Abstract
Geminiviruses are the largest family of plant viruses that cause severe diseases and devastating yield losses of economically important crops worldwide. In response to geminivirus infection, plants have evolved ingenious defense mechanisms to diminish or eliminate invading viral pathogens. However, increasing evidence shows that geminiviruses can interfere with plant defense response and create a suitable cell environment by hijacking host plant machinery to achieve successful infections. In this review, we discuss recent findings about plant defense and viral counter-defense during plant–geminivirus interactions.
1. Introduction
Plants pathogens, including viruses, are responsible for many diseases and cause significant losses of agricultural production [1,2,3]. Geminiviridae is one of the largest and most important families of plant viruses with small circular, single-stranded DNA that are 2.7–5.2 kb in size. These viruses infect a wide range of plant species and are a major threat to almost all economically important crops and food security. Viruses of the family Geminiviridae are divided into 14 genera based on their genome organization, host range, and insect vectors (ictv.global/report/geminiviridae). Currently, the family Geminiviridae includes more than 500 species. The genome of geminivirus can be either monopartite (a single DNA component) or bipartite (two DNA components: DNA A and DNA B). For effective infection, geminivirus encodes 6-8 multifunctional proteins, which are required for viral replication, the assembly of virus particles, cell-to-cell movement, and viral symptom induction. The replication initiator protein (Rep) encoded by ORF AC1/C1 (also called AL1/L1) is essential in virus rolling-circle replication, stimulates virus transcription and suppresses host gene silencing (transcriptional gene silencing). The geminiviral transcriptional activator protein (TrAP) encoded by ORF AC2/C2 acts as a silencing suppressor (both transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS)) and involved in symptom development, suppression of HR and inhibition of hormone-mediated defense. ORF AC3/C3 encodes a replication enhancer protein (REn), which interacts with Rep and enhances viral DNA accumulation and symptom development. AC4/C4 ORF contained entirely within the AC1 ORF, but in a different frame, encodes a multifunctional protein called AC4/C4. Geminiviral AC4/C4 proteins are critical in the suppression of gene silencing (both TGS and PTGS) and HR, regulation of cell cycle and cell division, symptom development and viral systemic movement. Coat protein (CP) is encoded by ORF AV1/V1, is a structural protein to geminiviral particles and it has been associated with virus genome packaging, insect transmission and the cell-to-cell and systemic spread of viruses. It also serves as a nucleocytoplasmic shuttling protein in monopartite viruses. The AV2/V2 protein is a pathogenicity determination factor, a silencing suppressor (both TGS and PTGS) and a movement protein of geminiviruses. The DNA-B component contains two genes, BC1 and BV1, that encode two proteins, MP and NSP, respectively, which are involved in the intercellular and intracellular movement of viral particles. Geminiviruses are often associated with additional small circular single-stranded DNA molecule referred to as satellites. Satellites are approximately half the size of geminivirus DNA genomes. Alpha and deltasatellites are associated with both monopartite and bipartite begomoviruses, whereas betasatellites are associated with monopartite begomoviruses only. Alphasatellites encode replication initiator proteins and have not been shown to play a crucial role in symptom development or pathogenicity. Betasatellites are pathogenicity determinants and depend completely on their helper virus for replication and encapsidation. The only protein encoded by betasatellites is βC1, which is essential in pathogenicity determination, silencing suppression (both transcriptional gene silencing and post-transcriptional gene silencing), systemic movement and suppressing host defense. Deltasatellites do not encode any proteins but some of them affect viral DNA accumulation and symptomatology (genus: Begomovirus, ICTV). Additional small proteins AC5/C5 or V3 from geminiviruses are identified as symptom inducers and silencing suppressors. These proteins also reprogram plant cell cycle and transcriptional control, inhibit cell death pathways, interfere with cell signaling and protein turnover and suppress plant defense.
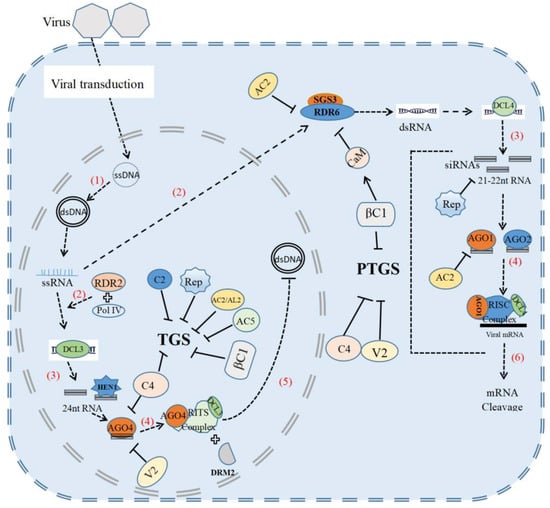
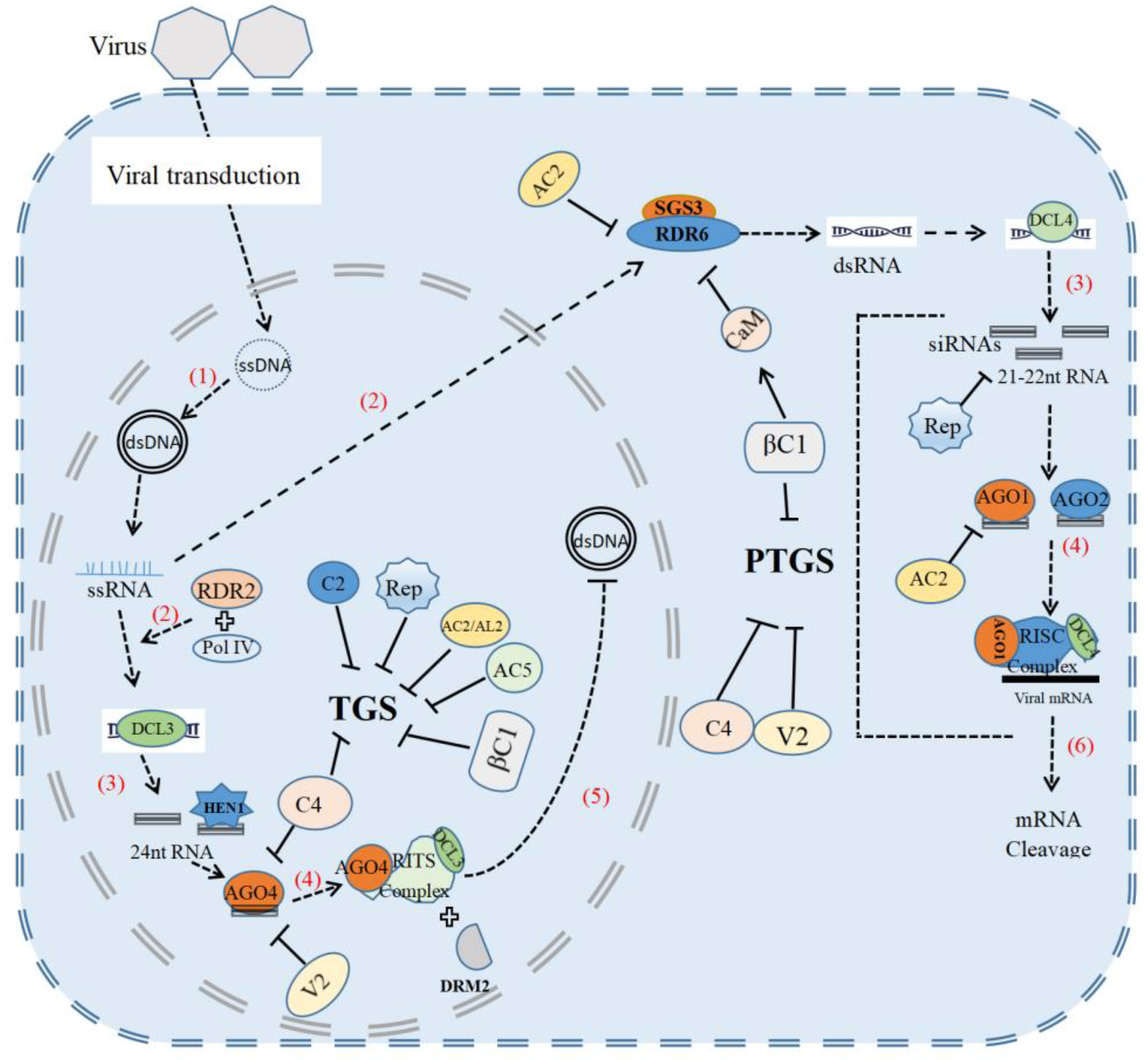
In the course of co-evolution, plants have evolved multilayered antiviral immune systems, including RNA silencing, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI). RNA silencing triggered by geminivirus infection can target either viral RNAs for degradation at the post-transcriptional level or viral DNAs for epigenetic modification at the transcriptional level to inhibit viral replication and pathogenicity [4]. To counter plant defense, geminiviruses can encode different viral proteins, such as AC1/C1, AC2/C2, AC4/C4, V2, AC5/C5 and βC1, to inhibit various steps in post-transcriptional gene silencing and transcriptional gene silencing pathways [5,6,7,8,9]. Beside RNA silencing, plants also develop protein-kinase-mediated antiviral immunity, effector-triggered immunity, autophagy-mediated antiviral defense, a ubiquitin-proteasomal protein-degradation system and hormone-mediated defense to defeat geminivirus [10]. However, geminiviruses can also evade or subvert these plant defense mechanisms for their own benefits.
