the 21st Century: Synthetic Polymers in Drug Delivery Applications. Prog. Polym. Sci. 2018, 87, 107–164. [Google Scholar] [CrossRef]
- Cabedo, L.; Feijoo, J.L.; Villanueva, M.P.; Lagarón, J.M.; Giménez, E. Optimization of Biodegradable Nanocomposites Based on APLA/PCL Blends for Food Packaging Applications. Macromol. Symp. 2006, 233, 191–197. [Google Scholar] [CrossRef]
- Grossen, P.; Witzigmann, D.; Sieber, S.; Huwyler, J. PEG-PCL-Based Nanomedicines: A Biodegradable Drug Delivery System and Its Application. J. Control. Release 2017, 260, 46–60. [Google Scholar] [CrossRef]
- Siddiqui, N.; Asawa, S.; Birru, B.; Baadhe, R.; Rao, S. PCL-Based Composite Scaffold Matrices for Tissue Engineering Applications. Mol. Biotechnol. 2018, 60, 506–532. [Google Scholar] [CrossRef]
- Pant, H.R.; Neupane, M.P.; Pant, B.; Panthi, G.; Oh, H.-J.; Lee, M.H.; Kim, H.Y. Fabrication of Highly Porous Poly (ɛ-Caprolactone) Fibers for Novel Tissue Scaffold via Water-Bath Electrospinning. Colloids Surf. B Biointerfaces 2011, 88, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Malikmammadov, E.; Tanir, T.E.; Kiziltay, A.; Hasirci, V.; Hasirci, N. PCL and PCL-Based Materials in Biomedical Applications. J. Biomater. Sci. Polym. Ed. 2018, 29, 863–893. [Google Scholar] [CrossRef] [PubMed]
- Sivasankaran, S.; Jonnalagadda, S. Advances in Controlled Release Hormonal Technologies for Contraception: A Review of Existing Devices, Underlying Mechanisms, and Future Directions. J. Control. Release 2021, 330, 797–811. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.A.; Snedeker, J.G.; Meyer, D.C. Two-Month Longitudinal Study of Mechanical Properties of Absorbable Sutures Used in Orthopedic Surgery. J. Orthop. Surg. Res. 2016, 11, 111. [Google Scholar] [CrossRef]
- Neves, A.; Godina, R.; Azevedo, S.G.; Matias, J.C.O. A Comprehensive Review of Industrial Symbiosis. J. Clean. Prod. 2020, 247, 119113. [Google Scholar] [CrossRef]
- Ganesh, M.; Gross, R.A. Embedded Enzymatic Biomaterial Degradation: Flow Conditions & Relative Humidity. Polymer 2012, 53, 3454–3461. [Google Scholar] [CrossRef]
- Pitt, C.G.; Chasalow, F.I.; Hibionada, Y.M.; Klimas, D.M.; Schindler, A. Aliphatic Polyesters. I. The Degradation of Poly(ε-Caprolactone) in vivo. J. Appl. Polym. Sci. 1981, 26, 3779–3787. [Google Scholar] [CrossRef]
- Bosworth, L.A.; Downes, S. Physicochemical Characterisation of Degrading Polycaprolactone Scaffolds. Polym. Degrad. Stab. 2010, 95, 2269–2276. [Google Scholar] [CrossRef]
- Li, F.; Yu, D.; Lin, X.; Liu, D.; Xia, H.; Chen, S. Biodegradation of Poly(ε-Caprolactone) (PCL) by a New Penicillium Oxalicum Strain DSYD05-1. World J. Microbiol. Biotechnol. 2012, 28, 2929–2935. [Google Scholar] [CrossRef]
- Shi, K.; Jing, J.; Song, L.; Su, T.; Wang, Z. Enzymatic Hydrolysis of Polyester: Degradation of Poly(ε-Caprolactone) by Candida Antarctica Lipase and Fusarium Solani Cutinase. Int. J. Biol. Macromol. 2020, 144, 183–189. [Google Scholar] [CrossRef]
- Ganesh, M.; Dave, R.N.; Amoreaux, W.L.; Gross, R. a Embedded Enzymatic Biomaterial Degradation. Macromolecules 2009, 42, 6836. [Google Scholar] [CrossRef]
