Abstract
Congenital heart disease (CHD) represents a multifaceted medical condition that requires early detection and diagnosis for effective management, given its diverse presentations and subtle symptoms that manifest from birth. This research article introduces a groundbreaking healthcare application, the Machine Learning-based Congenital Heart Disease Prediction Method (ML-CHDPM), tailored to address these challenges and expedite the timely identification and classification of CHD in pregnant women. The ML-CHDPM model leverages state-of-the-art machine learning techniques to categorize CHD cases, taking into account pertinent clinical and demographic factors. Trained on a comprehensive dataset, the model captures intricate patterns and relationships, resulting in precise predictions and classifications. The evaluation of the model’s performance encompasses sensitivity, specificity, accuracy, and the area under the receiver operating characteristic curve. Remarkably, the findings underscore the ML-CHDPM’s superiority across six pivotal metrics: accuracy, precision, recall, specificity, false positive rate (FPR), and false negative rate (FNR). The method achieves an average accuracy rate of 94.28%, precision of 87.54%, recall rate of 96.25%, specificity rate of 91.74%, FPR of 8.26%, and FNR of 3.75%. These outcomes distinctly demonstrate the ML-CHDPM’s effectiveness in reliably predicting and classifying CHD cases. This research marks a significant stride toward early detection and diagnosis, harnessing advanced machine learning techniques within the realm of ECG signal processing, specifically tailored to pregnant women.
1. Introduction
Cardiovascular disease is a significant medical condition that affects heart performance and leads to complications such as coronary artery disease and impaired vascular function [1]. These difficulties can result in myocardial infarction and cerebrovascular accidents. According to a survey, heart disease annually impacts an estimated 620,000 individuals in the United States [2]. While heart disease can affect both genders, males are more vulnerable. Statistics from 2010 show that a quarter of all fatalities were attributed to heart disease. In the United States, there are approximately 738,000 cases of heart attacks, with 528,000 of these cases being initial occurrences. The remaining 220,000 individuals experience subsequent episodes. Symptoms of heart disease include chest tightness, pain and discomfort, shortness of breath, ankle swelling, neck and abdominal pain, rapid heartbeat, dizziness, cardiac arrest, fainting, changes in skin color, ankle irritation, weight loss, and fatigue. The manifestation of symptoms depends on the type of cardiovascular ailment, which include but are not limited to arrhythmia, myocardial infarction, heart failure, congenital coronary artery disease, mitral valve insufficiency, and dilated cardiomyopathy.
Congenital heart disease (CHD) refers to a group of structural abnormalities in the heart that occur during the prenatal stage of development [3]. These congenital abnormalities manifest during the prenatal period and affect the morphology and physiology of the heart, leading to various cardiovascular complications. CHD is a common congenital anomaly around the world, imposing a significant health burden on affected individuals and medical systems alike. The global incidence of CHD shows significant variation, with an estimated occurrence of approximately 1% of all live births [4]. In the United States, it is believed that around 40,000 newborns are affected by CHD each year. The severity of the condition can vary, ranging from individuals who experience minimal to no symptoms to those who require immediate medical attention. CHD encompasses a diverse range of anomalies, including structural malformations in the heart valves, walls, and vasculature. Examples of frequently encountered instances of CHD include atrial septal defects, ventricular septal defects, and Tetralogy of Fallot [5].
It is widely recommended that all pregnant women worldwide undergo fetal evaluation and ultrasound between 18 and 24 weeks of gestation [6]. This procedure involves detailed imaging, including ultrasound scans of the heart, which has the potential to identify over 90% of severe congenital heart conditions. However, despite the widespread use of fetal ultrasound technology, the prevalence of fetal detection for genetic cardiovascular diseases within the community ranges from 30 to 50% [7]. The hypothesis suggests that the main reason for the significant disparity in diagnoses is insufficient and inconsistent proficiency in analyzing fetal cardiac images [8]. This is primarily due to the complexity of detecting a small and rapidly beating fetal heart as well as the relatively low awareness of congenital coronary artery disease among healthcare providers, given its low incidence. Although clinical quality assurance efforts focused on a single center and conducted on a small scale have shown promising results in improving CHD detection rates by up to 100%, the sustainability and scalability of such programs present significant challenges. To address this, experiments were conducted to determine if utilizing machine learning (ML) analysis of images could improve the evaluation rates typically observed in community medicine [9]. This was achieved by training the ML model on data from a limited number of clinically relevant imaging studies.
