Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal
Abstract
:1. Background
2. Material and Methods
2.1. Study Design, Study Site, and Sample Population
2.2. Sample Size and Sample Type
2.3. Sample Collection and Transportation
2.4. Laboratory Processing of the Specimens
2.4.1. For Blood Samples
2.4.2. For Urine Samples
2.4.3. For Sputum, Wound Swab, Pus, Pericardial Fluids, Other Body Fluids, and Valve Tissues
2.5. Identification of the Isolates
2.6. Antibiotic Susceptibility Test of Isolated Organisms
2.7. Screening of Multidrug Resistant (MDR) and Potential ESBL Producers
2.8. Phenotypic Confirmation of ESBL Production
2.9. Preservation of the Isolates
2.10. Plasmid DNA Extraction and Amplification
2.11. DNA Amplification and Detection
2.12. Quality Control
2.13. Statistical Analysis
3. Results
3.1. Distribution of Culture-Positive Bacterial Isolates
3.2. Antibiotic Susceptibility Pattern of Isolated Gram-Negative Bacteria
3.3. Multidrug Resistance (MDR) among Gram-Negative Organisms
3.4. Distribution of ESBL Producers ESBL-Producing E. coli
3.5. Antibiotic Susceptibility Pattern of ESBL-Producing Escherichia coli
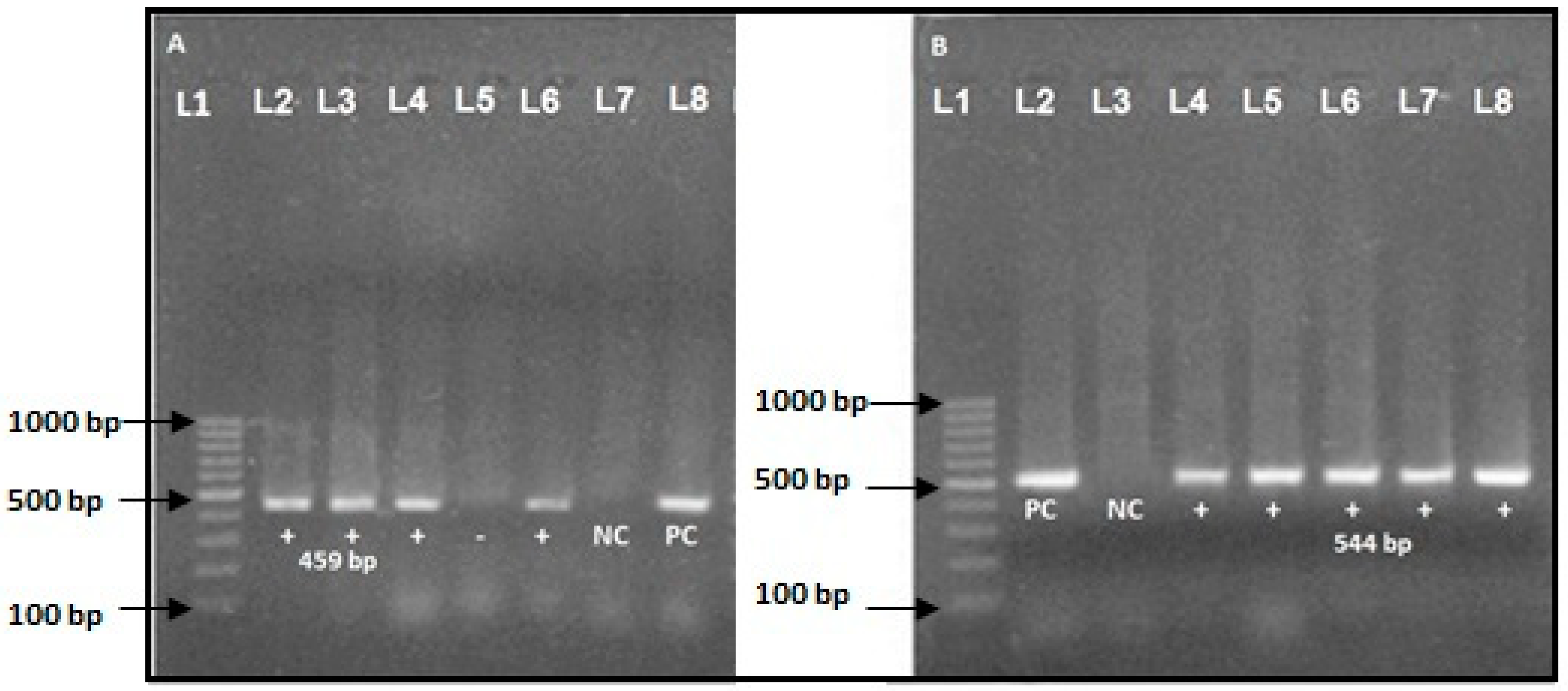
3.6. Molecular Detection of ESBL Producer Genes
4. Discussion
4.1. Overall Findings
4.2. Antibiotic Resistance and Multidrug Resistance
4.3. ESBL Producers and Acquisition of Resistant Genotypes
5. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| AST | Antimicrobial susceptibility test |
| ATCC | American Type Culture Collection |
| BA | Blood Agar |
| AZ | Ceftazidime |
| CAC | Ceftazidime plus Clavulanic Acid |
| CDC | Center for Disease Control |
| CEC | Cefotaxime plus Clavulanic Acid |
| CLSI | Clinical Laboratory and Standards Institute |
| CTX-M | Cefotaxime, Munich |