4. Protein-Kinase-Mediated Immunity
Protein kinases regulate the biological activity of many proteins by phosphorylation, and they play important roles in various plant biological processes, including defense [66]. Some protein kinases are reported to regulate plant defense against geminiviruses [10,67,68,69]. Sucrose non-fermenting1-related protein kinase 1 (SnRK1) is a Ser/Thr kinase, widely recognized as a key regulator of plant responses to various physiological processes, operating multi-organ crosstalk and potentially regulating downstream transcription factors to maintain cellular homeostasis [70]. SnRK1 belongs to the conserved kinase family and consists of a α catalytic subunit and β and γ regulatory subunits [71]. The overexpression of SnRK1 makes plant more resistant to geminivirus infection [72,73]. Geminiviral Rep interacts with Rep-interacting kinase (GRIK), an upstream activator of SnRK1, and their interaction stabilizes GRIK accumulation and activates SnRK1 to phosphorylate Rep [74,75,76]. SnRK1 interacts with βC1 encoded by TYLCCNB and CLCuMuB to reduce viral DNA accumulation and viral symptom severity by phosphorylating βC1. Phosphorylated βC1 fails to decrease DNA methylation and to upregulate rgs-CaM, thus impairing the suppression of both TGS and PTGS [68,77,78,79]. SnRK1 also phosphorylates the AL2/C2 protein to limit geminivirus infection [73]. Geminiviral C2 inactivates host SnRK1 and adenosine kinases through protein–protein interactions [48,80]. SnRK1 and ADK form a complex in plants, and alterations in either one may influence the others’ activity [81]. SnRK1 also inhibits translation by phosphorylating the cap-binding proteins eIF4E and eIFiso4E to condition antiviral defense. It is also inhibited by geminivirus pathogenicity factors [82]. These results suggest that SnRK1 interacts with and phosphorylates multiple viral proteins to control geminivirus infection.
Mitogen-activated protein kinases (MAPKs) play a crucial role in defense against diverse pathogens, including geminiviruses. MAPKs are activated during geminiviral infection and restrict geminiviral pathogenicity [83,84,85]. TYLCCNV infection activates MPK6/MPK3 and MPK4, although viral βC1 limits MAPK cascade-regulated defense by inhibiting MKK2 and MPK4 kinase activity [69]. Recently, TLCYnV C4 has been reported to interfere with MAPKs-mediated defense responses by inhibiting the dissociation of the ERECTA/BKI1 complex [86]. These findings illustrate the vital role of MAPK cascade in plant defense against geminiviruses.
Receptor-like kinases (RLKs) regulate cell differentiation, development and innate immunity [87]. Several NSP-interacting RLKs (NIKs) interact with NSPs from distinct geminiviruses [88,89]. NIK confers a broad-spectrum tolerance to begomoviruses by suppressing viral translation [90,91]. Deficiency of NIK displays increased susceptibility to geminiviral infection [90,91]. However, NSP suppresses NIK activity to prevail over NIK-mediated resistance against geminivirus [88,92]. The TYLCV C4 protein interacts with many plant RLKs, including CLV1, FLS2, BRI1 and two plasma-membrane- and plasmodesmata-localized barely any meristem (BAM) 1 and 2 [93,94]. BSCTV C4 interacts with CLV1, which regulates the expression of an antiviral factor (WUSCHEL) [95]. In addition, C4 may suppress PTGS by interacting with BAM1/2 [96].
Several geminiviral genes, such as C4/AC4, are reported to interact with many Shaggy-like protein kinases [97]. Shaggy-like protein kinase SKη negatively regulates brassinosteroid (BR) signaling [98]. C4–SKη interactions are critical for C4 multifunctions, including viral symptom induction, RNA silencing suppression, cell cycle and BR signaling regulation, the induction of hyperplasia and cell division [99,100]. These findings demonstrate that there are different protein kinases pivotal in plant defense against geminiviruses, and geminiviruses exploit various strategies to suppress protein-kinase-mediated defense for effective infection.
5. Effector-Triggered Immunity (ETI)
Plant immune systems have evolved multilayer receptor systems to sense and induce pathogen defense responses. ETI restricts the pathogen at the site of infection (local resistance) by inducing programmed cell death (PCD), a phenomenon known as hypersensitive response (HR). Geminiviral proteins are both the inducers and suppressors of HR. Rep, C2 and V2 proteins are able to induce HR, meanwhile C4 and C2 are reported to antagonize HR [101,102,103,104]. These findings suggest that there exist natural antiviral R genes that confer resistance against geminiviruses. Indeed, CYR1 encodes 1176 amino-acid-resistant proteins with a coiled structure at the N-terminus, central nucleotide-binding site (NBS) and C-terminal leucine-rich repeats (LRRs), conferring resistance against MYMIV by recognizing viral coat protein in Vigna mungo. Tomato Ty-2 also encodes a CC-NB-LRR R protein, which confers resistance against TYLCV by recognizing the TYLCV Rep/C1 protein [105,106,107].