Machine learning has been demonstrated to be proficient in detecting intricate image patterns and successfully applied in adult cardiovascular ultrasound technology [10]. It has even surpassed the performance of physicians in tasks involving the classification of views, using small and downsampled databases. However, despite its widespread usage across various domains, the application of machine learning in the context of CHD or fetal ultrasound still requires further refinement. The use of deep learning in medical scenarios that are inherently rare presents inherent limitations, irrespective of the volume of training data available. The hypothesis posits that by utilizing input data curated based on clinical recommendations, specifically by selecting five exclusive cardiac examination viewpoints, the algorithms would be capable of identifying diagnostic indicators even when dealing with databases of limited size. The identified research gaps are as follows:
- There is a need for further research on the utilization of data derived from the Internet of Medical Things (IoMT) in diagnosing cardiovascular disease.
- More investigation is needed to explore the potential of utilizing Long Short-Term Memory (LSTM) architecture and attention systems for the detection of cardiovascular disease.
- There is a lack of comprehensive research on the concurrent utilization of CNN-BiLSTM-AM (Convolutional Neural Networks, Bidirectional Long Short-Term Memory, Attention Mechanisms) to achieve precise diagnostic outcomes in the context of cardiovascular disease.
- It is necessary to conduct a thorough assessment and comparison of various models for identifying signs of cardiovascular disease using the Heart Disease UCI and Cardiovascular Disease Dataset databases.
The research presented in this study makes significant contributions to the field of cardiovascular disease diagnosis by leveraging data from the Internet of Medical Things (IoMT). Firstly, it addresses the crucial challenge of timely identification and detection of cardiovascular diseases by utilizing IoMT data, aiming to enhance the accuracy and efficiency of diagnosis, which in turn enables prompt intervention and improves patient outcomes. Secondly, the study explores the potential of leveraging IoMT data for accurate diagnosis, providing insights into the utilization of this emerging technology in the field. Thirdly, it investigates the integration of Long Short-Term Memory (LSTM) design and Attention Mechanisms to enhance disease detection, paving the way for advanced techniques to improve diagnostic accuracy. Moreover, the study introduces a novel diagnostic model that combines Convolutional Neural Network (CNN), Bidirectional Long Short-Term Memory (BiLSTM), and Attention Mechanism (AM) techniques, offering a comprehensive approach to enhance the classification accuracy of cardiovascular diseases. Lastly, a comprehensive assessment and comparison of various models is conducted using widely recognized databases such as the Heart Disease UCI and Cardiovascular Disease Dataset, contributing to the understanding and advancement of IoMT data, LSTM design, and advanced neural network models in the field of cardiovascular disease diagnosis, with the ultimate goal of improving patient outcomes.
The research paper is structured into several sections to provide a clear and organized presentation of the study. Section 2 focuses on the literature review, discussing the current issues and challenges associated with the classification of congenital heart disease (CHD). Moving on to Section 3, the Machine Learning-based Congenital Heart Disease Prediction Method (ML-CHDPM) is introduced, outlining the methodology, algorithms, and presenting the results obtained from applying ML-CHDPM to CHD datasets. Section 4 then presents the software results and performance assessments, analyzing the software implementation of ML-CHDPM and evaluating its performance through metrics such as accuracy, precision, recall, and F1-score. Finally, Section 5 offers concluding remarks, summarizing the key findings of the study and highlighting potential directions for future research in CHD detection methods. By following this structured format, the research paper ensures a coherent flow of information, enabling readers to easily navigate through the literature review, methodology, results, and conclusion, thereby comprehending the contributions and implications of the study.