| ESBL | Extended Spectrum β-lactamase |
| kDa | Kilo-Dalton |
| MA | MacConkey Agar |
| MDR | Multidrug resistant |
| MHA | Muller Hinton Agar |
| MIC | Minimum inhibitory concentration |
| MR/VP | Methyl Red/Voges Proskauer |
| OXA | Oxacillin hydrolyzing |
| PCR | Polymerase Chain Reaction |
| PER | Pseudomonas Extended Resistant |
| SPSS | Statistical Package for Social Science |
| SHV | Sulfhydryl Variable |
| SIM | Sulphide, Indole, Motility (Medium) |
| TSI | Triple Sugar Iron |
| TEM | Temocillin hydrolyzing |
| VEB | Vietnamese Extended Spectrum β-lactamases |
| WHO | World Health Organization |
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Kibret, M.; Abera, B. Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr. Health Sci. 2011, 11, S40–S45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayastha, K.; Dhungel, B.; Karki, S.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Extended-Spectrum be-ta-Lactamase-Producing Escherichia coli and Klebsiella species in Pediatric Patients Visiting International Friendship Children’s Hospital, Kathmandu, Nepal. Infect. Dis. (Auckl.) 2020, 13, 1178633720909798. [Google Scholar] [PubMed] [Green Version]
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial use in food animals and human health: Time to implement ‘One Health’ approach. Antimicrob. Resist. Infect. Control. 2020, 9, 1–5. [Google Scholar] [CrossRef]
- Raut, S.; Adhikari, B. ESBL and their identification in peripheral laboratories of Nepal. Nepal. Med. Coll. J. 2015, 17, 176–181. [Google Scholar]
- World Health Organization (WHO). WHO Global Strategy for Containment of Antimicrobial Resistance. Available online: http://www.who.int/csr/resources/publications/drugresist/en/EGlobal_Strat.pdf (accessed on 14 August 2020).
