Genetic Analysis of Peroxisomal Genes Required for Longevity in a Yeast Model of Citrin Deficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primers and Plasmids
2.2. Yeast Strains and Disruption of AGC1
2.3. Yeast Transformation
2.4. Culture Conditions and Longevity Assay
2.5. Peroxisomal and Cytosolic NADH Analysis, Peroxisome Proliferation Analysis, and Confocal Fluorescence Microscopy
2.6. Neutral Lipid Staining and Spectrofluorometry
2.7. Data Analysis
3. Results
3.1. Deletion of AGC1 Inhibits Fat Utilization in the Stationary Phase
3.2. Deletion of AGC1 Increases Peroxisomal NADH Signals, but Decreases Cytosolic NADH Signals in the Stationary Phase
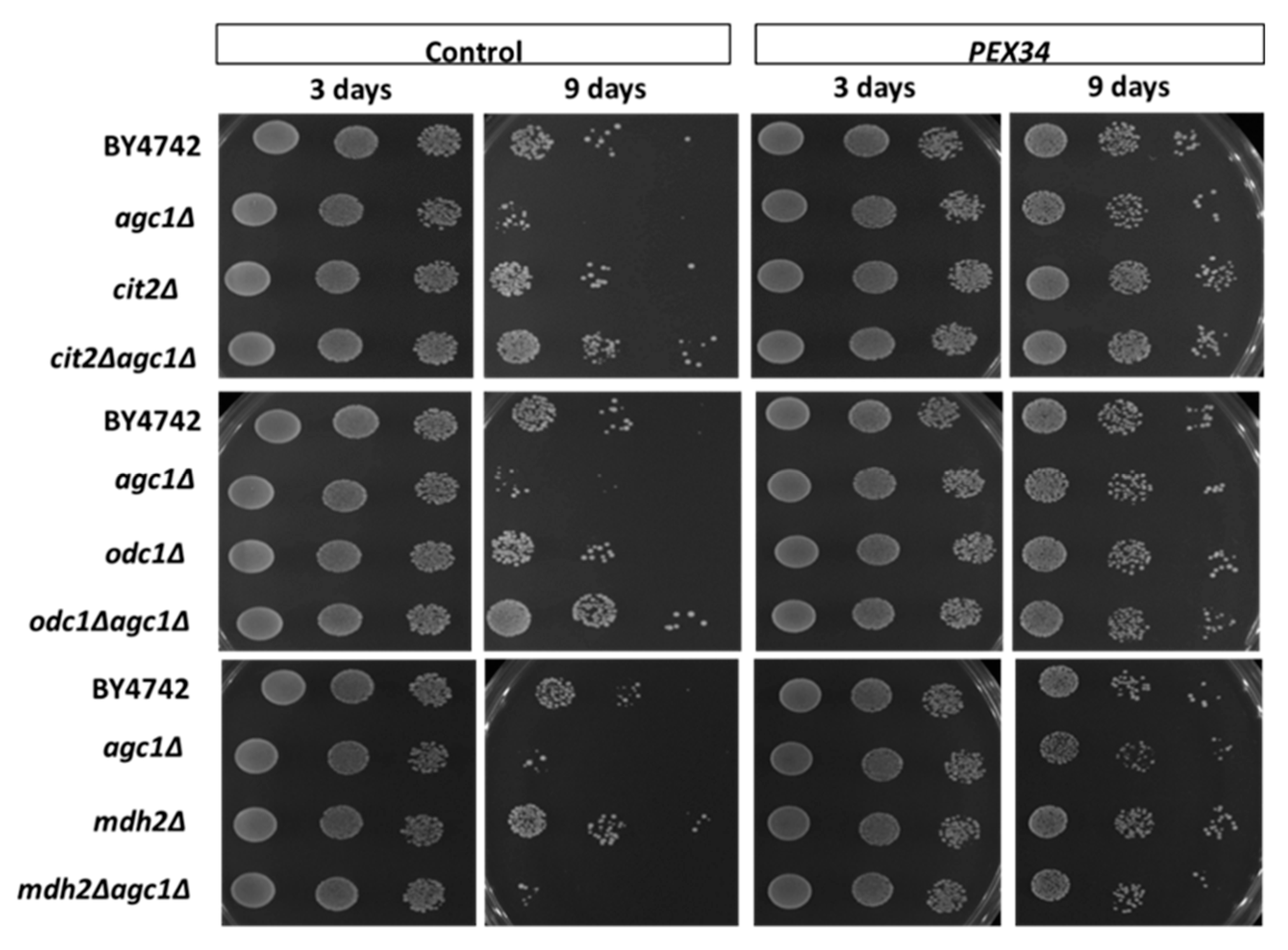
3.3. Over-Expression of PEX34, MDH3, GPD1 Increases the Longevity of agc1Δ Yeasts in the Stationary Phase
3.4. PEX34-Mediated Chronological Longevity Requires GPD1 but not PEX25 and PEX27
3.5. Role of Malate–Oxaloacetate NADH Perosixomal shuttle in Chronological Longevity of agc1Δ Yeast
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Komatsu, M.; Kimura, T.; Yazaki, M.; Tanaka, N.; Yang, Y.; Nakajima, T.; Horiuchi, A.; Fang, Z.Z.; Joshita, S.; Matsumoto, A.; et al. Steatogenesis in adult-onset type II citrullinemia is associated with down-regulation of PPARalpha. Biochim. Biophys. Acta 2015, 1852, 473–481. [Google Scholar] [CrossRef] [Green Version]
- Shigematsu, Y.; Hirano, S.; Hata, I.; Tanaka, Y.; Sudo, M.; Sakura, N.; Tajima, T.; Yamaguchi, S. Newborn mass screening and selective screening using electrospray tandem mass spectrometry in Japan. J. Chromatogr. B 2002, 776, 39–48. [Google Scholar] [CrossRef]
- Wongkittichote, P.; Sukasem, C.; Kikuchi, A.; Aekplakorn, W.; Jensen, L.T.; Kure, S.; Wattanasirichaigoon, D. Screening of SLC25A13 mutation in the Thai population. World J. Gastroenterol. 2013, 19, 7735–7742. [Google Scholar] [CrossRef]
- Kobayashi, K.; Sinasac, D.S.; Iijima, M.; Boright, A.P.; Begum, L.; Lee, J.R.; Yasuda, T.; Ikeda, S.; Hirano, R.; Terazono, H.; et al. The gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat. Genet. 1999, 22, 159–163. [Google Scholar] [CrossRef]
- Palmieri, L.; Pardo, B.; Lasorsa, F.M.; del Arco, A.; Kobayashi, K.; Iijima, M.; Runswick, M.J.; Walker, J.E.; Saheki, T.; Satrustegui, J.; et al. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. The EMBO J. 2001, 20, 5060–5069. [Google Scholar] [CrossRef]
- Saheki, T.; Kobayashi, K.; Iijima, M.; Horiuchi, M.; Begum, L.; Jalil, M.A.; Li, M.X.; Lu, Y.B.; Ushikai, M.; Tabata, A.; et al. Adult-onset type II citrullinemia and idiopathic neonatal hepatitis caused by citrin deficiency: Involvement of the aspartate glutamate carrier for urea synthesis and maintenance of the urea cycle. Mol. Genet. Metab. 2004, 81, 20–26. [Google Scholar] [CrossRef]