2. Background and Literature Survey
Various conventional techniques in machine learning have been employed to address the challenges associated with manually analyzing electrocardiogram (ECG) signals in Coronary Heart Disease (CHD). The conventional machine learning approach involves several steps, including preprocessing, feature extraction, feature selection, and categorization processes. Differentiating between normal and CHD signals based on their distinctive characteristics is a time and resource-intensive task. The robustness of the features obtained is significantly impacted by the quality of the underlying data. Preprocessing steps, such as noise elimination and R-peak identification, are essential to extract crucial attributes needed for effective categorization. This research suggests leveraging machine learning to improve the efficiency of an automated CHD diagnosis method, aiming to overcome the limitations associated with traditional machine learning approaches. Machine learning algorithms play a crucial role in acquiring and recognizing unique features from input ECG signals. The goal is to enhance the accuracy and effectiveness of the diagnostic process for CHD through the utilization of advanced machine learning techniques.
In their study, Xu et al. [11] presented a novel methodology for the automated classification of pediatric Congenital Heart Disease (CHD) through the analysis of heartbeats. The researchers conducted an extensive extraction of diverse features from normal heart signals, encompassing characteristics derived from the time domain, frequency domain, and wavelets. Employing machine learning methodologies, particularly random forest and support vector machines, the proposed approach demonstrated promising outcomes. The results revealed a commendable accuracy rate of 87.5% in effectively categorizing CHD cases. Additionally, the specificity and sensitivity values were noteworthy, standing at 89.7% and 85.2%, respectively. These findings underscore the efficacy of the devised method in reliably identifying pediatric CHD through the analysis of heartbeats, showcasing its potential as a valuable diagnostic tool in this medical context.
Ng et al. [12] designed an automated framework aimed at classifying perioperative hazards in patients with complex Congenital Heart Disease (CHD) by leveraging retinal images. The authors introduced an innovative feature extraction method that harnessed both color-based and texture-based characteristics obtained from retinal images. Subsequently, these extracted features were employed in risk classification through the application of machine learning, specifically utilizing a support vector machine. Results from the implemented framework demonstrated a notable predictive accuracy, achieving an impressive rate of 84.9% in effectively identifying perioperative risks in patients diagnosed with complex congenital heart disease. This research highlights the potential of utilizing retinal images and advanced machine learning techniques as a valuable tool for automating the identification of perioperative hazards, thereby contributing to enhanced patient care and risk management in the context of complex CHD cases.
Kobel et al. [13] conducted a thorough assessment of the Apple Watch iECG’s effectiveness in detecting Congenital Heart Disease (CHD) in children. The study involved obtaining iECG measurements from pediatric patients, including those with and without CHD. A meticulous comparative analysis was performed by juxtaposing the iECG data against conventional ECG records. The outcomes of this investigation suggest a promising role for the Apple Watch iECG as a potential screening tool for CHD in children, revealing a sensitivity of 92% and an accuracy of 93% in identifying the condition. In a separate study led by van Genuchten and colleagues [14], the physical capacity of children diagnosed with CHD was evaluated. A cohort of pediatric patients with CHD underwent exercise tests to assess their peak oxygen uptake (VO2peak). The research findings unveiled a significant revelation—children with CHD exhibited diminished exercise capacity compared to their healthy counterparts, as evidenced by lower VO2peak measurements. These results underscore the considerable impact of CHD on the ability of pediatric populations to partake in physical activities, shedding light on the broader implications of the condition on the overall well-being of affected individuals.
Kavitha et al. [15] introduced an innovative approach termed Multilayer Deep Detection Perceptron (MLDDP) for the identification of testicular deviations, both with and without Congenital Heart Disease (CHD). This practical method utilized a Multilayer Deep Learning framework that incorporated multiple layers of perceptrons to discern anomalies associated with CHD. Upon evaluating the proposed MLDDP on a provided dataset, it demonstrated an exceptional detection accuracy of 95.4% in precisely identifying testicular deviations, irrespective of the presence of CHD. The success of MLDDP underscores the potential of machine learning techniques in advancing the diagnosis of CHD-related conditions. In a distinct study, Liu et al. [16] concentrated on the computer-aided analysis of heart sounds in pediatric patients diagnosed with left-to-right shunt CHD. The researchers introduced a methodology leveraging machine learning techniques, specifically employing a Convolutional Neural Network (CNN), to extract pertinent features from heart sound signals and accurately classify the presence of left-to-right shunt CHD. The outcomes were noteworthy, with a precision rate of 90.8% and a region under the receiver operating characteristics curve of 0.935, indicating the method’s efficacy in identifying and categorizing CHD with left-to-right shunt. These findings underscore the potential of machine learning-based analyses in supporting medical professionals in diagnosing specific types of CHD, showcasing the promising intersection of technology and healthcare.