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33. [Google Scholar] [CrossRef]
- Gajdács, M.; Albericio, F. Antibiotic Resistance: From the Bench to Patients. Antibiotics 2019, 8, 129. [Google Scholar] [CrossRef] [Green Version]
- Raut, S.; Rijal, K.R.; Khatiwada, S.; Karna, S.; Khanal, R.; Adhikari, J.; Adhikari, B. Trend and Characteristics of Acinetobacter baumannii Infections in Patients Attending Universal College of Medical Sciences, Bhairahawa, Western Nepal: A Longitudinal Study of 2018. Infect. Drug Resist. 2020, 13, 1631–1641. [Google Scholar] [CrossRef]
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob. Health 2019, 4, e002104. [Google Scholar] [CrossRef]
- Raut, S.; Adhikari, B. Global leadership against antimicrobial resistance ought to include developing countries. Lancet Infect. Dis. 2016, 16, 775. [Google Scholar] [CrossRef] [Green Version]
- Pokharel, S.; Adhikari, B. Antimicrobial resistance and over the counter use of drugs in Nepal. J. Glob. Health 2020, 10, 010360. [Google Scholar] [CrossRef]
- Chander, A.; Shrestha, C.D. Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in a tertiary care hospital in Kathmandu, Nepal. BMC Res. Notes 2013, 6, 487. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, U.T.; Shrestha, S.; Adhikari, N.; Rijal, K.R.; Shrestha, B.; Adhikari, B.; Banjara, M.R.; Ghimire, P. Plasmid Profiling and Occurrence of beta-Lactamase Enzymes in Multidrug-Resistant Uropathogenic Escherichia coli in Kathmandu, Nepal. Infect. Drug Resist. 2020, 13, 1905–1917. [Google Scholar] [CrossRef]
- Pandit, R.; Awal, B.; Shrestha, S.S.; Joshi, G.; Rijal, B.; Parajuli, N.P. Extended-Spectrum β-Lactamase (ESBL) Genotypes among Multidrug-Resistant Uropathogenic Escherichia coli Clinical Isolates from a Teaching Hospital of Nepal. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raya, G.B.; Dhoubhadel, B.G.; Shrestha, D.; Raya, S.; Laghu, U.; Shah, A.; Raya, B.B.; Kafle, R.; Parry, C.M.; Ariyoshi, K. Multi-drug-resistant and extended-spectrum β-lactamase-producing uropathogens in children in Bhaktapur, Nepal. Trop. Med. Health 2020, 48, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, N.P.; Maharjan, P.; Parajuli, H.; Joshi, G.; Paudel, D.; Sayami, S.; Khanal, P.R. High rates of multidrug resistance among uropathogenic Escherichia coli in children and analyses of ESBL producers from Nepal. Antimicrob. Resist. Infect. Control. 2017, 6, 9. [Google Scholar] [CrossRef] [Green Version]
- Acharya, K.P.; Wilson, R.T. Antimicrobial Resistance in Nepal. Front. Med. 2019, 6, 105. [Google Scholar] [CrossRef] [Green Version]
- Guragain, N.; Pradhan, A.; Dhungel, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Extended spectrum β-lactamase producing Gram negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. TUJM 2019, 6, 26–31. [Google Scholar] [CrossRef]
- Forbes, B.A.; Daniel, S.F.; Weissfelt, S.A. Bailey and Scott’s Diagnostic Microbiology; Mosby Publication: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Chesbrough, M. District Laboratory Practice in Tropical Countries, Part II, 2nd ed.; Cambridge University Press: New York, NY, USA, 2006. [Google Scholar]