- Gupta, V.; Bhavanasi, S.; Quadir, M.; Singh, K.; Ghosh, G.; Vasamreddy, K.; Ghosh, A.; Siahaan, T.J.; Banerjee, S.; Banerjee, S.K. Protein PEGylation for Cancer Therapy: Bench to Bedside. J. Cell Commun. Signal. 2019, 13, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kitayama, H.; Gotoh, Y. High Strength Ultrafine Cellulose Fibers Generated by Solution Blow Spinning. Eur. Polym. J. 2020, 125, 109513. [Google Scholar] [CrossRef]
- Li, X.; Teng, S.; Xu, X.; Wang, H.; Dong, F.; Zhuang, X.; Cheng, B. Solution Blowing of Polyacrylonitrile Nanofiber Mats Containing Fluoropolymer for Protective Applications. Fibers Polym. 2018, 19, 775–781. [Google Scholar] [CrossRef]
- Kolbasov, A.; Sinha-Ray, S.; Joijode, A.; Hassan, M.A.; Brown, D.; Maze, B.; Pourdeyhimi, B.; Yarin, A.L. Industrial-Scale Solution Blowing of Soy Protein Nanofibers. Ind. Eng. Chem. Res. 2016, 55, 323–333. [Google Scholar] [CrossRef]
- Sinha-Ray, S.; Sinha-Ray, S.; Yarin, A.L.; Pourdeyhimi, B. Theoretical and Experimental Investigation of Physical Mechanisms Responsible for Polymer Nanofiber Formation in Solution Blowing. Polymer 2015, 56, 452–463. [Google Scholar] [CrossRef]
- Asaduzzaman, F.; Salmon, S. Protease Immobilization in Solution-Blown Poly(Ethylene Oxide) Nanofibrous Nonwoven Webs. ACS Appl. Eng. Mater. 2023, 1, 447–457. [Google Scholar] [CrossRef]
- Kumar, A.; Sinha-Ray, S. A Review on Biopolymer-Based Fibers via Electrospinning and Solution Blowing and Their Applications. Fibers 2018, 6, 45. [Google Scholar] [CrossRef]
- Pant, B.; Park, M.; Park, S.J. Drug Delivery Applications of Core-Sheath Nanofibers Prepared by Coaxial Electrospinning: A Review. Pharmaceutics 2019, 11, 305. [Google Scholar] [CrossRef]
- Miranda, K.W.E.; Mattoso, L.H.C.; Bresolin, J.D.; Hubinger, S.Z.; Medeiros, E.S.; de Oliveira, J.E. Polystyrene Bioactive Nanofibers Using Orange Oil as an Ecofriendly Solvent. J. Appl. Polym. Sci. 2019, 136, 47337. [Google Scholar] [CrossRef]
- Uyar, T.; Besenbacher, F. Electrospinning of Uniform Polystyrene Fibers: The Effect of Solvent Conductivity. Polymer 2008, 49, 5336–5343. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, C.; Yang, Z.; Cai, Z.; Deng, Z.; Wu, D. Polycaprolactone/Polyvinyl Pyrrolidone Nanofibers Developed by Solution Blow Spinning for Encapsulation of Chlorogenic Acid. Food Qual. Saf. 2022, 6, fyac014. [Google Scholar] [CrossRef]
- Li, R.; Li, Z.; Yang, R.; Yin, X.; Lv, J.; Zhu, L.; Yang, R. Polycaprolactone/Poly(L-Lactic Acid) Composite Micro/Nanofibrous Membrane Prepared through Solution Blow Spinning for Oil Adsorption. Mater. Chem. Phys. 2020, 241, 122338. [Google Scholar] [CrossRef]
- Lorente, M.A.; Corral, A.; González-Benito, J. PCL/Collagen Blends Prepared by Solution Blow Spinning as Potential Materials for Skin Regeneration. J. Appl. Polym. Sci. 2021, 138, 50493. [Google Scholar] [CrossRef]
- Lv, J.; Yin, X.; Li, R.; Chen, J.; Lin, Q.; Zhu, L. Superhydrophobic PCL/PS Composite Nanofibrous Membranes Prepared through Solution Blow Spinning with an Airbrush for Oil Adsorption. Polym. Eng. Sci. 2019, 59, E171–E181. [Google Scholar] [CrossRef]