- Saheki, T.; Song, Y.Z. Citrin Deficiency. In GeneReviews ®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, Seattle, WA, USA, 2017. [Google Scholar]
- Mutoh, K.; Kurokawa, K.; Kobayashi, K.; Saheki, T. Treatment of a citrin-deficient patient at the early stage of adult-onset type II citrullinaemia with arginine and sodium pyruvate. J. Inherit. Metab. Dis. 2008, 31, 343–347. [Google Scholar] [CrossRef]
- Tenreiro, S.; Outeiro, T.F. Simple is good: Yeast models of neurodegeneration. FEMS Yeast Res. 2010, 10, 970–979. [Google Scholar] [CrossRef]
- Wongkittichote, P.; Tungpradabkul, S.; Wattanasirichaigoon, D.; Jensen, L.T. Prediction of the functional effect of novel SLC25A13 variants using a S. cerevisiae model of AGC2 deficiency. J. Inherit. Metab. Dis. 2013, 36, 821–830. [Google Scholar] [CrossRef]
- Forsburg, S.L. The art and design of genetic screens: Yeast. Nat. Rev. Genet. 2001, 2, 659–668. [Google Scholar] [CrossRef]
- Cavero, S.; Vozza, A.; del Arco, A.; Palmieri, L.; Villa, A.; Blanco, E.; Runswick, M.J.; Walker, J.E.; Cerdan, S.; Palmieri, F.; et al. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 2003, 50, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Koch, J.; Kragler, F.; Brocard, C.; Hartig, A. A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 2012, 13, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Jang, J.W.; Kim, K.J.; Maeng, P.J. TCA cycle-independent acetate metabolism via the glyoxylate cycle in Saccharomyces cerevisiae. Yeast 2011, 28, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Ahuir, A.; Manzanares-Estreder, S.; Proft, M. Pro- and Antioxidant Functions of the Peroxisome-Mitochondria Connection and Its Impact on Aging and Disease. Oxidative Med. Cell. Longev. 2017, 2017, 9860841. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.J.; Aitchison, J.D. Peroxisomes take shape. Nat. Rev. Mol. Cell Boil. 2013, 14, 803–817. [Google Scholar] [CrossRef] [Green Version]
- Lefevre, S.D.; van Roermund, C.W.; Wanders, R.J.; Veenhuis, M.; van der Klei, I.J. The significance of peroxisome function in chronological aging of Saccharomyces cerevisiae. Aging Cell 2013, 12, 784–793. [Google Scholar] [CrossRef] [Green Version]
- van Roermund, C.W.; Waterham, H.R.; Ijlst, L.; Wanders, R.J. Fatty acid metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. CMLS 2003, 60, 1838–1851. [Google Scholar] [CrossRef]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar]
- Sullivan, J.A.; Lewis, M.J.; Nikko, E.; Pelham, H.R. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol. Biol. Cell 2007, 18, 2429–2440. [Google Scholar] [CrossRef] [Green Version]