- American Society for Microbiology. Manual of Clinical Microbiology, 2nd ed.; ASM Press: Hoboken, NJ, USA, 2016. [Google Scholar]
- Collee, J.G.; Mackie, T.J.; McCartney, J.E. Mackie and McCartney Practical Medical Microbiology, 14th ed.; Churchill Livingstone: New York, NY, USA, 1996; pp. 131–149. [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI). M 100 Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; An Informational Supplement: Wayne, PA, USA, 2017. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute. Performance of Standards for Antimicrobial Susceptibility Testing; M100; Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2018. [Google Scholar]
- Gurung, S.; Kafle, S.; Dhungel, B.; Adhikari, N.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Detection of OXA-48 Gene in Carbapenem-Resistant Escherichia coli and Klebsiella pneumoniae from Urine Samples. Infect. Drug Resist. 2020, 13, 2311–2321. [Google Scholar] [CrossRef]
- Muktan, B.; Thapa Shrestha, U.; Dhungel, B.; Mishra, B.C.; Shrestha, N.; Adhikari, N.; Banjara, M.R.; Adhikari, B.; Rijal, K.R.; Ghimire, P. Plasmid mediated colistin resistant mcr-1 and co-existence of OXA-48 among Escherichia coli from clinical and poultry isolates: First report from Nepal. Gut Pathog. 2020, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Lab Press: New York, NY, USA, 2001. [Google Scholar]
- Edelstein, M.V.; Pimkin, M.; Palagin, I.; Stratchounski, L.S. Prevalence and Molecular Epidemiology of CTX-M Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae in Russian Hospitals. Antimicrob. Agents Chemother. 2003, 47, 3724–3732. [Google Scholar] [CrossRef] [Green Version]
- Hyun, H.S.; Kim, J.H.; Cho, M.H.; Park, E.; Ha, I.S.; Cheong, H.I.; Kang, H.G. Low relapse rate of urinary tract infections from extended-spectrum β-lactamase-producing bacteria in young children. Pediatr. Nephrol. 2019, 34, 2399–2407. [Google Scholar] [CrossRef]
- Dhungana, K.; Awal, B.K.; Dhungel, B.; Sharma, S.; Banjara, M.R.; Rijal, K.R. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo β-lactamase (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. ASMI 2019, 2, 60–69. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Ward, M.J. Microbiology. Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef]
- Thakur, P.; Ghimire, P.; Rijal, K.R.; Singh, G.K. Antimicrobial resistance pattern of Escherichia coli isolated from urine samples in patients visiting tertiary health care centre in eastern Nepal. Sunsari Tech. Coll. J. 2012, 1, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Tamang, K.; Shrestha, P.; Koirala, A.; Khadka, J.; Gautam, N.; Rijal, K.R. Prevalence of bacterial uropathogens among diabetic patients attending Padma Nursing Hospital of Western Nepal. HiJOST 2017, 1, 15–19. [Google Scholar] [CrossRef]
- Upadhyaya, G.; Bhattarai, A.; Rijal, K.R.; Ghimire, P.; Upadhyaya, B. Urinary tract infections in Kidney transplant patients of Kathmandu Valley. Int. J. Microbiol. Res. Rev. 2013, 3, 1–6. [Google Scholar]
