Characterization of Human Norovirus Nonstructural Protein NS1.2 Involved in the Induction of the Filamentous Endoplasmic Reticulum, Enlarged Lipid Droplets, LC3 Recruitment, and Interaction with NTPase and NS4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Plasmids
2.3. Immunoprecipitation and Immunoblot Analysis
2.4. Immunofluorescence Staining
2.5. Quantification of Lipid Droplets
2.6. Statistical Analysis
3. Results
3.1. Analysis of the NS1.2 Domain Required for Membrane Targeting and ER Reorganization
3.2. Analysis of the NS1.2 Domain Required for the Localization of LDs and Induction of Enlarged LDs
3.3. HuNoV GII.4 NS1.2 Recruited LC3 to the ER Membrane in an Autophagy-Independent Pathway
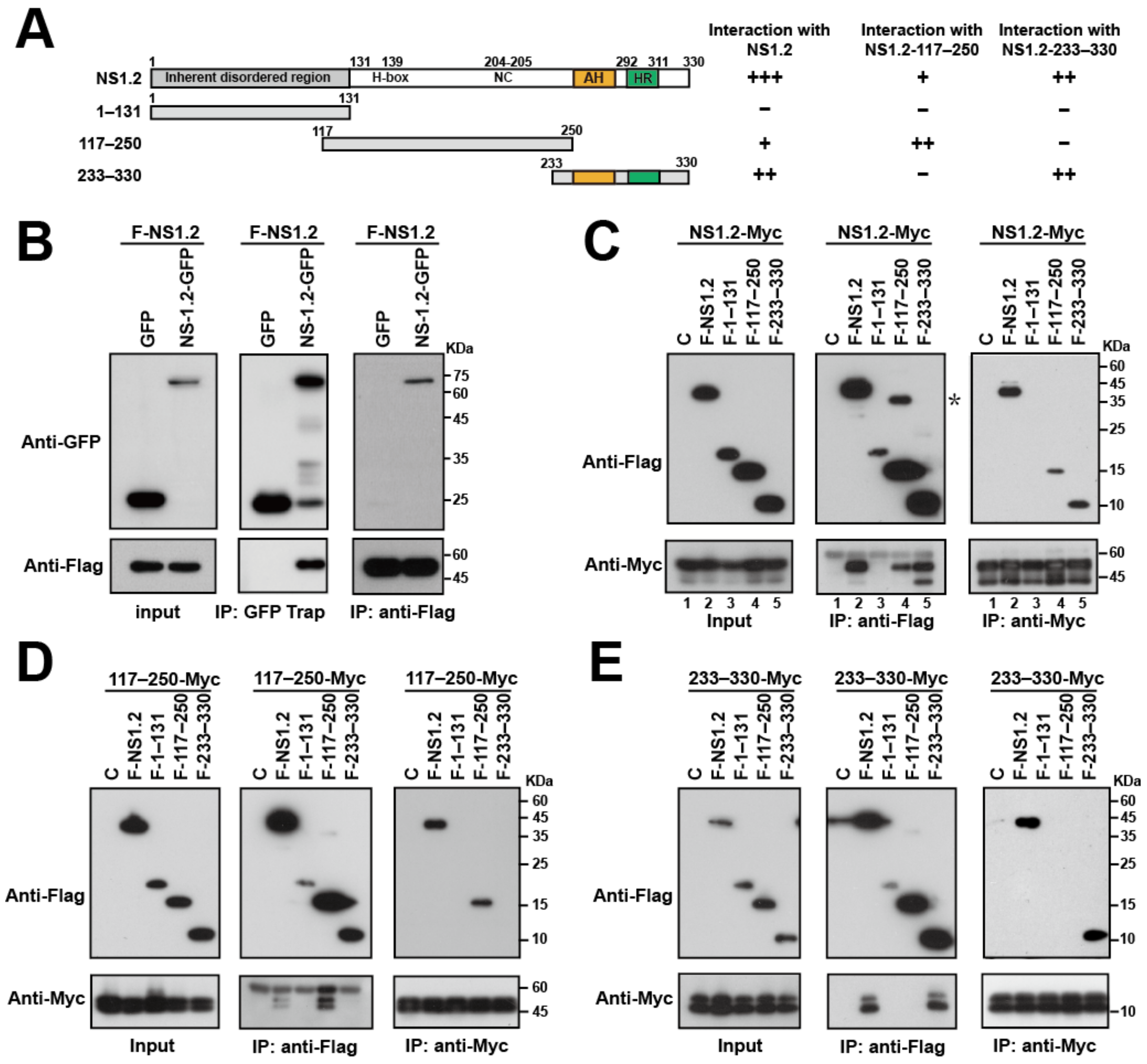
3.4. Analysis of the NS1.2 Domain Required for the Self-Interaction of NS1.2
3.5. NS1.2 Forms Heterodimer/Oligomer with NTPase and NS4
3.6. NS1.2-NTPase-NS4 Complex Expressed from a cDNA Expression System of HuNoV GII.4 Was Colocalized with LC3 and LDs
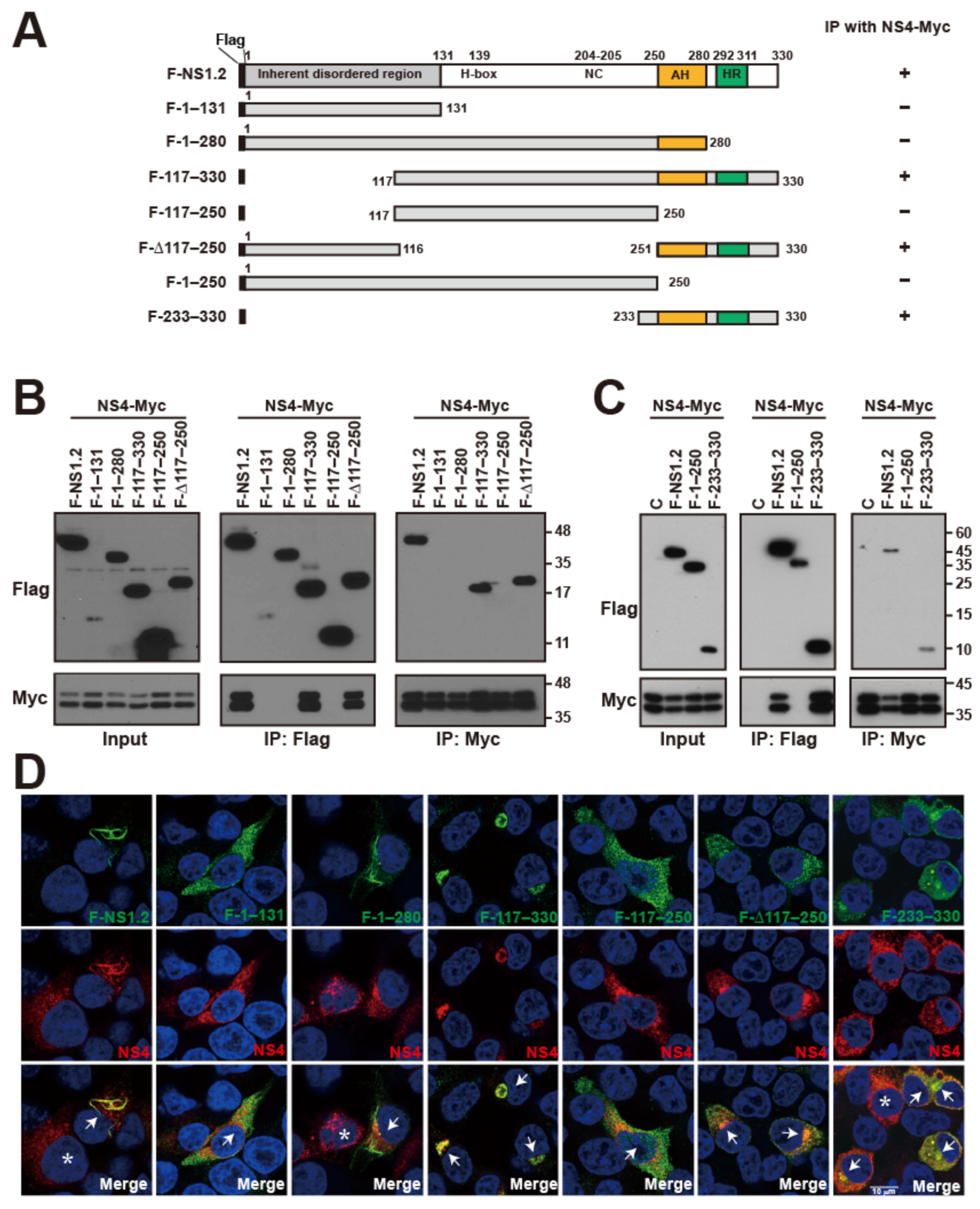
3.7. Analysis of the NS1.2 Domain Involved in the Interaction with NS4
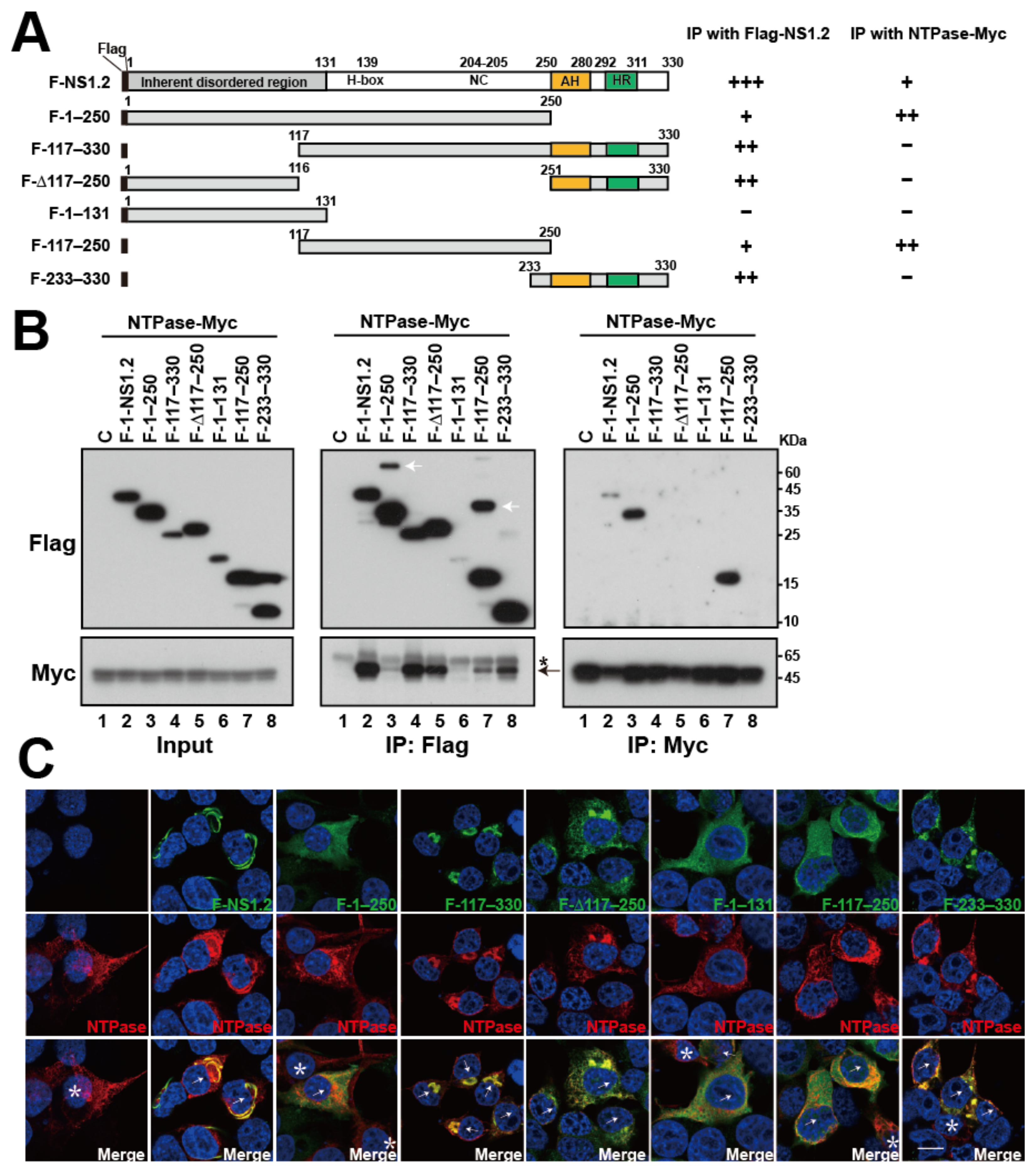
3.8. Analysis of the NS1.2 Domain Involved in the Interaction with NTPase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.J.; Eisenbart, V.G.; Etingue, A.L.; Gould, L.H.; Lopman, B.A.; Parashar, U.D. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg. Infect. Dis. 2012, 18, 1566–1573. [Google Scholar] [CrossRef]
- Ohe, M. A “blind spot” regarding the norovirus infection pathway. Tohoku J. Exp. Med. 2013, 229, 125–128. [Google Scholar]
- Bartsch, S.M.; Lopman, B.A.; Ozawa, S.; Hall, A.J.; Lee, B.Y. Global Economic Burden of Norovirus Gastroenteritis. PLoS ONE 2016, 11, e0151219. [Google Scholar] [CrossRef] [Green Version]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- Kroneman, A.; Vega, E.; Vennema, H.; Vinje, J.; White, P.A.; Hansman, G.; Green, K.; Martella, V.; Katayama, K.; Koopmans, M. Proposal for a unified norovirus nomenclature and genotyping. Arch. Virol. 2013, 158, 2059–2068. [Google Scholar] [CrossRef] [Green Version]