8. Hormone-Mediated Defense against Geminivirus
Plant hormones are small, structurally unrelated molecules that not only regulate plant growth and development, but are also essential in plant defense against viral pathogens [126,127]. Several studies have highlighted the involvement of various phytohormones, such as salicylic acid (SA), jasmonic acid (JA), ethylene, auxin, cytokinin, gibberellic acid, brassinosteroids and abscisic acid, in plant–geminivirus infection [10,128]. The use of exogenous SA and JA improves resistance to TYLCV infection in plants [129]. SA, ethylene and cytokinin pathways genes are upregulated within geminivirus infections [85,130,131,132,133]. Whereas, the genes in JA and auxin pathways are differentially regulated in geminivirus infections [130,134,135,136]. Geminiviral C2 interacts with CSN5 and alters the derubylation activity of the CSN complex, which affects downstream signaling pathways, such as those of auxin, gibberellic acid (GA), ethylene (ET), salicylic acid (SA) and JA [117]. The C2 protein of geminivirus has also been shown to downregulate the expression of certain defense genes in the JA-mediated signaling pathway [137]. Geminiviral βC1 suppress JA-mediated defense by repressing JA downstream markers or by interacting with MYC2 andAS1 [125,138]. Furthermore, the ßC1 protein encoded by TYLCCB suppresses JA-dependent plant terpene biosynthesis to subvert plant resistance [138]. Geminiviral C4 interacts with auxin biosynthetic enzymes and disrupts endogenous auxin content [139]. The relationship between plant hormone pathways and geminiviruses has previously been well reviewed [128,140].
9. Conclusions
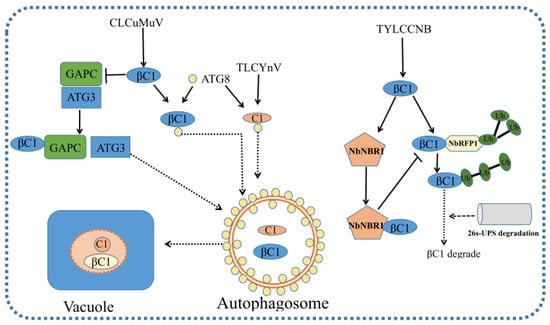
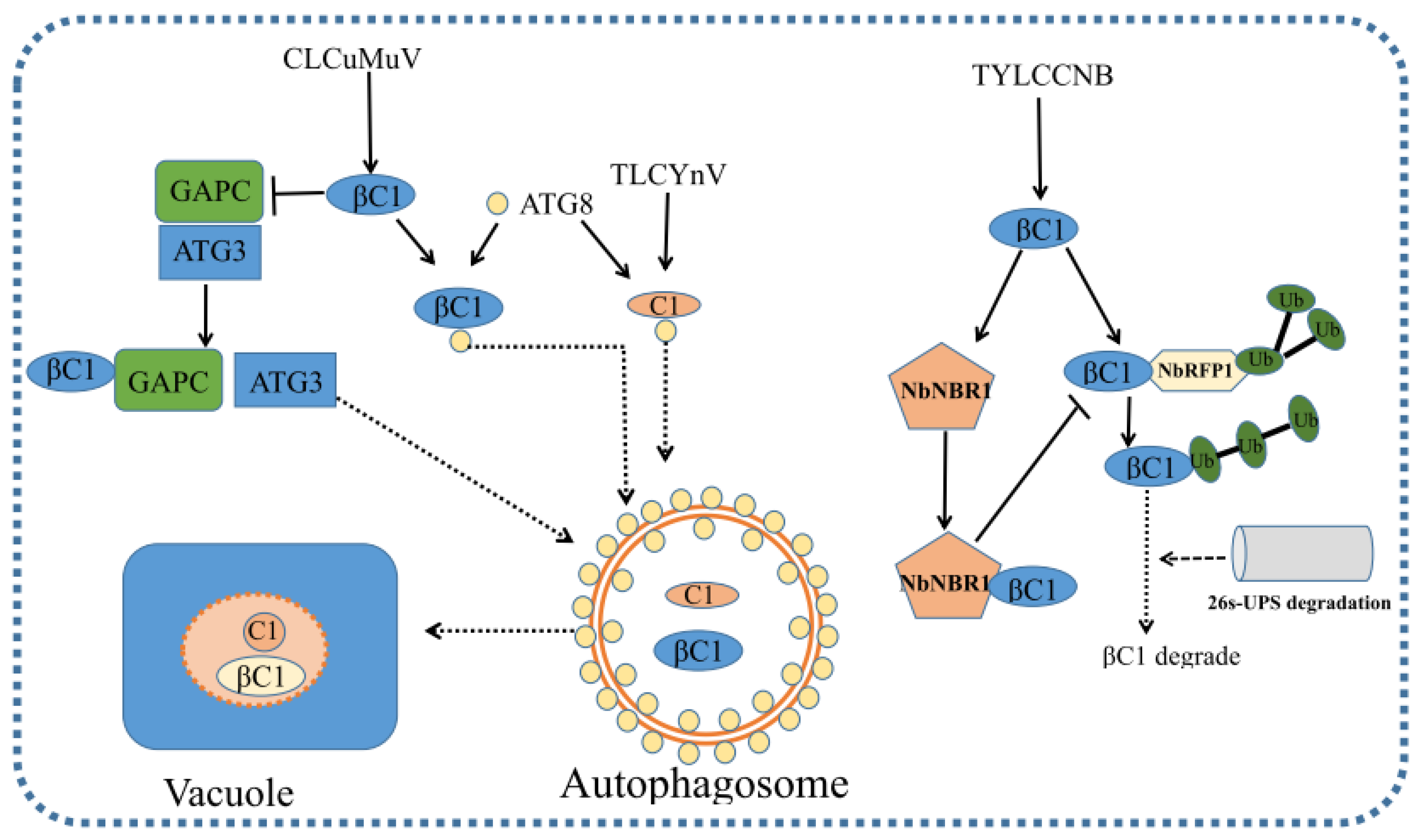
The Geminiviridae family is one of the largest families of DNA viruses infecting numerous crops and weeds (dicots and monocots). It also causes severe yield losses worldwide. Plants pose multilayered and comprehensive antiviral strategies to manipulate virus, such as RNA silencing, plant signaling, hormone signaling, protein degradation and so on. To make the microenvironment suitable for geminivirus infection, geminiviruses encode various proteins to interfere with host antiviral mechanisms, including the manipulation of the cell cycle, DNA replication, intra- and inter-cellular movement and the suppression of gene silencing and other antiviral defenses, such as the response to defense-related hormones. Viruses also usurp host-protein-degradation processes in order to reduce host defense, reduce cell death and promote viral replication. Geminiviruses co-evolve in long term plant–virus infection, and defense and counter-defense mechanisms in plant–geminivirus interactions are perplexing. Recently, a CRISPR/Cas9 system has emerged as a great tool to integrate geminivirus resistance [141,142]. Cas9-mediated immunity in tobacco enhanced resistance to cotton leaf curl disease (CLCuD) and African cassava mosaic virus (ACMV) [143,144]. In addition, the CRISPR/Cas9 system enhances resistance to TYLCV in tomato [145]. Plants could also possess other defense pathways against geminivirus, in addition to the defense pathways described above [146,147,148,149,150]. For examples, plants CMD1, CMD2 and CMD3 confer phenotypic disease tolerance to geminivirus with unknown mechanisms [151,152,153,154]. The Ty-5 gene encodes the mRNA surveillance factor Pelota, and its loss-of-function allele impairs viral translation, leading to viral tolerance, indicating that the Pelota gene is a susceptibility gene for multiple geminiviruses, including TYLCV [155,156]. In addition, plants recognize Ca2+ flux triggered by injuries to plant cells as the common molecular pattern of different viral infections to prime antiviral RNAi defense [38]. Recently, Yang et al. (2021) found that vacuolar acidification is required for plant antiviral defense against a positive-strand RNA virus–barley stripe mosaic virus (BSMV). Meanwhile, BSMV replicase γa inhibits the acidification of vacuolar lumen and suppresses autophagic degradation to promote viral infection by interacting with the V-ATPase catalytic subunit [157]. Many plant RNA viruses have evolved to suppress or manipulate host autophagy to promote viral infection [158,159]. Whether geminiviruses suppress or manipulate autophagy and how its underlying mechanisms work need to be of further concern. The identification of new host factors involved in virus infection that interact directly or indirectly with virus-encoded proteins is essential for the establishment of novel antiviral strategies.