- Shrestha, L.B.; Baral, R.; Poudel, P.; Khanal, B. Clinical, etiological and antimicrobial susceptibility profile of pediatric urinary tract infections in a tertiary care hospital of Nepal. BMC Pediatr. 2019, 19, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, R.K.; Rijal, K.R.; Neupane, B.; Lekhak, B.; Kumar, V. Bacterial profile and drug susceptibility pattern of urinary tract infection. IJBPR 2014, 5, 812–819. [Google Scholar]
- van Duin, D.; Cober, E.; Richter, S.S.; Perez, F.; Kalayjian, R.C.; Salata, R.A.; Evans, S.; Fowler, V.G.; Jr Kaye, K.S.; Bonomo, R.A. Impact of therapy and strain type on outcomes in urinary tract infections caused by carbapenem-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 1203–1211. [Google Scholar] [CrossRef] [Green Version]
- Nepal, K.; Pant, N.D.; Neupane, B.; Belbase, A.; Baidhya, R.; Shrestha, R.K.; Lekhak, B.; Bhatta, D.R.; Jha, B. Extended spectrum β-lactamase and metallo β-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 62. [Google Scholar] [CrossRef] [Green Version]
- Walson, J.L.; Marshall, B.; Pokhrel, B.M.; Kafle, K.K.; Levy, S.B. Carriage of Antibiotic-Resistant Fecal Bacteria in Nepal Reflects Proximity to Kathmandu. J. Infect. Dis. 2001, 184, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, R.H.; Thapa, B.; Kafle, R.; Shah, P.K.; Tribuddharat, C. Co-existence of βlactamases in clinical isolates of Escherichia coli from Kathmandu, Nepal. BMC Res. Notes 2014, 7, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiratisin, P.; Apisarnthanarak, A.; Laesripa, C.; Saifon, P. Molecular characterization and epidemiology of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob. Agents Chemother. 2008, 52, 2818–2824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Pathak, S.; Srivastava, P. Prevalence and antibiogram of Extended Spectrum β-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin. Diagn. Res. 2013, 7, 2173–2177. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, N.P.; Maharjan, P.; Joshi, G.; Khanal, P.R. Emerging perils of extended spectrum β-Lactamase producing enterobacteriaceae clinical isolates in a teaching hospital of Nepal. Biomed. Res. Int. 2016, 2016, 1782835. [Google Scholar] [CrossRef]
- Lee, S.Y.; Park, Y.J.; Yu, J.K.; Jung, S.; Kim, Y.; Jeong, S.H.; Arakawa, Y. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 2012, 67, 2843–2847. [Google Scholar] [CrossRef] [Green Version]
- Raut, S.; Gokhale, S.; Adhikari, B. Prevalence of Extended Spectrum Beta-Lactamases among Escherichia coli and Klebsiella spp isolates in Manipal Teaching Hospital, Pokhara, Nepal. J. Microbiol. Infect. Dis. 2015, 5, 69–75. [Google Scholar] [CrossRef]
- Ansari, S.; Nepal, H.P.; Gautam, R.; Shrestha, S.; Neopane, P.; Gurung, G.; Chapagain, M.L. Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care center of Nepal. Antimicrob. Resist. Infect. Control. 2015, 4, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimal, U.; Thapa, S.; Maharjan, R. Prevalence of Extended Spectrum Beta-Lactamase Producing Escherichia coli and Klebsiella species from Urinary Specimens of Children attending Friendship International Children’s Hospital. Nepal J. Biotechnol. 2017, 5, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Ghimire, A.; Acharya, B.; Tuladhar, R. Extended spectrum β-lactamases (ESBL) producing MDR Gram negative bacteria from various clinical specimens of patients visiting a tertiary care hospital. TUJM 2017, 4, 1–8. [Google Scholar]