- Barclay, L.; Wikswo, M.; Gregoricus, N.; Vinjé, J.; Lopman, B.; Parashar, U.; Hall, A.; Leshem, E. Emergence of new norovirus strain GII.4 Syndey-United State. MMWR Morb. Mortal. Wkly. Rep. 2012, 62, 55. [Google Scholar]
- van Beek, J.; Ambert-Balay, K.; Botteldoorn, N.; Eden, J.S.; Fonager, J.; Hewitt, J.; Iritani, N.; Kroneman, A.; Vennema, H.; Vinje, J.; et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Eurosurveillance 2013, 18, 8–9. [Google Scholar]
- Jin, M.; Zhou, Y.K.; Xie, H.P.; Fu, J.G.; He, Y.Q.; Zhang, S.; Jing, H.B.; Kong, X.Y.; Sun, X.M.; Li, H.Y.; et al. Characterization of the new GII.17 norovirus variant that emerged recently as the predominant strain in China. J. Gen. Virol. 2016, 97, 2620–2632. [Google Scholar] [CrossRef] [Green Version]
- Alhatlani, B.; Vashist, S.; Goodfellow, I. Functions of the 5′ and 3′ ends of calicivirus genomes. Virus Res. 2015, 206, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Clarke, I.N.; Lambden, P.R. The molecular biology of caliciviruses. J. Gen. Virol. 1997, 78 Pt 2, 291–301. [Google Scholar] [CrossRef]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [Green Version]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [Green Version]
- Wobus, C.E.; Karst, S.M.; Thackray, L.B.; Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Krug, A.; Mackenzie, J.M.; Green, K.Y.; Virgin, H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004, 2, e432. [Google Scholar] [CrossRef]
- Chaudhry, Y.; Skinner, M.A.; Goodfellow, I.G. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 2007, 88 Pt 8, 2091–2100. [Google Scholar] [CrossRef]
- Hwang, S.; Alhatlani, B.; Arias, A.; Caddy, S.L.; Christodoulou, C.; Cunha, J.B.; Emmott, E.; Gonzalez-Hernandez, M.; Kolawole, A.; Lu, J.; et al. Murine norovirus: Propagation, quantification, and genetic manipulation. Curr. Protoc. Microbiol. 2014, 33, 15K.2.1–15K.2.61. [Google Scholar] [CrossRef] [Green Version]
- den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic viral replication complexes. Cell. Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Wolff, G.; Melia, C.E.; Snijder, E.J.; Barcena, M. Double-Membrane Vesicles as Platforms for Viral Replication. Trends Microbiol. 2020, 28, 1022–1033. [Google Scholar] [CrossRef]