Author Contributions
A.I. and J.Z. conceived the manuscript. All authors contributed to different sections and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31920103013, 32130086) and the National Key R&D Program of China (2021YFD1400400 and 2022YFD1400800).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.S.; Martin, G.B. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011, 16, 132–140. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Dangl, J.L. Arabidopsis and the plant immune system. Plant J. 2010, 61, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Negrete, E.A.; Carrillo-Tripp, J.; Rivera-Bustamante, R.F. RNA silencing against geminivirus: Complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J. Virol. 2009, 83, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Veluthambi, K.; Sunitha, S. Targets and Mechanisms of Geminivirus Silencing Suppressor Protein AC2. Front. Microbiol. 2021, 12, 645419. [Google Scholar] [CrossRef]
- Chen, K.; Khatabi, B.; Fondong, V.N. The AC4 Protein of a Cassava Geminivirus Is Required for Virus Infection. Mol. Plant-Microbe Interact. 2019, 32, 865–875. [Google Scholar] [CrossRef]
- Li, F.; Xu, X.; Huang, C.; Gu, Z.; Cao, L.; Hu, T.; Ding, M.; Li, Z.; Zhou, X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef]
- Li, F.; Yang, X.; Bisaro, D.M.; Zhou, X. The βC1 Protein of Geminivirus-Betasatellite Complexes: A Target and Repressor of Host Defenses. Mol. Plant 2018, 11, 1424–1426. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Gong, Q.; Ismayil, A.; Yuan, Y.; Lian, B.; Jia, Q.; Han, M.; Deng, H.; Hong, Y.; et al. Geminiviral V2 Protein Suppresses Transcriptional Gene Silencing through Interaction with AGO4. J. Virol. 2019, 93, e01675-18. [Google Scholar] [CrossRef]
- Gupta, N.; Reddy, K.; Bhattacharyya, D.; Chakraborty, S. Plant responses to geminivirus infection: Guardians of the plant immunity. Virol. J. 2021, 18, 143. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Llave, C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Raja, P.; Wolf, J.N.; Bisaro, D.M. RNA silencing directed against geminiviruses: Post-transcriptional and epigenetic components. Biochim. Biophys. Acta 2010, 1799, 337–351. [Google Scholar] [CrossRef]
- Huang, C.J.; Zhang, T.; Li, F.F.; Zhang, X.Y.; Zhou, X.P. Development and application of an efficient virus-induced gene silencing system in Nicotiana tabacum using geminivirus alphasatellite. J. Zhejiang Univ. Sci. B 2011, 12, 83–92. [Google Scholar] [CrossRef]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef]
- Ismayil, A.; Haxim, Y.; Wang, Y.; Li, H.; Qian, L.; Han, T.; Chen, T.; Jia, Q.; Yihao Liu, A.; Zhu, S.; et al. Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 2018, 14, e1007282. [Google Scholar] [CrossRef]
- Mei, Y.; Wang, Y.; Li, F.; Zhou, X. The C4 protein encoded by tomato leaf curl Yunnan virus reverses transcriptional gene silencing by interacting with NbDRM2 and impairing its DNA-binding ability. PLoS Pathog. 2020, 16, e1008829. [Google Scholar] [CrossRef] [PubMed]
- Raja, P.; Jackel, J.N.; Li, S.; Heard, I.M.; Bisaro, D.M. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J. Virol. 2014, 88, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163, Erratum in Nat. Plants 2016, 3, 16211. [Google Scholar] [CrossRef]
- Matzke, M.A.; Kanno, T.; Matzke, A.J. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef]
- Wendte, J.M.; Pikaard, C.S. The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 140–148. [Google Scholar] [CrossRef]
- Blevins, T.; Podicheti, R.; Mishra, V.; Marasco, M.; Wang, J.; Rusch, D.; Tang, H.; Pikaard, C.S. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 2015, 4, e09591. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, P.; Vanitharani, R.; Pita, J.; Fauquet, C.M. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 2004, 78, 7465–7477, Erratum in J. Virol. 2006, 80, 1064. [Google Scholar] [CrossRef]
- Voorburg, C.M.; Bai, Y.; Kormelink, R. Small RNA Profiling of Susceptible and Resistant Ty-1 Encoding Tomato Plants Upon Tomato Yellow Leaf Curl Virus Infection. Front. Plant Sci. 2021, 12, 757165. [Google Scholar] [CrossRef]
- Sun, Y.W.; Tee, C.S.; Ma, Y.H.; Wang, G.; Yao, X.M.; Ye, J. Attenuation of Histone Methyltransferase KRYPTONITE-mediated transcriptional gene silencing by Geminivirus. Sci. Rep. 2015, 5, 16476. [Google Scholar] [CrossRef]
- Castillo-González, C.; Liu, X.; Huang, C.; Zhao, C.; Ma, Z.; Hu, T.; Sun, F.; Zhou, Y.; Zhou, X.; Wang, X.J.; et al. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 2015, 4, e06671. [Google Scholar] [CrossRef]
- Coursey, T.; Milutinovic, M.; Regedanz, E.; Brkljacic, J.; Bisaro, D.M. Arabidopsis Histone Reader EMSY-LIKE 1 Binds H3K36 and Suppresses Geminivirus Infection. J. Virol. 2018, 92, e00219-18. [Google Scholar] [CrossRef]
- Betti, F.; Ladera-Carmona, M.J.; Weits, D.A.; Ferri, G.; Iacopino, S.; Novi, G.; Svezia, B.; Kunkowska, A.B.; Santaniello, A.; Piaggesi, A.; et al. Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 2021, 7, 1379–1388. [Google Scholar] [CrossRef]
- Aregger, M.; Borah, B.K.; Seguin, J.; Rajeswaran, R.; Gubaeva, E.G.; Zvereva, A.S.; Windels, D.; Vazquez, F.; Blevins, T.; Farinelli, L.; et al. Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 2012, 8, e1002941. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.M.; Ferreira, M.A.; Raimundo, G.A.S.; Fontes, E.P.B. Geminiviral Triggers and Suppressors of Plant Antiviral Immunity. Microorganisms 2021, 9, 775. [Google Scholar] [CrossRef]
- Ali, S.; Ahmad Nasir, I.; Rafiq, M.; Javed Butt, S.; Ihsan, F.; Qayyum Rao, A.; Husnain, T. Sugarcane Mosaic Virus-Based Gene Silencing in Nicotiana benthamiana. Iran. J. Biotechnol. 2017, 15, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef]