- Bilan, D.S.; Matlashov, M.E.; Gorokhovatsky, A.Y.; Schultz, C.; Enikolopov, G.; Belousov, V.V. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochim. Biophys. Acta 2014, 1840, 951–957. [Google Scholar] [CrossRef] [Green Version]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 1991, 7, 253–263. [Google Scholar] [CrossRef]
- Sherman, F.; Fink, G.R.; Lawrence, C.W. Methods in Yeast Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1978; pp. 178–179. [Google Scholar]
- Zipor, G.; Haim-Vilmovsky, L.; Gelin-Licht, R.; Gadir, N.; Brocard, C.; Gerst, J.E. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc. National Acad. Sci. USA 2009, 106, 19848–19853. [Google Scholar] [CrossRef] [Green Version]
- Al-Saryi, N.A.; Al-Hejjaj, M.Y.; van Roermund, C.W.T.; Hulmes, G.E.; Ekal, L.; Payton, C.; Wanders, R.J.A.; Hettema, E.H. Two NAD-linked redox shuttles maintain the peroxisomal redox balance in Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 11868. [Google Scholar] [CrossRef] [Green Version]
- Luttik, M.A.; Overkamp, K.M.; Kotter, P.; de Vries, S.; van Dijken, J.P.; Pronk, J.T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 1998, 273, 24529–24534. [Google Scholar] [CrossRef] [Green Version]
- Tower, R.J.; Fagarasanu, A.; Aitchison, J.D.; Rachubinski, R.A. The peroxin Pex34p functions with the Pex11 family of peroxisomal divisional proteins to regulate the peroxisome population in yeast. Mol. Boil. Cell 2011, 22, 1727–1738. [Google Scholar] [CrossRef]
- Lewin, A.S.; Hines, V.; Small, G.M. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol. Cell. Boil. 1990, 10, 1399–1405. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, L.; Agrimi, G.; Runswick, M.J.; Fearnley, I.M.; Palmieri, F.; Walker, J.E. Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem. 2001, 276, 1916–1922. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Siewers, V.; Nielsen, J. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e42475. [Google Scholar] [CrossRef] [Green Version]
- Saheki, T.; Iijima, M.; Li, M.X.; Kobayashi, K.; Horiuchi, M.; Ushikai, M.; Okumura, F.; Meng, X.J.; Inoue, I.; Tajima, A.; et al. Citrin/mitochondrial glycerol-3-phosphate dehydrogenase double knock-out mice recapitulate features of human citrin deficiency. J. Biol. Chem. 2007, 282, 25041–25052. [Google Scholar] [CrossRef] [Green Version]
- Kaeberlein, M.; Andalis, A.A.; Fink, G.R.; Guarente, L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Boil. 2002, 22, 8056–8066. [Google Scholar] [CrossRef] [Green Version]
- Torres-Quiroz, F.; Garcia-Marques, S.; Coria, R.; Randez-Gil, F.; Prieto, J.A. The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J. Biol. Chem. 2010, 285, 20088–20096. [Google Scholar] [CrossRef] [Green Version]