- Shulla, A.; Randall, G. (+) RNA virus replication compartments: A safe home for (most) viral replication. Curr. Opin. Microbiol. 2016, 32, 82–88. [Google Scholar] [CrossRef]
- Hyde, J.L.; Sosnovtsev, S.V.; Green, K.Y.; Wobus, C.; Virgin, H.W.; Mackenzie, J.M. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 2009, 83, 9709–9719. [Google Scholar] [CrossRef] [Green Version]
- Doerflinger, S.Y.; Cortese, M.; Romero-Brey, I.; Menne, Z.; Tubiana, T.; Schenk, C.; White, P.A.; Bartenschlager, R.; Bressanelli, S.; Hansman, G.S.; et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017, 13, e1006705. [Google Scholar] [CrossRef] [Green Version]
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Hughes, P.J.; Stanway, G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000, 81 Pt 1, 201–207. [Google Scholar] [CrossRef]
- Baker, E.S.; Luckner, S.R.; Krause, K.L.; Lambden, P.R.; Clarke, I.N.; Ward, V.K. Inherent structural disorder and dimerisation of murine norovirus NS1-2 protein. PLoS ONE 2012, 7, e30534. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Vega, V.; Sosnovtsev, S.V.; Belliot, G.; King, A.D.; Mitra, T.; Gorbalenya, A.; Green, K.Y. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 2004, 78, 4827–4837. [Google Scholar] [CrossRef] [Green Version]
- Bailey, D.; Kaiser, W.J.; Hollinshead, M.; Moffat, K.; Chaudhry, Y.; Wileman, T.; Sosnovtsev, S.V.; Goodfellow, I.G. Feline calicivirus p32, p39 and p30 proteins localize to the endoplasmic reticulum to initiate replication complex formation. J. Gen. Virol. 2010, 91 Pt 3, 739–749. [Google Scholar] [CrossRef]
- Ettayebi, K.; Hardy, M.E. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 2003, 77, 11790–11797. [Google Scholar]
- Sosnovtsev, S.V.; Belliot, G.; Chang, K.O.; Prikhodko, V.G.; Thackray, L.B.; Wobus, C.E.; Karst, S.M.; Virgin, H.W.; Green, K.Y. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 2006, 80, 7816–7831. [Google Scholar] [CrossRef] [Green Version]
