Neonatal Hypoxic-Ischemic Encephalopathy Grading from Multi-Channel EEG Time-Series Data Using a Fully Convolutional Neural Network
Abstract
:1. Introduction
2. Method
2.1. Dataset
2.2. Fully Convolutional Neural Network
2.2.1. Pre-Processing
2.2.2. FCN Architecture
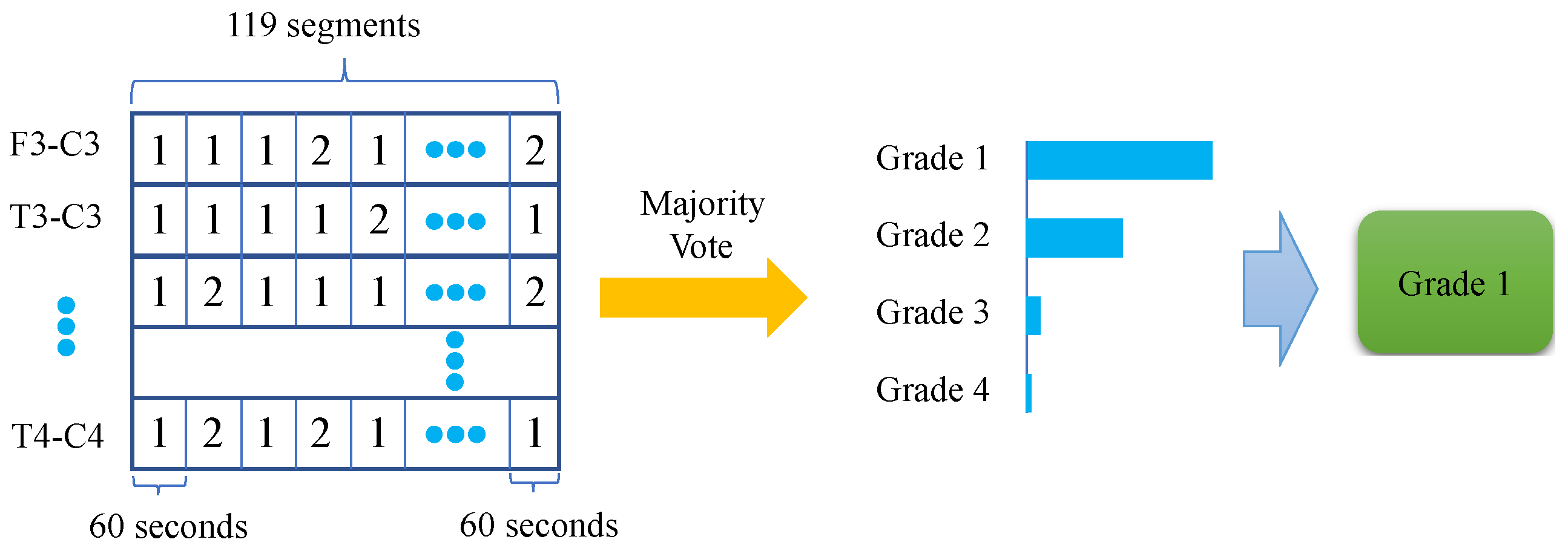
2.2.3. Post-Processing
2.2.4. Metrics
2.2.5. Visualization
3. Results
4. Discussion
4.1. Comparison with CNN Baseline
4.2. Receptive Field
4.3. UMAP Visualization
4.4. UMAP Dimension Reduction
4.5. Weak Labels
4.6. Clinical Implementation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, M.-C.; Yang, S.-N. Perinatal hypoxic-ischemic encephalopathy. BioMed Res. Int. 2011, 2011, 609813. [Google Scholar] [CrossRef] [PubMed]
- Perlman, J.M. Summary proceedings from the neurology group on hypoxic-ischemic encephalopathy. Pediatrics 2006, 117 (Suppl. S1), S28–S33. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, R.C. Hypoxic-ischemic encephalopathy. Am. J. Perinatol. 2000, 17, 113–120. [Google Scholar] [CrossRef]
- Hagberg, B.; Hagberg, G.; Beckung, E.; Uvebrant, P. Changing panorama of cerebral palsy in Sweden. VIII. Prevalence and origin in the birth year period 1991–1994. Acta Paediatr. 2001, 90, 271–277. [Google Scholar] [CrossRef]
- Douglas-Escobar, M.; Weiss, M.D. Hypoxic-ischemic encephalopathy: A review for the clinician. JAMA Pediatr. 2015, 169, 397–403. [Google Scholar] [CrossRef]
- Azzopardi, D.V.; Strohm, B.; Edwards, A.D.; Dyet, L.; Halliday, H.L.; Juszczak, E.; Kapellou, O.; Levene, M.; Marlow, N.; Porter, E.; et al. Moderate hypothermia to treat perinatal asphyxial encephalopathy. N. Engl. J. Med. 2009, 361, 1349–1358. [Google Scholar] [CrossRef]
- Gluckman, P.D.; Wyatt, J.S.; Azzopardi, D.; Ballard, R.; Edwards, A.D.; Ferriero, D.M.; Polin, R.A.; Robertson, C.M.; Thoresen, M.; Whitelaw, A.; et al. Selective head cooling with mild systemic hypothermia after neonatal encephalopathy: Multicentre randomised trial. Lancet 2005, 365, 663–670. [Google Scholar] [CrossRef]
- Imanishi, T.; Shimizu, M.; Sumiya, W.; Kanno, C.; Kanno, M.; Kanno, M.; Kawabata, K. Brain injury following mild hypoxic-ischemic encephalopathy in neonates–ten-year experience in a tertiary perinatal center. J. Perinatol. 2022, 42, 1630–1636. [Google Scholar] [CrossRef]
- Goswami, I.R.; Whyte, H.; Wintermark, P.; Mohammad, K.; Shivananda, S.; Louis, D.; Yoon, E.W.; Shah, P.S. Characteristics and short-term outcomes of neonates with mild hypoxic-ischemic encephalopathy treated with hypothermia. J. Perinatol. 2020, 40, 275–283. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Chaban, B.; Montaldo, P.; Thayyil, S. Predictive value of amplitude-integrated eeg (aeeg) after rescue hypothermic neuroprotection for hypoxic ischemic encephalopathy: A meta-analysis. J. Perinatol. 2017, 37, 684–689. [Google Scholar] [CrossRef]