- Muangsan, N.; Beclin, C.; Vaucheret, H.; Robertson, D. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 2004, 38, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gong, Q.; Wu, Y.; Huang, F.; Ismayil, A.; Zhang, D.; Li, H.; Gu, H.; Ludman, M.; Fátyol, K.; et al. A calmodulin-binding transcription factor links calcium signaling to antiviral RNAi defense in plants. Cell Host Microbe 2021, 29, 1393–1406.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Zhou, X. SGS3 Cooperates with RDR6 in Triggering Geminivirus-Induced Gene Silencing and in Suppressing Geminivirus Infection in Nicotiana benthamiana. Viruses 2017, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.S.; Vazquez, F.; Robertson, D.; Meins FJr Hohn, T.; Pooggin, M.M. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef]
- Luna, A.P.; Morilla, G.; Voinnet, O.; Bejarano, E.R. Functional analysis of gene-silencing suppressors from tomato yellow leaf curl disease viruses. Mol. Plant-Microbe Interact. 2012, 25, 1294–1306. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005, 6, 206–220. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Sahu, P.P.; Prasad, M.; Praveen, S.; Pappu, H.R. Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race. Viruses 2017, 9, 256. [Google Scholar] [CrossRef]
- Bisaro, D.M. Silencing suppression by geminivirus proteins. Virology 2006, 344, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dang, M.; Hou, H.; Mei, Y.; Qian, Y.; Zhou, X. Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 2014, 95, 2082–2088. [Google Scholar] [CrossRef]
- Rodríguez-Negrete, E.; Lozano-Durán, R.; Piedra-Aguilera, A.; Cruzado, L.; Bejarano, E.R.; Castillo, A.G. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene silencing. New Phytol. 2013, 199, 464–475. [Google Scholar] [CrossRef]
- Wang, H.; Buckley, K.J.; Yang, X.; Buchmann, R.C.; Bisaro, D.M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005, 79, 7410–7418. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, L.; Shung, C.Y.; Sunter, G.; Bisaro, D.M. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 2003, 15, 3020–3032. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef]
- Vinutha, T.; Kumar, G.; Garg, V.; Canto, T.; Palukaitis, P.; Ramesh, S.V.; Praveen, S. Tomato geminivirus encoded RNAi suppressor protein, AC4 interacts with host AGO4 and precludes viral DNA methylation. Gene 2018, 678, 184–195. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants. J. Virol. 2018, 92, e00036-18. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Liu, C.; Qi, Y.; Zhou, X. Geminiviruses employ host DNA glycosylases to subvert DNA methylation-mediated defense. Nat. Commun. 2022, 13, 575. [Google Scholar] [CrossRef]
- Yong Chung, H.; Lacatus, G.; Sunter, G. Geminivirus AL2 protein induces expression of, and interacts with, a calmodulin-like gene, an endogenous regulator of gene silencing. Virology 2014, 460–461, 108–118. [Google Scholar] [CrossRef]
- Trinks, D.; Rajeswaran, R.; Shivaprasad, P.V.; Akbergenov, R.; Oakeley, E.J.; Veluthambi, K.; Hohn, T.; Pooggin, M.M. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005, 79, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Mishra, S.K.; Rahman, J.; Taneja, J.; Sundaresan, G.; Mishra, N.S.; Mukherjee, S.K. Mungbean yellow mosaic Indian virus encoded AC2 protein suppresses RNA silencing by inhibiting Arabidopsis RDR6 and AGO1 activities. Virology 2015, 486, 158–172. [Google Scholar] [CrossRef]
- Sunitha, S.; Shanmugapriya, G.; Balamani, V.; Veluthambi, K. Mungbean yellow mosaic virus (MYMV) AC4 suppresses post-transcriptional gene silencing and an AC4 hairpin RNA gene reduces MYMV DNA accumulation in transgenic tobacco. Virus Genes 2013, 46, 496–504. [Google Scholar] [CrossRef]
- Luna, A.P.; Rodríguez-Negrete, E.A.; Morilla, G.; Wang, L.; Lozano-Durán, R.; Castillo, A.G.; Bejarano, E.R. V2 from a curtovirus is a suppressor of post-transcriptional gene silencing. J. Gen. Virol. 2017, 98, 2607–2614. [Google Scholar] [CrossRef] [PubMed]
- Glick, E.; Zrachya, A.; Levy, Y.; Mett, A.; Gidoni, D.; Belausov, E.; Citovsky, V.; Gafni, Y. Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. USA 2008, 105, 157–161, Erratum in Proc. Natl. Acad. Sci. USA 2009, 106, 4571. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, J.; Xu, Y.; Wu, J. V2 protein encoded by Tomato yellow leaf curl China virus is an RNA silencing suppressor. Virus Res. 2012, 163, 51–58. [Google Scholar] [CrossRef]
- Amin, I.; Hussain, K.; Akbergenov, R.; Yadav, J.S.; Qazi, J.; Mansoor, S.; Hohn, T.; Fauquet, C.M.; Briddon, R.W. Suppressors of RNA silencing encoded by the components of the cotton leaf curl begomovirus-betasatellite complex. Mol. Plant-Microbe Interact. 2011, 24, 973–983. [Google Scholar] [CrossRef]
- Eini, O. A Betasatellite-Encoded Protein Regulates Key Components of Gene Silencing System in Plants. Mol. Biol. 2017, 51, 656–663. (In Russian) [Google Scholar] [CrossRef]
- Li, F.; Huang, C.; Li, Z.; Zhou, X. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 2014, 10, e1003921. [Google Scholar] [CrossRef]
- Kamal, H.; Minhas, F.A.; Tripathi, D.; Abbasi, W.A.; Hamza, M.; Mustafa, R.; Khan, M.Z.; Mansoor, S.; Pappu, H.R.; Amin, I. βC1, pathogenicity determinant encoded by Cotton leaf curl Multan betasatellite, interacts with calmodulin-like protein 11 (Gh-CML11) in Gossypium hirsutum. PLoS ONE 2019, 14, e0225876. [Google Scholar] [CrossRef]
- Gong, P.; Zhao, S.; Liu, H.; Chang, Z.; Li, F.; Zhou, X. Tomato yellow leaf curl virus V3 protein traffics along microfilaments to plasmodesmata to promote virus cell-to-cell movement. Sci. China Life Sci. 2022, 65, 1046–1049. [Google Scholar] [CrossRef]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Durán, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef] [PubMed]
- Breiden, M.; Simon, R. Q&A: How does peptide signaling direct plant development? BMC Biol. 2016, 14, 58. [Google Scholar] [CrossRef]