- Nice, T.J.; Strong, D.W.; McCune, B.T.; Pohl, C.S.; Virgin, H.W. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 2013, 87, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Wilen, C.B.; Orvedahl, A.; McCune, B.T.; Kim, K.W.; Orchard, R.C.; Peterson, S.T.; Nice, T.J.; Baldridge, M.T.; Virgin, H.W. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell. Host Microbe 2017, 22, 449–459.e4. [Google Scholar] [CrossRef] [Green Version]
- Robinson, B.A.; Van Winkle, J.A.; McCune, B.T.; Peters, A.M.; Nice, T.J. Caspase-mediated cleavage of murine norovirus NS1/2 potentiates apoptosis and is required for persistent infection of intestinal epithelial cells. PLoS Pathog. 2019, 15, e1007940. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Liu, H.; Wilen, C.B.; Sychev, Z.E.; Desai, C.; Hykes, B.L., Jr.; Orchard, R.C.; McCune, B.T.; Kim, K.W.; Nice, T.J.; et al. A Secreted Viral Nonstructural Protein Determines Intestinal Norovirus Pathogenesis. Cell. Host Microbe 2019, 25, 845–857.e5. [Google Scholar] [CrossRef]
- Yen, J.B.; Wei, L.H.; Chen, L.W.; Chen, L.Y.; Hung, C.H.; Wang, S.S.; Chang, P.J. Subcellular Localization and Functional Characterization of GII.4 Norovirus-Encoded NTPase. J. Virol. 2018, 92, e01824-17. [Google Scholar] [CrossRef] [Green Version]
- Bissa, B.; Deretic, V. Autophagosome Formation: Cutting the Gordian Knot at the ER. Curr. Biol. 2018, 28, R347–R349. [Google Scholar] [CrossRef] [Green Version]
- Biering, S.B.; Choi, J.; Halstrom, R.A.; Brown, H.M.; Beatty, W.L.; Lee, S.; McCune, B.T.; Dominici, E.; Williams, L.E.; Orchard, R.C.; et al. Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell. Host Microbe 2017, 22, 74–85.e7. [Google Scholar] [CrossRef] [Green Version]
- Monastyrska, I.; Ulasli, M.; Rottier, P.J.; Guan, J.L.; Reggiori, F.; de Haan, C.A. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy 2013, 9, 164–174. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Bhattacharyya, S.; Nain, M.; Kaur, M.; Sood, V.; Gupta, V.; Khasa, R.; Abdin, M.Z.; Vrati, S.; Kalia, M. Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes. Autophagy 2014, 10, 1637–1651. [Google Scholar] [CrossRef] [Green Version]