- Chalermwat, C.; Thosapornvichai, T.; Wongkittichote, P.; Phillips, J.D.; Cox, J.E.; Jensen, A.N.; Wattanasirichaigoon, D.; Jensen, L.T. Over-expression of the peroxin Pex34p suppresses impaired acetate utilization in yeast lacking the mitochondrial aspartate/glutamate carrier Agc1p. FEMS Yeast Res. 2019, 19, foz078. [Google Scholar] [CrossRef]
- Breitenbach, M.; Rinnerthaler, M.; Hartl, J.; Stincone, A.; Vowinckel, J.; Breitenbach-Koller, H.; Ralser, M. Mitochondria in ageing: There is metabolism beyond the ROS. FEMS Yeast Res. 2014, 14, 198–212. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Butow, R.A. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006, 40, 159–185. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Denis, S.; Faust, P.L.; Wanders, R.J. Bile acids: The role of peroxisomes. J. Lipid Res. 2009, 50, 2139–2147. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Chen, C.Y.; Chou, Y.Y.; Chiu, H.C.; Tsai, W.L.; Shiesh, S.C. Bile acid profiles in neonatal intrahepatic cholestasis caused by citrin deficiency. Clin. Chim. Acta 2017, 475, 28–35. [Google Scholar] [CrossRef]
- Gronemeyer, T.; Wiese, S.; Ofman, R.; Bunse, C.; Pawlas, M.; Hayen, H.; Eisenacher, M.; Stephan, C.; Meyer, H.E.; Waterham, H.R.; et al. The proteome of human liver peroxisomes: Identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS ONE 2013, 8, e57395. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Hooiveld, G.; Muller, M.; Kersten, S. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS ONE 2009, 4, e6796. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalermwat, C.; Thosapornvichai, T.; Jensen, L.T.; Wattanasirichaigoon, D. Genetic Analysis of Peroxisomal Genes Required for Longevity in a Yeast Model of Citrin Deficiency. Diseases 2020, 8, 2. https://doi.org/10.3390/diseases8010002
Chalermwat C, Thosapornvichai T, Jensen LT, Wattanasirichaigoon D. Genetic Analysis of Peroxisomal Genes Required for Longevity in a Yeast Model of Citrin Deficiency. Diseases. 2020; 8(1):2. https://doi.org/10.3390/diseases8010002
Chicago/Turabian StyleChalermwat, Chalongchai, Thitipa Thosapornvichai, Laran T. Jensen, and Duangrurdee Wattanasirichaigoon. 2020. "Genetic Analysis of Peroxisomal Genes Required for Longevity in a Yeast Model of Citrin Deficiency" Diseases 8, no. 1: 2. https://doi.org/10.3390/diseases8010002
APA StyleChalermwat, C., Thosapornvichai, T., Jensen, L. T., & Wattanasirichaigoon, D. (2020). Genetic Analysis of Peroxisomal Genes Required for Longevity in a Yeast Model of Citrin Deficiency. Diseases, 8(1), 2. https://doi.org/10.3390/diseases8010002