- Shen, C.; Cao, Y.; Rao, J.; Zou, Y.; Zhang, H.; Wu, D.; Chen, K. Application of Solution Blow Spinning to Rapidly Fabricate Natamycin-Loaded Gelatin/Zein/Polyurethane Antimicrobial Nanofibers for Food Packaging. Food Packag. Shelf Life 2021, 29, 100721. [Google Scholar] [CrossRef]
- Ferreira, K.N.; Oliveira, R.R.; Castellano, L.R.C.; Bonan, P.R.F.; Carvalho, O.V.; Pena, L.; Souza, J.R.; Oliveira, J.E.; Medeiros, E.S. Controlled Release and Antiviral Activity of Acyclovir-Loaded PLA/PEG Nanofibers Produced by Solution Blow Spinning. Biomater. Adv. 2022, 136, 212785. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, G.; Zhuang, X.; Li, S.; Shi, L.; Kang, W.; Cheng, B.; Xu, X. Solution Blown Nylon 6 Nanofibrous Membrane as Scaffold for Nanofiltration. Polymers 2019, 11, 364. [Google Scholar] [CrossRef]
- Wang, Y.; Chao, G.; Li, X.; Dong, F.; Zhuang, X.; Shi, L.; Cheng, B.; Xu, X. Hierarchical Fibrous Microfiltration Membranes by Self-Assembling DBS Nanofibrils in Solution-Blown Nanofibers. Soft Matter 2018, 14, 8879–8882. [Google Scholar] [CrossRef]
- Popkov, A.V.; Kulbakin, D.E.; Popkov, D.A.; Gorbach, E.N.; Kononovich, N.A.; Danilenko, N.V.; Stankevich, K.S.; Choynzonov, E.L.; Zheravin, A.A.; Khlusov, I.A.; et al. Solution Blow Spinning of PLLA/Hydroxyapatite Composite Scaffolds for Bone Tissue Engineering. Biomed. Mater. 2021, 16, 055005. [Google Scholar] [CrossRef]
- Behrens, A.M.; Casey, B.J.; Sikorski, M.J.; Wu, K.L.; Tutak, W.; Sandler, A.D.; Kofinas, P. In Situ Deposition of PLGA Nanofibers via Solution Blow Spinning. ACS Macro Lett. 2014, 3, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Tomecka, E.; Wojasinski, M.; Jastrzebska, E.; Chudy, M.; Ciach, T.; Brzozka, Z. Poly(L-Lactic Acid) and Polyurethane Nanofibers Fabricated by Solution Blow Spinning as Potential Substrates for Cardiac Cell Culture. Mater. Sci. Eng. C 2017, 75, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Asaduzzaman, F.; Salmon, S. Enzyme Immobilization: Polymer–Solvent–Enzyme Compatibility. Mol. Syst. Des. Eng. 2022, 7, 1385–1414. [Google Scholar] [CrossRef]
- Shen, J.; Yuan, Y.; Salmon, S. Carbonic Anhydrase Immobilized on Textile Structured Packing Using Chitosan Entrapment for CO 2 Capture. ACS Sustain. Chem. Eng. 2022, 10, 7772–7785. [Google Scholar] [CrossRef]
- Pereira, A.D.S.; Diniz, M.M.; De Jong, G.; Gama Filho, H.S.; dos Anjos, M.J.; Finotelli, P.V.; Fontes-Sant’Ana, G.C.; Amaral, P.F.F. Chitosan-Alginate Beads as Encapsulating Agents for Yarrowia Lipolytica Lipase: Morphological, Physico-Chemical and Kinetic Characteristics. Int. J. Biol. Macromol. 2019, 139, 621–630. [Google Scholar] [CrossRef]
- Festag, R.; Alexandratos, S.D.; Joy, D.C.; Wunderlich, B.; Annis, B.; Cook, K.D. Effects of Molecular Entanglements during Electrospray of High Molecular Weight Polymers. J. Am. Soc. Mass Spectrom. 1998, 9, 299–304. [Google Scholar] [CrossRef]
- Annabi, N.; Fathi, A.; Mithieux, S.M.; Weiss, A.S.; Dehghani, F. Fabrication of Porous PCL/Elastin Composite Scaffolds for Tissue Engineering Applications. J. Supercrit. Fluids 2011, 59, 157–167. [Google Scholar] [CrossRef]