- Reggiori, F.; Monastyrska, I.; Verheije, M.H.; Cali, T.; Ulasli, M.; Bianchi, S.; Bernasconi, R.; de Haan, C.A.; Molinari, M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell. Host Microbe 2010, 7, 500–508. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zou, Z.; Jiang, Z.; Huang, X.; Liu, Q. Biological Function and Application of Picornaviral 2B Protein: A New Target for Antiviral Drug Development. Viruses 2019, 11, 510. [Google Scholar] [CrossRef] [Green Version]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef]
- Strtak, A.C.; Perry, J.L.; Sharp, M.N.; Chang-Graham, A.L.; Farkas, T.; Hyser, J.M. Recovirus NS1-2 Has Viroporin Activity That Induces Aberrant Cellular Calcium Signaling to Facilitate Virus Replication. mSphere 2019, 4, e00506-19. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Galea, A.; Sytnyk, V.; Crossley, M. Controlling the size of lipid droplets: Lipid and protein factors. Curr. Opin. Cell Biol. 2012, 24, 509–516. [Google Scholar] [CrossRef]
- Xu, Q.; Rawlings, N.D.; Chiu, H.J.; Jaroszewski, L.; Klock, H.E.; Knuth, M.W.; Miller, M.D.; Elsliger, M.A.; Deacon, A.M.; Godzik, A.; et al. Structural analysis of papain-like NlpC/P60 superfamily enzymes with a circularly permuted topology reveals potential lipid binding sites. PLoS ONE 2011, 6, e22013. [Google Scholar] [CrossRef] [Green Version]
- Uyama, T.; Morishita, J.; Jin, X.H.; Okamoto, Y.; Tsuboi, K.; Ueda, N. The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J. Lipid Res. 2009, 50, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Yen, J.B.; Chen, L.W.; Wei, L.H.; Hung, C.H.; Wang, S.S.; Lin, C.L.; Chang, P.J. Identification and Characterization of Human Norovirus NTPase Regions Required for Lipid Droplet Localization, Cellular Apoptosis, and Interaction with the Viral P22 Protein. Microbiol. Spectr. 2021, 9, e0042221. [Google Scholar] [CrossRef]
- Katayama, K.; Murakami, K.; Sharp, T.M.; Guix, S.; Oka, T.; Takai-Todaka, R.; Nakanishi, A.; Crawford, S.E.; Atmar, R.L.; Estes, M.K. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc. Natl. Acad. Sci. USA 2014, 111, E4043–E4052. [Google Scholar] [CrossRef] [Green Version]
- Filipe, A.; McLauchlan, J. Hepatitis C virus and lipid droplets: Finding a niche. Trends Mol. Med. 2015, 21, 34–42. [Google Scholar] [CrossRef]