- Roth, S.; Baudin, J.; Cady, E.; Johal, K.; Townsend, J.; Wyatt, J.S.; Reynolds, E.R.; Stewart, A.L. Relation of deranged neonatal cerebral oxidative metabolism with neurodevelopmental outcome and head circumference at 4 years. Dev. Med. Child Neurol. 1997, 39, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Hrachovy, R.A.; Mizrahi, E.M. Atlas of Neonatal Electroencephalography; Springer Publishing Company: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Biagioni, E.; Bartalena, L.; Boldrini, A.; Pieri, R.; Cioni, G. Constantly discontinuous EEG patterns in full-term neonates with hypoxic-ischaemic encephalopathy. Clin. Neurophysiol. 1999, 110, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Matić, V.; Cherian, P.J.; Jansen, K.; Koolen, N.; Naulaers, G.; Swarte, R.M.; Govaert, P.; Van Huffel, S.; De Vos, M. Improving reliability of monitoring background EEG dynamics in asphyxiated infants. IEEE Trans. Biomed. Eng. 2015, 63, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Korotchikova, I.; Stevenson, N.; Walsh, B.; Murray, D.; Boylan, G. Quantitative EEG analysis in neonatal hypoxic ischaemic encephalopathy. Clin. Neurophysiol. 2011, 122, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Lacan, L.; Betrouni, N.; Lamblin, M.D.; Chaton, L.; Delval, A.; Bourriez, J.L.; Storme, L.; Derambure, P.; Tich, S.N. Quantitative approach to early neonatal EEG visual analysis in hypoxic-ischemic encephalopathy severity: Bridging the gap between eyes and machine. Neurophysiol. Clin. 2021, 51, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, N.; Korotchikova, I.; Temko, A.; Lightbody, G.; Marnane, W.; Boylan, G. An automated system for grading EEG abnormality in term neonates with hypoxic-ischaemic encephalopathy. Ann. Biomed. Eng. 2013, 41, 775–785. [Google Scholar] [CrossRef]
- Ahmed, R.; Temko, A.; Marnane, W.; Lightbody, G.; Boylan, G. Grading hypoxic–ischemic encephalopathy severity in neonatal EEG using GMM supervectors and the support vector machine. Clin. Neurophysiol. 2016, 127, 297–309. [Google Scholar] [CrossRef]
- Raurale, S.A.; Boylan, G.B.; Mathieson, S.R.; Marnane, W.P.; Lightbody, G.; O’Toole, J.M. Grading hypoxic-ischemic encephalopathy in neonatal EEG with convolutional neural networks and quadratic time–frequency distributions. J. Neural Eng. 2021, 18, 046007. [Google Scholar] [CrossRef]
- O’Sullivan, M.E.; Lightbody, G.; Mathieson, S.R.; Marnane, W.P.; Boylan, G.B.; O’Toole, J.M. Development of an EEG artefact detection algorithm and its application in grading neonatal hypoxic-ischemic encephalopathy. Expert Syst. Appl. 2023, 213, 118917. [Google Scholar] [CrossRef]
- O’Shea, A.; Lightbody, G.; Boylan, G.; Temko, A. Neonatal seizure detection from raw multi-channel EEG using a fully convolutional architecture. Neural Netw. 2020, 123, 12–25. [Google Scholar] [CrossRef]