- Santos, A.A.; Carvalho, C.M.; Florentino, L.H.; Ramos, H.J.; Fontes, E.P. Conserved threonine residues within the A-loop of the receptor NIK differentially regulate the kinase function required for antiviral signaling. PLoS ONE 2009, 4, e5781. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Liu, Z.; Song, F.; Xie, Q.; Hanley-Bowdoin, L.; Zhou, X. Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol. 2011, 157, 1394–1406. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Huang, C.; He, Y.; Castillo-González, C.; Gui, X.; Wang, Y.; Zhang, X.; Zhou, X. βC1 protein encoded in geminivirus satellite concertedly targets MKK2 and MPK4 to counter host defense. PLoS Pathog. 2019, 15, e1007728. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Hanley-Bowdoin, L. SnRK1: A versatile plant protein kinase that limits geminivirus infection. Curr. Opin. Virol. 2021, 47, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Dallas, M.B.; Goshe, M.B.; Hanley-Bowdoin, L. SnRK1 phosphorylation of AL2 delays Cabbage leaf curl virus infection in Arabidopsis. J. Virol. 2014, 88, 10598–10612. [Google Scholar] [CrossRef]
- Kong, L.J.; Hanley-Bowdoin, L. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 2002, 14, 1817–1832. [Google Scholar] [CrossRef]
- Shen, W.; Hanley-Bowdoin, L. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 2006, 142, 1642–1655. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Bobay, B.G.; Greeley, L.A.; Reyes, M.I.; Rajabu, C.A.; Blackburn, R.K.; Dallas, M.B.; Goshe, M.B.; Ascencio-Ibáñez, J.T.; Hanley-Bowdoin, L. Sucrose Nonfermenting 1-Related Protein Kinase 1 Phosphorylates a Geminivirus Rep Protein to Impair Viral Replication and Infection. Plant Physiol. 2018, 178, 372–389. [Google Scholar] [CrossRef]
- Shen, Q.; Bao, M.; Zhou, X. A plant kinase plays roles in defense response against geminivirus by phosphorylation of a viral pathogenesis protein. Plant Signal Behav. 2012, 7, 888–892. [Google Scholar] [CrossRef]
- Kamal, H.; Minhas, F.A.; Farooq, M.; Tripathi, D.; Hamza, M.; Mustafa, R.; Khan, M.Z.; Mansoor, S.; Pappu, H.R.; Amin, I. In silico Prediction and Validations of Domains Involved in Gossypium hirsutum SnRK1 Protein Interaction with Cotton Leaf Curl Multan Betasatellite Encoded βC1. Front. Plant Sci. 2019, 10, 656. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Cao, L.; Wang, Y.; Xie, Y.; Zhou, X. Mimic Phosphorylation of a βC1 Protein Encoded by TYLCCNB Impairs Its Functions as a Viral Suppressor of RNA Silencing and a Symptom Determinant. J. Virol. 2017, 91, e00300-17. [Google Scholar] [CrossRef]
- Hao, L.; Wang, H.; Sunter, G.; Bisaro, D.M. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 2003, 15, 1034–1048. [Google Scholar] [CrossRef]
- Mohannath, G.; Jackel, J.N.; Lee, Y.H.; Buchmann, R.C.; Wang, H.; Patil, V.; Adams, A.K.; Bisaro, D.M. A complex containing SNF1-related kinase (SnRK1) and adenosine kinase in Arabidopsis. PLoS ONE 2014, 9, e87592. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.N.; Li, S.; Mohannath, G.; Bisaro, D.M. Phosphorylation of Arabidopsis eIF4E and eIFiso4E by SnRK1 inhibits translation. FEBS J. 2019, 286, 3778–3796. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Dey, N.; Chaudhuri, S.; Pal, A. Molecular and biochemical characterization of a Vigna mungo MAP kinase associated with Mungbean Yellow Mosaic India Virus infection and deciphering its role in restricting the virus multiplication. Plant Sci. 2017, 262, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qin, L.; Zhao, J.; Muhammad, T.; Cao, H.; Li, H.; Zhang, Y.; Liang, Y. SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum). PLoS ONE 2017, 12, e0172466. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Mei, Y.; Wang, Y.; Hu, T.; He, Z.; Zhou, X. The C4 protein encoded by Tomato leaf curl Yunnan virus interferes with mitogen-activated protein kinase cascade-related defense responses through inhibiting the dissociation of the ERECTA/BKI1 complex. New Phytol. 2021, 231, 747–762. [Google Scholar] [CrossRef]
- Yang, X.; Deng, F.; Ramonell, K.M. Receptor-like kinases and receptor-like proteins: Keys to pathogen recognition and defense signaling in plant innate immunity. Front. Biol. 2012, 7, 155–166. [Google Scholar] [CrossRef]
- Fontes, E.P.; Santos, A.A.; Luz, D.F.; Waclawovsky, A.J.; Chory, J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004, 18, 2545–2556. [Google Scholar] [CrossRef]
- Mariano, A.C.; Andrade, M.O.; Santos, A.A.; Carolino, S.M.; Oliveira, M.L.; Baracat-Pereira, M.C.; Brommonshenkel, S.H.; Fontes, E.P. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 2004, 318, 24–31. [Google Scholar] [CrossRef]
- Brustolini, O.J.B.; Machado, J.P.B.; Condori-Apfata, J.A.; Coco, D.; Deguchi, M.; Loriato, V.A.P.; Pereira, W.A.; Alfenas-Zerbini, P.; Zerbini, F.M.; Inoue-Nagata, A.K.; et al. Sustained NIK-mediated antiviral signalling confers broad-spectrum tolerance to begomoviruses in cultivated plants. Plant Biotechnol. J. 2015, 13, 1300–1311. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; Machado, J.P.; Lopes, K.V.; Nascimento, K.J.; Pereira, W.A.; Brustolini, O.J.; Reis, P.A.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef]
- Martins, L.G.C.; Raimundo, G.A.S.; Ribeiro, N.G.A.; Silva, J.C.F.; Euclydes, N.C.; Loriato, V.A.P.; Duarte, C.E.M.; Fontes, E.P.B. A Begomovirus Nuclear Shuttle Protein-Interacting Immune Hub: Hijacking Host Transport Activities and Suppressing Incompatible Functions. Front. Plant Sci. 2020, 11, 398. [Google Scholar] [CrossRef] [PubMed]
- Garnelo Gómez, B.; Zhang, D.; Rosas-Díaz, T.; Wei, Y.; Macho, A.P.; Lozano-Durán, R. The C4 Protein from Tomato Yellow Leaf Curl Virus Can Broadly Interact with Plant Receptor-Like Kinases. Viruses 2019, 11, 1009. [Google Scholar] [CrossRef]
- Fan, P.; Aguilar, E.; Bradai, M.; Xue, H.; Wang, H.; Rosas-Diaz, T.; Tang, W.; Wolf, S.; Zhang, H.; Xu, L.; et al. The receptor-like kinases BAM1 and BAM2 are required for root xylem patterning. Proc. Natl. Acad. Sci. USA 2021, 118, e2022547118. [Google Scholar] [CrossRef]
- Li, H.; Zeng, R.; Chen, Z.; Liu, X.; Cao, Z.; Xie, Q.; Yang, C.; Lai, J. S-acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J. Exp. Bot. 2018, 69, 4459–4468. [Google Scholar] [CrossRef]