- Pathak, P.J.N.; Yadav, B.K.; Shah, P.K. Prevalence of Extended spectrum β-lactamases (ESBL) and Metallo beta lactamases (MBL) mediated resistance in Gram negative bacterial pathogen. TUJM 2017, 4, 49–54. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Character/Bacterial Isolates | Number | Culture Positive | p-Value | |
|---|---|---|---|---|
| Number | Percentage | |||
| Gender | ||||
| Male | 480 | 88 | 46.3 | 0.7 |
| Female | 585 | 102 | 53.7 | |
| Age groups (in years) | ||||
| 0–15 | 180 | 31 | 16.3 | 0.3 |
| 15–45 | 490 | 78 | 41.1 | |
| >46 | 395 | 81 | 42.6 | |
| Type of specimens | ||||
| Blood | 246 | 13 | 6.8 | 1.8 |
| Urine | 304 | 92 | 48.4 | |
| Sputum | 280 | 29 | 15.3 | |
| Pus/wound swab | 151 | 40 | 21.1 | |
| Catheter tips | 59 | 10 | 5.3 | |
| Body fluids | 25 | 6 | 3.1 | |
| Type of bacteria | ||||
| Gram-negative bacteria | 109 | 57.4 | ||
| E. coli | 44 | 40.4 | ||
| Klebsiella pneumoniae | 33 | 30.3 | ||
| Acinetobacter baumannii | 12 | 11.1 | ||
| Pseudomonas aeruginosa | 9 | 8.3 | ||
| Serratia marcescens | 7 | 6.3 | ||
| Citrobacter spp. | 2 | 1.8 | ||
| Proteus mirabilis | 2 | 1.8 | ||
| Gram-positive bacteria | 81 | 42.6 | ||
| Staphylococcus aureus | 39 | 48.2 | ||
| CONS | 19 | 23.5 | ||
| Enterococcus spp. | 15 | 18.5 | ||
| Streptococcus spp. | 8 | 9.8 | ||
| Antibiotics | E. coli (n = 44) | K. pneumoniae (n = 33) | A. baumannii (n = 12) | P. aeruginosa (n = 9) | S. marcescens (n = 7) | C. freundii (n = 2) | P. mirabilis (n = 2) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | |
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | |
| Ampicillin (10 µg) | 11 (25.0) | 33 (75.0) | 10 (30.4) | 23 (69.6) | 3 (42.9) | 4 (57.1) | 0 | 2 (100) | 1 (50.0) | 1 (50.0) | ||||
| Amikacin (30 µg) | 29 (66.0) | 15 (34.0) | 15 (45.5) | 18 (54.5) | 6 (50) | 6 (50) | 6 (66.7) | 3 (33.3) | 6 (85.8) | 1 (14.2) | 1 (50.0) | 1 (50.0) | 2 (100%) | 0 |
| Cotrimoxazole (25 µg) | 16 (36.4) | 28 (63.6) | 13 (39.4) | 20 (60.6) | 5 (71.5) | 2 (28.5) | 1 (50.0) | 1 (50.0) | 2 (100%) | 0 | ||||
| Nitrofurantoin (300 µg) | 18 (41.0) | 26 (59.0) | 12 (36.4) | 21 (63.6) | - | - | 1 (50.0) | 1 (50.0) | 2 (100%) | 0 | ||||
| Nalidixic acid (30 µg) | 17 (38.7) | 27 (61.3) | 11 (33.4) | 22 (66.6) | - | - | 1 (50.0) | 1 (50.0) | 0 | 2 (100) | ||||
| Norfloxacin (5 µg) | 16 (36.4) | 28 (63.6) | - | - | - | - | 1 (50.0) | 1 (50.0) | 0 | 2 (100) | ||||
| Gentamicin (30 µg) | 32 (72.8) | 12 (27.2) | 17 (51.6) | 16 (48.4) | 6 (50) | 6 (50) | 7 (77.8) | 2 (22.2) | 7 (100) | 0 | 2 (100) | 0 | 2 (100) | 0 |
| Ceftazidime (30 µg) | 22 (50.0) | 22 (50.0) | 21 (63.7) | 12 (36.3) | 7 (58.4) | 5 (41.6) | 7 (77.8) | 2 (22.2) | 7 (100) | 0 | 0 | 2 (100) | 2 (100%) | 0 |
| Cefotaxime (30 µg) | 20 (45.5) | 24 (54.5) | 22 (66.7) | 11 (33.3) | 7 (100) | 0 | 0 | 2 (100) | 2 (100) | 0 | ||||
| Cefepime (30 µg) | 24 (54.6) | 20 (45.4) | 23 (69.7 | 10 (30.3) | 7 (58.4) | 5 (41.6) | 6 (66.7) | 3 (33.3) | 7 (100) | 0 | 0 | 2 (100) | 2 (100%) | 0 |
| Imipenem (10 µg) | 38 (86.4) | 6 (13.6) | 29 (87.9) | 4 (12.1) | 8 (66.7) | 4 (33.3) | 9 (100) | 0 (0) | - | - | 2 (100) | 0 | - | - |