- Ansari, A.H.; De Wel, O.; Pillay, K.; Dereymaeker, A.; Jansen, K.; Van Huffel, S.; Naulaers, G.; De Vos, M. A convolutional neural network outperforming state-of-the-art sleep staging algorithms for both preterm and term infants. J. Neural Eng. 2020, 17, 016028. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1–11. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2020; pp. 11976–11986. [Google Scholar]
- Lim, W.; Sourina, O.; Wang, L.P. STEW: Simultaneous task EEG workload data set. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2106–2114. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. Deap: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Tsuchida, T.N.; Wusthoff, C.J.; Shellhaas, R.A.; Abend, N.S.; Hahn, C.D.; Sullivan, J.E.; Nguyen, S.; Weinstein, S.; Scher, M.S.; Riviello, J.J.; et al. American clinical neurophysiology society standardized EEG terminology and categorization for the description of continuous EEG monitoring in neonates: Report of the American Clinical Neurophysiology Society critical care monitoring committee. J. Clin. Neurophysiol. 2013, 30, 161–173. [Google Scholar] [CrossRef]
- Vanhatalo, S.; Palva, J.M.; Andersson, S.; Rivera, C.; Voipio, J.; Kaila, K. Slow endogenous activity transients and developmental expression of K+–Cl- cotransporter 2 in the immature human cortex. Eur. J. Neurosci. 2005, 22, 2799–2804. [Google Scholar] [CrossRef]
- Stewart, A.X.; Nuthmann, A.; Sanguinetti, G. Single-trial classification of EEG in a visual object task using ICA and machine learning. J. Neurosci. Methods 2014, 228, 1–14. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning. PMLR, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Sainburg, T.; McInnes, L.; Gentner, T.Q. Parametric umap embeddings for representation and semisupervised learning. Neural Comput. 2021, 33, 2881–2907. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N.; Grossberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Stikic, M.; Johnson, R.R.; Tan, V.; Berka, C. EEG-based classification of positive and negative affective states. Brain-Comput. Interfaces 2014, 1, 99–112. [Google Scholar] [CrossRef]
- Lan, Z.; Sourina, O.; Wang, L.; Scherer, R.; Müller-Putz, G.R. Domain adaptation techniques for EEG-based emotion recognition: A comparative study on two public datasets. IEEE Trans. Cogn. Dev. Syst. 2018, 11, 85–94. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Diaz-Papkovich, A.; Anderson-Trocmé, L.; Gravel, S. A review of UMAP in population genetics. J. Hum. Genet. 2021, 66, 85–91. [Google Scholar] [CrossRef]
- Wattenberg, M.; Viégas, F.; Johnson, I. How to Use t-SNE Effectively. Distill 2016, 1, e2. [Google Scholar] [CrossRef]
- Ansari, A.H.; Cherian, P.J.; Caicedo, A.; Naulaers, G.; De Vos, M.; Van Huffel, S. Neonatal seizure detection using deep convolutional neural networks. Int. J. Neural Syst. 2019, 29, 1850011. [Google Scholar] [CrossRef]
- Li, R.; Wang, L.; Suganthan, P.; Sourina, O. Sample-Based Data Augmentation Based on Electroencephalogram Intrinsic Characteristics. IEEE J. Biomed. Health Inform. 2022, 26, 4996–5003. [Google Scholar] [CrossRef]