- Janarthanan, G.; Kim, I.G.; Chung, E.J.; Noh, I. Comparative Studies on Thin Polycaprolactone-Tricalcium Phosphate Composite Scaffolds and Its Interaction with Mesenchymal Stem Cells. Biomater. Res. 2019, 23, 1. [Google Scholar] [CrossRef]
- Islam, M.M.; Zaman, A.; Islam, M.S.; Khan, M.A.; Rahman, M.M. Physico-Chemical Characteristics of Gamma-Irradiated Gelatin. Prog. Biomater. 2014, 3, 21. [Google Scholar] [CrossRef]
- Arjunan, P.; Umland, T.; Dyda, F.; Swaminathan, S.; Furey, W.; Sax, M.; Farrenkopf, B.; Gao, Y.; Zhang, D.; Jordan, F. Crystal Structure of the Thiamin Diphosphate-Dependent Enzyme Pyruvate Decarboxylase from the Yeast Saccharomyces Cerevisiae at 2.3 Å Resolution. J. Mol. Biol. 1996, 256, 590–600. [Google Scholar] [CrossRef]
- Pelton, J.T.; McLean, L.R. Spectroscopic Methods for Analysis of Protein Secondary Structure. Anal. Biochem. 2000, 277, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Sóti, P.L.; Weiser, D.; Vigh, T.; Nagy, Z.K.; Poppe, L.; Marosi, G. Electrospun Polylactic Acid and Polyvinyl Alcohol Fibers as Efficient and Stable Nanomaterials for Immobilization of Lipases. Bioprocess Biosyst. Eng. 2016, 39, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Padilla, R.Y.; Lisboa, M.C.; Fricks, A.T.; Franceschi, E.; Lima, A.S.; Silva, D.P.; Soares, C.M.F. Immobilization of Candida Rugosa Lipase on Poly(3-Hydroxybutyrate-Co-Hydroxyvalerate): A New Eco-Friendly Support. J. Ind. Microbiol. Biotechnol. 2012, 39, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Park, J.-M.; Um, H.-J.; Lee, D.-H.; Lee, K.-H.; Kobayashi, F.; Iwasaka, Y.; Hong, C.-S.; Min, J.; Kim, Y.-H. Immobilization of Cross-Linked Lipase Aggregates onto Magnetic Beads for Enzymatic Degradation of Polycaprolactone. J. Basic Microbiol. 2010, 50, 218–226. [Google Scholar] [CrossRef]









| Sample | Incubation Medium | Equivalent Protein, mg Protein Loading/mL Buffer | Weight Loss, % |
|---|---|---|---|
| SBN-PCL | Buffer-only | 0 | 0 |
| PCL bead | Buffer-only | 0 | 0 |
| 1.30% CALB -EFSBN-PCL | Buffer-only | 0.238 | 100 |
| SBN-PCL | Free CALB in buffer | 0.232 | 100 |
| PCL bead | Free CALB in buffer | 0.232 | 7.2 |
| PCL bead | Free CALB in buffer | 1.192 | 6.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asaduzzaman, F.; Salmon, S. Controllable Water-Triggered Degradation of PCL Solution-Blown Nanofibrous Webs Made Possible by Lipase Enzyme Entrapment. Fibers 2023, 11, 49. https://doi.org/10.3390/fib11060049
Asaduzzaman F, Salmon S. Controllable Water-Triggered Degradation of PCL Solution-Blown Nanofibrous Webs Made Possible by Lipase Enzyme Entrapment. Fibers. 2023; 11(6):49. https://doi.org/10.3390/fib11060049
Chicago/Turabian StyleAsaduzzaman, Fnu, and Sonja Salmon. 2023. "Controllable Water-Triggered Degradation of PCL Solution-Blown Nanofibrous Webs Made Possible by Lipase Enzyme Entrapment" Fibers 11, no. 6: 49. https://doi.org/10.3390/fib11060049
APA StyleAsaduzzaman, F., & Salmon, S. (2023). Controllable Water-Triggered Degradation of PCL Solution-Blown Nanofibrous Webs Made Possible by Lipase Enzyme Entrapment. Fibers, 11(6), 49. https://doi.org/10.3390/fib11060049