Ge et al. proposed an innovative method for identifying Pulmonary Hypertension (PH) associated with Congenital Heart Disease (CHD) by incorporating time–frequency domain analysis and machine learning (ML) characteristics [17]. The researchers integrated time–frequency analysis techniques into an ML framework to extract relevant features from echocardiographic data. Through their approach, the method achieved an impressive precision level of 91.6% in accurately identifying pulmonary hypertension linked to congenital heart disease. This study demonstrates the efficacy of combining advanced signal processing techniques with machine learning approaches to enhance the identification and characterization of specific cardiac conditions, specifically focusing on the challenging context of pulmonary hypertension in the presence of congenital heart disease.
Steeden et al. [18] delved into the exploration of utilizing artificial intelligence (AI) in the assessment of Congenital Heart Disease (CHD). The authors undertook a thorough examination of AI-based methodologies, specifically machine learning (ML), applied to tasks such as image analysis, risk estimation, and detection within the realm of CHD. Their comprehensive analysis provided insightful perspectives on the potential of AI in augmenting the assessment and treatment of individuals with CHD. By shedding light on the various applications of AI in the context of CHD, the study contributes to the evolving landscape of medical technology and its role in advancing cardiac care and diagnostics.
Alici-Karaca et al. introduced a Convolutional Neural Network (CNN) with a lightweight architecture designed to precisely classify cases of radiation-induced liver disease, as detailed in their research publication [19]. While their study does not directly focus on congenital heart disease, it underscores the application of machine learning in medical image analysis. The featured lightweight CNN successfully achieved an impressive classification accuracy rate of 93.1% when tasked with identifying radiation-induced liver disease. This result highlights the versatile capacity of machine learning to be applied across diverse medical conditions, showcasing its potential in aiding accurate diagnoses beyond the specific context of congenital heart disease.
Qiao et al. presented the Residual Learning based Diagnostic System (RLDS), an innovative diagnostic system employing residual learning, designed for cases of fetal Congenital Heart Disease (CHD) [20]. This system utilized residual learning, a type of machine learning, to extract distinctive features from images of fetal echocardiography for discrimination. Remarkably, the diagnostic accuracy of the RLDS for fetal CHD reached an impressive 96.5%. Additionally, the system offered interpretability by generating attention maps and assigning importance scores to features, enhancing the understanding of the diagnostic process for medical professionals. This research underscores the potential of incorporating machine learning, specifically residual learning, in creating advanced diagnostic tools for fetal CHD with the added benefit of interpretability.
The following Table 1 summarizes key findings from various studies investigating the use of machine learning (ML) in cardiovascular health, focusing on congenital heart disease (CHD) and related conditions. Each entry includes the reference number, author, study objective, and identified limitations. This compilation offers a succinct overview of the objectives pursued by researchers, shedding light on both the potential and challenges associated with ML applications in the diagnosis and assessment of cardiovascular health.

Table 1.
Key Findings.
Based on recent research, several limitations have been identified within the domain of congestive heart failure detection and machine learning applications in electrocardiogram (ECG) diagnosis systems:
- Scope for Improvement in Existing Methods:
- -
- The current machine learning methods utilized for congestive heart failure detection are widely acknowledged to have room for enhancement. Further research and innovation are needed to refine and optimize these methodologies for improved accuracy and reliability.
- Reduction of Training Time for Feature Extraction:
- -
- Opportunities exist for streamlining the training process by reducing the time required for feature extraction and model training. Efforts should be directed towards developing more efficient algorithms to enhance the overall efficiency of machine learning models.
- Challenges in Fully Controlled ECG Multi-Class Categorization:
- -
- Despite achieving satisfactory levels of accuracy, reaching up to 98%, contemporary ECG diagnosis systems utilizing machine learning face challenges in developing a fully controlled ECG multi-class categorization diagnosis system. The quest for precision and reliability in a multi-class categorization system remains a formidable challenge that requires continued research and innovation.