- Samsa, M.M.; Mondotte, J.A.; Iglesias, N.G.; Assuncao-Miranda, I.; Barbosa-Lima, G.; Da Poian, A.T.; Bozza, P.T.; Gamarnik, A.V. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009, 5, e1000632. [Google Scholar] [CrossRef] [Green Version]
- Herker, E.; Ott, M. Emerging role of lipid droplets in host/pathogen interactions. J. Biol. Chem. 2012, 287, 2280–2287. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Dutra, F.S.; Teixeira, L.; de Souza Costa, M.F.; Bozza, P.T. Fat, fight, and beyond: The multiple roles of lipid droplets in infections and inflammation. J. Leukoc. Biol. 2019, 106, 563–580. [Google Scholar] [CrossRef]
- Boulant, S.; Targett-Adams, P.; McLauchlan, J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 2007, 88 Pt 8, 2204–2213. [Google Scholar] [CrossRef]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Bartenschlager, R.; Penin, F.; Lohmann, V.; Andre, P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011, 19, 95–103. [Google Scholar] [CrossRef]
- Laufman, O.; Perrino, J.; Andino, R. Viral Generated Inter-Organelle Contacts Redirect Lipid Flux for Genome Replication. Cell 2019, 178, 275–289.e16. [Google Scholar] [CrossRef]
- Miller, S.; Krijnse-Locker, J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008, 6, 363–374. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- McCune, B.T.; Tang, W.; Lu, J.; Eaglesham, J.B.; Thorne, L.; Mayer, A.E.; Condiff, E.; Nice, T.J.; Goodfellow, I.; Krezel, A.M.; et al. Noroviruses Co-opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine-Phenylalanine-Acidic-Tract-Motif Mimic in Nonstructural Viral Protein NS1/2. mBio 2017, 8, e00668-17. [Google Scholar] [CrossRef] [Green Version]
- Tu, H.; Gao, L.; Shi, S.T.; Taylor, D.R.; Yang, T.; Mircheff, A.K.; Wen, Y.; Gorbalenya, A.E.; Hwang, S.B.; Lai, M.M. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 1999, 263, 30–41. [Google Scholar] [CrossRef] [Green Version]
- Barajas, D.; Xu, K.; de Castro Martin, I.F.; Sasvari, Z.; Brandizzi, F.; Risco, C.; Nagy, P.D. Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog. 2014, 10, e1004388. [Google Scholar] [CrossRef] [Green Version]
- Reggiori, F.; de Haan, C.A.; Molinari, M. Unconventional use of LC3 by coronaviruses through the alleged subversion of the ERAD tuning pathway. Viruses 2011, 3, 1610–1623. [Google Scholar] [CrossRef]