- Li, Z.; Du, Z.; Tang, Y.; She, X.; Wang, X.; Zhu, Y.; Yu, L.; Lan, G.; He, Z. C4, the Pathogenic Determinant of Tomato Leaf Curl Guangdong Virus, May Suppress Post-transcriptional Gene Silencing by Interacting with BAM1 Protein. Front. Microbiol. 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Deom, C.M.; Mills-Lujan, K. Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signal Behav. 2015, 10, e1109758, Erratum in PLoS ONE 2015, 10, e0122356. https://doi.org/10.1371/journal.pone.0122356. [Google Scholar] [CrossRef] [PubMed]
- Zolkiewicz, K.; Gruszka, D. Glycogen synthase kinases in model and crop plants—From negative regulators of brassinosteroid signaling to multifaceted hubs of various signaling pathways and modulators of plant reproduction and yield. Front. Plant Sci. 2022, 13, 939487. [Google Scholar] [CrossRef]
- Dogra, S.C.; Eini, O.; Rezaian, M.A.; Randles, J.W. A novel shaggy-like kinase interacts with the Tomato leaf curl virus pathogenicity determinant C4 protein. Plant Mol. Biol. 2009, 71, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Mills-Lujan, K.; Andrews, D.L.; Chou, C.W.; Deom, C.M. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 2015, 10, e0122356. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Eini, O.; Koolivand, D.; Varrelmann, M. The Rep and C1 of Beet curly top Iran virus represent pathogenicity factors and induce hypersensitive response in Nicotiana benthamiana plants. Virus Genes 2022, 58, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Matić, S.; Pegoraro, M.; Noris, E. The C2 protein of tomato yellow leaf curl Sardinia virus acts as a pathogenicity determinant and a 16-amino acid domain is responsible for inducing a hypersensitive response in plants. Virus Res. 2016, 215, 12–19. [Google Scholar] [CrossRef]
- Mubin, M.; Amin, I.; Amrao, L.; Briddon, R.W.; Mansoor, S. The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 2010, 11, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Ma, Z.; Wang, Y.; Zhou, X. Geminivirus C4 antagonizes the HIR1-mediated hypersensitive response by inhibiting the HIR1 self-interaction and promoting degradation of the protein. New Phytol. 2020, 225, 1311–1326. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.A. The NLR Protein Encoded by the Resistance Gene Ty-2 Is Triggered by the Replication-Associated Protein Rep/C1 of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Maiti, S.; Paul, S.; Pal, A. Isolation, characterization, and structure analysis of a non-TIR-NBS-LRR encoding candidate gene from MYMIV-resistant Vigna mungo. Mol. Biotechnol. 2012, 52, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef]
- Li, F.; Zhang, M.; Zhang, C.; Zhou, X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020, 225, 1746–1761. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Haxim, Y.; Wang, Y.; Li, J.; Han, L.; Wang, Y.; Zheng, X.; Wei, X.; Nagalakshmi, U.; et al. Cotton leaf curl Multan virus βC1 Protein Induces Autophagy by Disrupting the Interaction of Autophagy-Related Protein 3 with Glyceraldehyde-3-Phosphate Dehydrogenases. Plant Cell. 2020, 32, 1124–1135. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Liu, Y. Role of autophagy during plant-virus interactions. Semin. Cell Dev. Biol. 2020, 101, 36–40. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, M.; Gong, P.; Li, F.; Zhou, X. Selective autophagic receptor NbNBR1 prevents NbRFP1-mediated UPS-dependent degradation of βC1 to promote geminivirus infection. PLoS Pathog. 2021, 17, e1009956. [Google Scholar] [CrossRef]
- Li, F.; Zhao, N.; Li, Z.; Xu, X.; Wang, Y.; Yang, X.; Liu, S.S.; Wang, A.; Zhou, X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017, 13, e1006213. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Wang, J.; Han, P.; Miao, S.; Zheng, X.; Han, M.; Shen, X.; Li, H.; Wu, M.; et al. Plant UVRAG interacts with ATG14 to regulate autophagosome maturation and geminivirus infection. New Phytol. 2022, 236, 1358–1374. [Google Scholar] [CrossRef] [PubMed]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Rosas-Díaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef]
- Eini, O.; Dogra, S.; Selth, L.A.; Dry, I.B.; Randles, J.W.; Rezaian, M.A. Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA beta satellite. Mol. Plant-Microbe Interact. 2009, 22, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Chen, H.; Teng, K.; Zhao, Q.; Zhang, Z.; Li, Y.; Liang, L.; Xia, R.; Wu, Y.; Guo, H.; et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2009, 57, 905–917. [Google Scholar] [CrossRef]
- Czosnek, H.; Eybishtz, A.; Sade, D.; Gorovits, R.; Sobol, I.; Bejarano, E.; Rosas-Díaz, T.; Lozano-Durán, R. Discovering host genes involved in the infection by the Tomato Yellow Leaf Curl Virus complex and in the establishment of resistance to the virus using Tobacco Rattle Virus-based post transcriptional gene silencing. Viruses 2013, 5, 998–1022. [Google Scholar] [CrossRef]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co- option of SCF-mediated ubiquitination. Plant Signal Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Rosas-Díaz, T.; Luna, A.P.; Bejarano, E.R. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE 2011, 6, e22383. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Hu, T.; Bao, M.; Cao, L.; Zhang, H.; Song, F.; Xie, Q.; Zhou, X. Tobacco RING E3 Ligase NtRFP1 Mediates Ubiquitination and Proteasomal Degradation of a Geminivirus-Encoded βC1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, N.; Xie, K.; Dai, Y.; Han, S.; Zhao, X.; Qian, L.; Wang, Y.; Zhao, J.; Gorovits, R.; et al. CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog. 2016, 12, e1005668. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.H. betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Ghosh, D.; Chakraborty, S. Molecular interplay between phytohormones and geminiviruses: A saga of a never-ending arms race. J. Exp. Bot. 2021, 72, 2903–2917. [Google Scholar] [CrossRef]
- Wang, P.; Sun, S.; Liu, K.; Peng, R.; Li, N.; Hu, B.; Wang, L.; Wang, H.; Afzal, A.J.; Geng, X. Physiological and transcriptomic analyses revealed gene networks involved in heightened resistance against tomato yellow leaf curl virus infection in salicylic acid and jasmonic acid treated tomato plants. Front. Microbiol. 2022, 13, 970139. [Google Scholar] [CrossRef]