3. Proposed Methodology
This research introduces an innovative automated detection methodology engineered to enhance the accuracy of congenital heart disease (CHD) detection. This novel approach leverages a sophisticated amalgamation of Convolutional Neural Networks (CNNs), Bidirectional Long Short-Term Memory (BiLSTM) networks, and Attention Mechanisms (AMs). Each component plays a pivotal role in fortifying the model’s ability to accurately identify CHD cases. The CNN component, a fundamental pillar of this methodology, is meticulously designed to focus on the salient characteristics inherent in the input data, with particular emphasis on capturing details within the line of sight. This feature engineering approach proves to be exceptionally advantageous in the context of CHD detection, as it empowers the model to discern intricate patterns and subtle nuances within the data, thus augmenting its diagnostic capabilities. Complementing the CNN, the BiLSTM component is seamlessly integrated into the model architecture. The BiLSTM networks are instrumental in capturing temporal dependencies within the data. Specifically, they conduct an in-depth analysis of preprocessed electrocardiogram (ECG) signals, which are pivotal in CHD diagnosis. By considering the temporal dynamics of the data, the BiLSTM networks enhance the model’s ability to adapt and make accurate predictions, especially when dealing with time-series data like ECG signals.
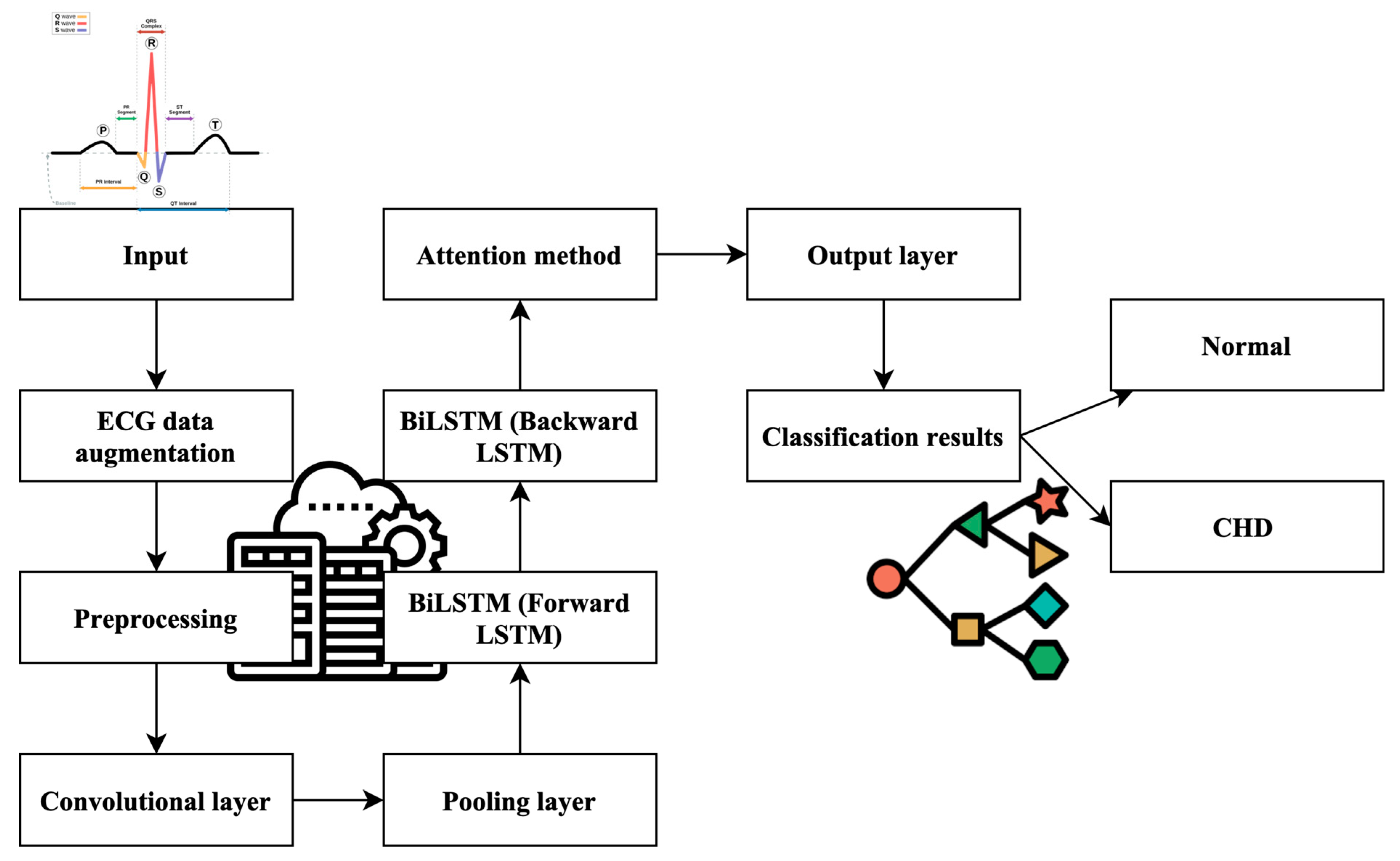