- Bernasconi, R.; Molinari, M. ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011, 23, 176–183. [Google Scholar] [CrossRef]
- Bernasconi, R.; Noack, J.; Molinari, M. Unconventional roles of nonlipidated LC3 in ERAD tuning and coronavirus infection. Autophagy 2012, 8, 1534–1536. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, T.B.; Hyde, J.L.; Mintern, J.D.; Mackenzie, J.M. Mouse Norovirus infection promotes autophagy induction to facilitate replication but prevents final autophagosome maturation. Virology 2016, 492, 130–139. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hung, C.-H.; Yen, J.-B.; Chang, P.-J.; Chen, L.-W.; Huang, T.-Y.; Tsai, W.-J.; Tsai, Y.-C. Characterization of Human Norovirus Nonstructural Protein NS1.2 Involved in the Induction of the Filamentous Endoplasmic Reticulum, Enlarged Lipid Droplets, LC3 Recruitment, and Interaction with NTPase and NS4. Viruses 2023, 15, 812. https://doi.org/10.3390/v15030812
Hung C-H, Yen J-B, Chang P-J, Chen L-W, Huang T-Y, Tsai W-J, Tsai Y-C. Characterization of Human Norovirus Nonstructural Protein NS1.2 Involved in the Induction of the Filamentous Endoplasmic Reticulum, Enlarged Lipid Droplets, LC3 Recruitment, and Interaction with NTPase and NS4. Viruses. 2023; 15(3):812. https://doi.org/10.3390/v15030812
Chicago/Turabian StyleHung, Chien-Hui, Ju-Bei Yen, Pey-Jium Chang, Lee-Wen Chen, Tsung-Yu Huang, Wan-Ju Tsai, and Yu-Chin Tsai. 2023. "Characterization of Human Norovirus Nonstructural Protein NS1.2 Involved in the Induction of the Filamentous Endoplasmic Reticulum, Enlarged Lipid Droplets, LC3 Recruitment, and Interaction with NTPase and NS4" Viruses 15, no. 3: 812. https://doi.org/10.3390/v15030812
APA StyleHung, C.-H., Yen, J.-B., Chang, P.-J., Chen, L.-W., Huang, T.-Y., Tsai, W.-J., & Tsai, Y.-C. (2023). Characterization of Human Norovirus Nonstructural Protein NS1.2 Involved in the Induction of the Filamentous Endoplasmic Reticulum, Enlarged Lipid Droplets, LC3 Recruitment, and Interaction with NTPase and NS4. Viruses, 15(3), 812. https://doi.org/10.3390/v15030812