- Li, K.; Wu, G.; Li, M.; Ma, M.; Du, J.; Sun, M.; Sun, X.; Qing, L. Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus. Virol. J. 2018, 15, 138. [Google Scholar] [CrossRef]
- Li, T.; Huang, Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. Salicylic acid-induced differential resistance to the Tomato yellow leaf curl virus among resistant and susceptible tomato cultivars. BMC Plant Biol. 2019, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.; Van Wees, S.C. Ethylene: Traffic Controller on Hormonal Crossroads to Defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef] [PubMed]
- Baliji, S.; Lacatus, G.; Sunter, G. The interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 2010, 402, 238–247. [Google Scholar] [CrossRef]
- Wu, X.; Ye, J. Manipulation of Jasmonate Signaling by Plant Viruses and Their Insect Vectors. Viruses 2020, 12, 148. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, X.; Yao, X.; Zhang, P.; Fang, R.; Ye, J. A 7-Amino-Acid Motif of Rep Protein Essential for Virulence Is Critical for Triggering Host Defense Against Sri Lankan Cassava Mosaic Virus. Mol. Plant-Microbe Interact. 2020, 33, 78–86. [Google Scholar] [CrossRef]
- Miozzi, L.; Napoli, C.; Sardo, L.; Accotto, G.P. Transcriptomics of the interaction between the monopartite phloem-limited geminivirus tomato yellow leaf curl Sardinia virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS ONE 2014, 9, e89951. [Google Scholar] [CrossRef]
- Rosas-Díaz, T.; Macho, A.P.; Beuzón, C.R.; Lozano-Durán, R.; Bejarano, E.R. The C2 Protein from the Geminivirus Tomato Yellow Leaf Curl Sardinia Virus Decreases Sensitivity to Jasmonates and Suppresses Jasmonate-Mediated Defences. Plants 2016, 5, 8. [Google Scholar] [CrossRef]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef] [PubMed]
- Vinutha, T.; Vanchinathan, S.; Bansal, N.; Kumar, G.; Permar, V.; Watts, A.; Ramesh, S.V.; Praveen, S. Tomato auxin biosynthesis/signaling is reprogrammed by the geminivirus to enhance its pathogenicity. Planta 2020, 252, 51. [Google Scholar] [CrossRef]
- Gupta, K.; Rishishwar, R.; Dasgupta, I. The interplay of plant hormonal pathways and geminiviral proteins: Partners in disease development. Virus Genes 2022, 58, 1–14. [Google Scholar] [CrossRef]
- Loriato, V.A.P.; Martins, L.G.C.; Euclydes, N.C.; Reis, P.A.B.; Duarte, C.E.M.; Fontes, E.P.B. Engineering resistance against geminiviruses: A review of suppressed natural defenses and the use of RNAi and the CRISPR/Cas system. Plant Sci. 2020, 292, 110410. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Han, T.; Liu, Y. Use of Geminivirus for Delivery of CRISPR/Cas9 Components to Tobacco by Agro-infiltration. Bio-Protoc. 2017, 7, e2209. [Google Scholar] [CrossRef]
- Binyameen, B.; Khan, Z.; Khan, S.H.; Ahmad, A.; Munawar, N.; Mubarik, M.S.; Riaz, H.; Ali, Z.; Khan, A.A.; Qusmani, A.T.; et al. Using Multiplexed CRISPR/Cas9 for Suppression of Cotton Leaf Curl Virus. Int. J. Mol. Sci. 2021, 22, 12543. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cao, B.; Zhang, H.; Fan, Z.; Zhou, X.; Li, F. Geminivirus satellite-encoded βC1 activates UPR, induces bZIP60 nuclear export, and manipulates the expression of bZIP60 downstream genes to benefit virus infection. Sci. China Life Sci. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jimenez-Gongora, T.; Krenz, B.; Lozano-Duran, R. Chloroplast clustering around the nucleus is a general response to pathogen perception in Nicotiana benthamiana. Mol. Plant Pathol. 2019, 20, 1298–1306. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; Ponnusamy, K.; Chakraborty, S. A geminivirus betasatellite encoded βC1 protein interacts with PsbP and subverts PsbP-mediated antiviral defence in plants. Mol. Plant Pathol. 2019, 20, 943–960. [Google Scholar] [CrossRef]
- Zhao, W.; Zhou, Y.; Zhou, X.; Wang, X.; Ji, Y. Host GRXC6 restricts Tomato yellow leaf curl virus infection by inhibiting the nuclear export of the V2 protein. PLoS Pathog. 2021, 17, e1009844. [Google Scholar] [CrossRef]
- Medina-Puche, L.; Tan, H.; Dogra, V.; Wu, M.; Rosas-Diaz, T.; Wang, L.; Ding, X.; Zhang, D.; Fu, X.; Kim, C.; et al. A Defense Pathway Linking Plasma Membrane and Chloroplasts and Co-opted by Pathogens. Cell 2020, 182, 1109–1124.e25. [Google Scholar] [CrossRef]
- Prasanna, H.; Sinha, D.P.; Rai, G.; Krishna, R.; Pratap Kashyap, S.; Singh, N.; Singh, M.; Malathi, V.G. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 2014, 64, 256–264. [Google Scholar] [CrossRef]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Rai, N.; Lima, J.; Singh, M.; Singh, S.; Kumar, S. Genetic and molecular characterisations of Tomato leaf curl virus resistance in tomato (Solanum lycopersicum L.). J. Hortic. Sci. Biotechnol. 2015, 90, 503–510. [Google Scholar] [CrossRef]
- Akano, O.; Dixon, O.; Mba, C.; Barrera, E.; Fregene, M. Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 2002, 105, 521–525. [Google Scholar] [CrossRef]
- Lapidot, M.; Karniel, U.; Gelbart, D.; Fogel, D.; Evenor, D.; Kutsher, Y.; Makhbash, Z.; Nahon, S.; Shlomo, H.; Chen, L.; et al. A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota. PLoS Genet. 2015, 11, e1005538. [Google Scholar] [CrossRef]
- Ren, Y.; Tao, X.; Li, D.; Yang, X.; Zhou, X. Ty-5 Confers Broad-Spectrum Resistance to Geminiviruses. Viruses 2022, 14, 1804. [Google Scholar] [CrossRef]
- Yang, M.; Ismayil, A.; Jiang, Z.; Wang, Y.; Zheng, X.; Yan, L.; Hong, Y.; Li, D.; Liu, Y. A viral protein disrupts vacuolar acidification to facilitate virus infection in plants. EMBO J. 2022, 41, e108713. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, Y.; Xie, X.; Yue, N.; Li, J.; Wang, X.B.; Han, C.; Yu, J.; Liu, Y.; Li, D. Barley stripe mosaic virus γb Protein Subverts Autophagy to Promote Viral Infection by Disrupting the ATG7-ATG8 Interaction. Plant Cell 2018, 30, 1582–1595. [Google Scholar] [CrossRef]
- Yang, M.; Ismayil, A.; Liu, Y. Autophagy in Plant-Virus Interactions. Annu. Rev. Virol. 2020, 7, 403–419. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).