Furthermore, the model incorporates the indispensable Attention Mechanisms (AMs), a pivotal element in refining its predictive accuracy. These AMs introduce a mechanism to emphasize the significance of past time series data and state information features, allowing the model to incorporate historical context into its final predictions. This strategic utilization of AMs significantly influences the model’s adaptive capabilities and contributes to more precise CHD predictions. The culmination of these components results in a robust CHD prediction model, driven by advanced machine learning techniques. Figure 1 visually encapsulates the architecture of this model, highlighting the distinctive roles played by a CNN, BiLSTM, and AM in the pursuit of accurate CHD detection. This comprehensive approach signifies a significant advancement in the field, with the potential to revolutionize the accuracy and efficiency of CHD diagnosis.
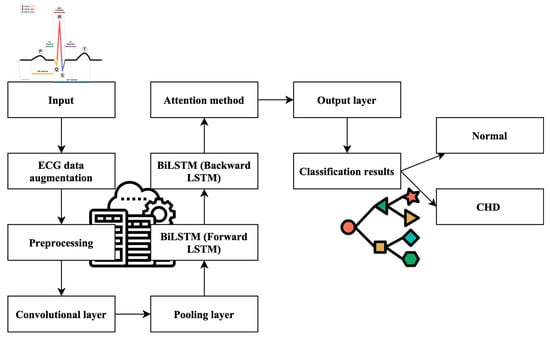
Figure 1.
Work process of the proposed ML-CHDPM. Geometric symbol for classification and line for process flow.
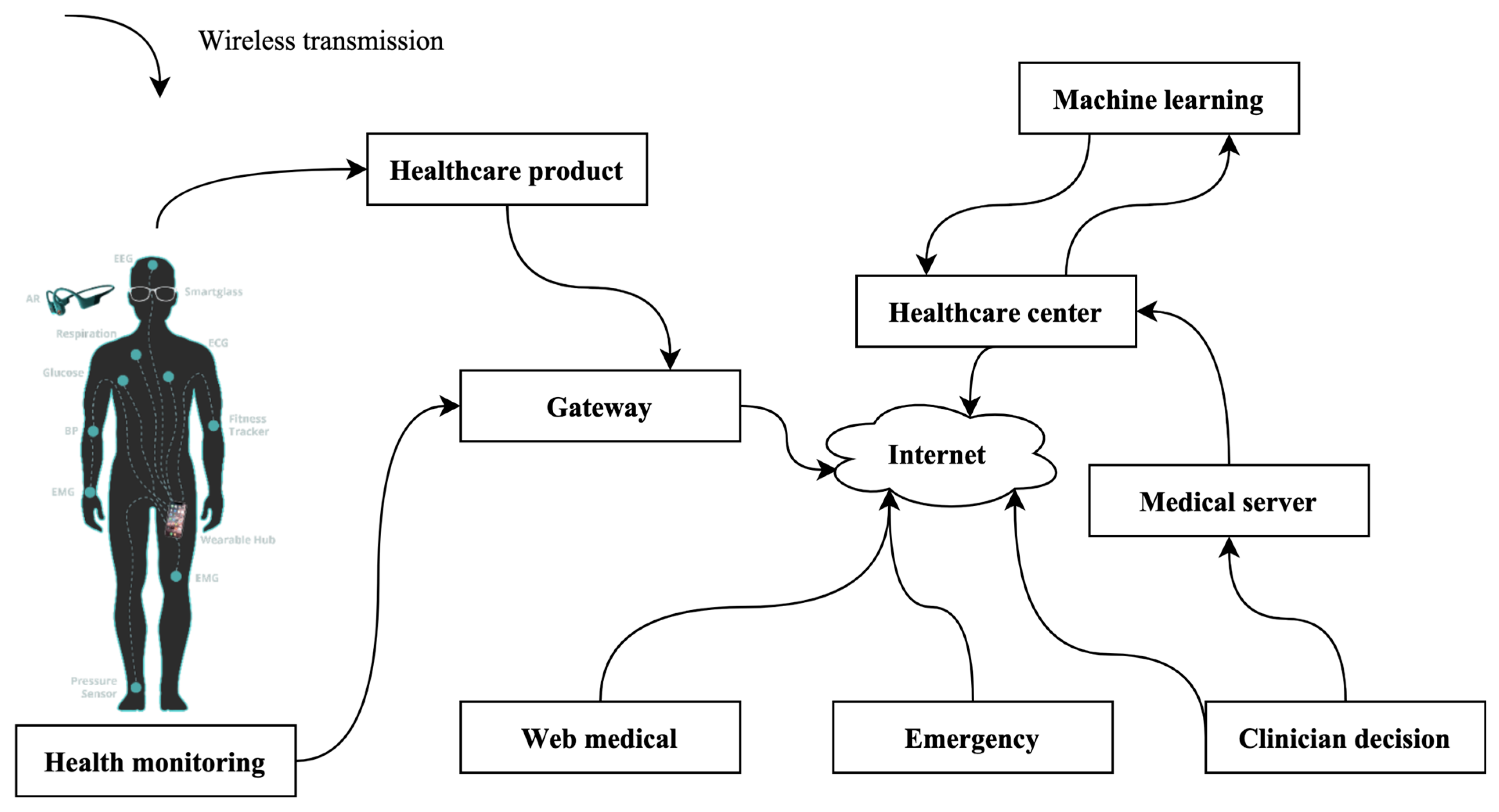
3.1. Data Source Collection from IoMT
Data for this study are drawn from the burgeoning field of the Internet of Medical Things (IoMT), a paradigm depicted in Figure 2. For this innovative approach, an array of sensors is strategically deployed and meticulously placed on the cardiac muscle, serving as vigilant sentinels to capture and scrutinize electrocardiogram (ECG) signals. Additionally, the