| Meropenem (10 µg) | 40 (91.0) | 4 (9.0) | 30 (91.0) | 3 (9.0) | 9 (75.0) | 3 (25.0) | 9 (100) | 0 (0) | - | - | 2 (100) | 0 | - | - |
| Piperacillin (100 µg) | 4 (33.4) | 8 (66.6) | 6 (66.7) | 3 (33.3) | ||||||||||
| Piperacillin-tazobactam (100 µg/10 µg) | 5 (41.7) | 7 (58.3) | 8 (89.0) | 1 (11.0) | ||||||||||
| Ciprofloxacin (5 µg) | 4 (44.5) | 5 (55.5) | ||||||||||||
| Character | MDR | Non MDR | p-Value | ||
|---|---|---|---|---|---|
| Number | % | Number | % | ||
| Age Group (in years) | |||||
| 0–15 | 9 | 16.1 | 7 | 13.2 | 0.5 |
| 16–45 | 24 | 42.9 | 19 | 35.8 | |
| >45 | 23 | 41.1 | 27 | 50.9 | |
| Gender | |||||
| Male | 22 | 39.3 | 31 | 58.5 | 0.05 |
| Female | 34 | 60.7 | 22 | 41.5 | |
| Types of specimens | |||||
| Blood | 7 | 12.5 | 1 | 1.9 | 0.02 |
| Urine | 38 | 67.8 | 24 | 45.3 | |
| Sputum | 3 | 5.4 | 11 | 20.7 | |
| Pus/wound swab | 5 | 8.9 | 15 | 28.3 | |
| Catheter tips | 3 | 5.4 | 2 | 3.8 | |
| Type of bacteria | |||||
| E. coli | 28 | 50 | 16 | 30.3 | 0.01 |
| Klebsiella pneumoniae | 18 | 32.1 | 15 | 28.3 | |
| A. baumannii | 7 | 12.4 | 5 | 9.4 | |
| Pseudomonas aeruginosa | 2 | 3.6 | 7 | 13.2 | |
| Citrobacter spp. | 1 | 1.9 | 1 | 1.8 | |
| Serratia marcescens | 0 | 0 | 7 | 13.2 | |
| Proteus mirabilis | 0 | 0 | 2 | 3.8 | |
| Character | ESBL Producer (n = 12) | blaCTX-M Gene (n = 7) | blaTEM Gene (n = 5) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | % | p-Value | Number | % | p-Value | Number | % | p-Value | |
| Age group (in years) | |||||||||
| 0–15 | 1 | 8.3 | 0.33 | 1 | 14.3 | 0.65 | 0 | 0.65 | |
| 16–45 | 7 | 58.3 | 4 | 57.1 | 3 | 60 | |||
| >45 | 4 | 33.3 | 2 | 28.6 | 2 | 40 | |||
| Gender | |||||||||
| Male | 4 | 33.3 | 0.53 | 4 | 57.1 | 0.08 | 1 | 20 | 0.57 |
| Female | 8 | 66.7 | 3 | 42.9 | 4 | 80 | |||
| Types of specimens | |||||||||
| Blood | 0 | 0 | 0.54 | ||||||
| Urine | 12 | 100 | 7 | 100 | 5 | 100 | |||
| Sputum | 0 | 0 | |||||||
| Pus/wound swab | 0 | 0 | |||||||
| Catheter tips | 0 | 0 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sah, R.S.P.; Dhungel, B.; Yadav, B.K.; Adhikari, N.; Thapa Shrestha, U.; Lekhak, B.; Banjara, M.R.; Adhikari, B.; Ghimire, P.; Rijal, K.R. Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases 2021, 9, 15. https://doi.org/10.3390/diseases9010015
Sah RSP, Dhungel B, Yadav BK, Adhikari N, Thapa Shrestha U, Lekhak B, Banjara MR, Adhikari B, Ghimire P, Rijal KR. Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases. 2021; 9(1):15. https://doi.org/10.3390/diseases9010015
Chicago/Turabian StyleSah, Ram Shankar Prasad, Binod Dhungel, Binod Kumar Yadav, Nabaraj Adhikari, Upendra Thapa Shrestha, Binod Lekhak, Megha Raj Banjara, Bipin Adhikari, Prakash Ghimire, and Komal Raj Rijal. 2021. "Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal" Diseases 9, no. 1: 15. https://doi.org/10.3390/diseases9010015
APA StyleSah, R. S. P., Dhungel, B., Yadav, B. K., Adhikari, N., Thapa Shrestha, U., Lekhak, B., Banjara, M. R., Adhikari, B., Ghimire, P., & Rijal, K. R. (2021). Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases, 9(1), 15. https://doi.org/10.3390/diseases9010015