| L | S1 | S2 | K | |
|---|---|---|---|---|
| FCN10 | 3 | 2 | 3 | 3 |
| FCN13 | 4 | 1 | 4 | 3 |
| FCN16 | 5 | 1 | 3 | 2 |
| True Label | Prediction | ||||||
| 1 | 2 | 3 | 4 | sensitivity | specificity | ||
| 1 | 181 | 9 | 0 | 0 | 0.953 | 0.838 | |
| 2 | 24 | 52 | 5 | 0 | 0.642 | 0.953 | |
| 3 | 0 | 3 | 32 | 1 | 0.889 | 0.967 | |
| 4 | 0 | 0 | 5 | 26 | 0.839 | 0.997 | |
| PPV | 0.883 | 0.813 | 0.762 | 0.963 | |||
| Dev. Set | Test Set | Dev. Method | Input | |
|---|---|---|---|---|
| TFD-CNN [19] | 54 neonates (Cork) | ANSeR1 | Leave one out | TFD Features |
| TFD-CNN-Reg [20] | ANSeR1&2 | ANSeR1&2 | 10-fold CV | TFD Features |
| Proposed | ANSeR2 | ANSeR1 | Internal CV | Raw EEG |
| TFD-CNN | TFD-CNN-Reg | FCN16(Ensembled) | |
|---|---|---|---|
| Accuracy | 69.5% | 82.8% | 86.09% |
| 95%CI | 65.3–73.6% | 80.5–85.2% | 82.41–89.78% |
| MCC | - | 0.722 | 0.7691 |
| Receptive Field | No. of Params | Test Acc. | True Negative Rate | Test AUC | Val. Acc. | Val. AUC | |
|---|---|---|---|---|---|---|---|
| FCN10 | 29.66 s | 25,540 | 0.8458 | 0.9348 | 0.8693 | 0.8005 | 0.8674 |
| FCN13 | 39.94 s | 34,916 | 0.8421 | 0.9360 | 0.8643 | 0.7917 | 0.8689 |
| FCN16 | 49.25 s | 44,292 | 0.8572 | 0.9387 | 0.8620 | 0.8199 | 0.8639 |
| IL | S3 | S4 | S5 | |
|---|---|---|---|---|
| FCN13_30s | 480 | 2 | 2 | 2 |
| FCN13_60s | 960 | 3 | 2 | 1 |
| FCN13_90s | 1440 | 4 | 1 | - |
| Receptive Field | MCC | Test Acc. | Test AUC | Val. Acc. | Val. AUC | |
|---|---|---|---|---|---|---|
| FCN13_30s | 10.5 s | 0.7261 | 0.8336 | 0.8784 | 0.6747 | 0.8609 |
| FCN13_60s | 32.69 s | 0.7692 | 0.8539 | 0.8694 | 0.7193 | 0.8693 |
| FCN13_90s | 79.88 s | 0.7773 | 0.8661 | 0.8793 | 0.7529 | 0.8893 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Marnane, W.P.; Boylan, G.B.; Lightbody, G. Neonatal Hypoxic-Ischemic Encephalopathy Grading from Multi-Channel EEG Time-Series Data Using a Fully Convolutional Neural Network. Technologies 2023, 11, 151. https://doi.org/10.3390/technologies11060151
Yu S, Marnane WP, Boylan GB, Lightbody G. Neonatal Hypoxic-Ischemic Encephalopathy Grading from Multi-Channel EEG Time-Series Data Using a Fully Convolutional Neural Network. Technologies. 2023; 11(6):151. https://doi.org/10.3390/technologies11060151
Chicago/Turabian StyleYu, Shuwen, William P. Marnane, Geraldine B. Boylan, and Gordon Lightbody. 2023. "Neonatal Hypoxic-Ischemic Encephalopathy Grading from Multi-Channel EEG Time-Series Data Using a Fully Convolutional Neural Network" Technologies 11, no. 6: 151. https://doi.org/10.3390/technologies11060151
APA StyleYu, S., Marnane, W. P., Boylan, G. B., & Lightbody, G. (2023). Neonatal Hypoxic-Ischemic Encephalopathy Grading from Multi-Channel EEG Time-Series Data Using a Fully Convolutional Neural Network. Technologies, 11(6), 151. https://doi.org/10.3390/technologies11060151