research explores the potential utilization of alternative sensors designed to gather electroencephalogram (EEG) indications. It is imperative to emphasize that this investigation maintains a laser focus on the automated identification of congenital heart disease (CHD). Consequently, the study exclusively relies on the rich dataset derived from ECG signals. While the availability of alternative sensors for EEG data collection is acknowledged, their utilization remains beyond the scope of the current research. This rigorous and specialized focus on ECG signals underscores the study’s commitment to delivering precise and effective solutions for CHD diagnosis.
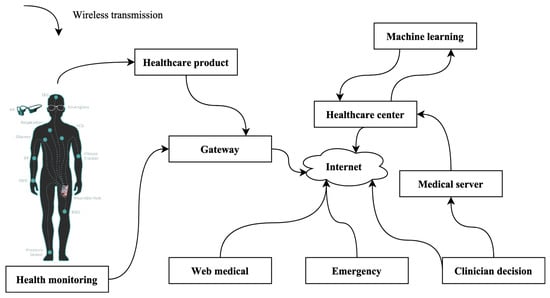
Figure 2.
IoMT-based environment.
In this study, our primary focus is on pregnant women, recognizing the exceptional significance of detecting congenital heart disease (CHD) during pregnancy. CHD is a complex and potentially life-threatening condition, and early detection is paramount for effective management. This importance is magnified during pregnancy, where the health and well-being of both the expectant mother and the developing fetus are intricately linked. Our research dataset comprises two distinct groups, both consisting exclusively of pregnant women. The first group includes 15 pregnant patients who have been medically diagnosed with CHD. The second group consists of 18 pregnant individuals who are considered to be in good health, as they exhibit normal sinus rhythm (NSR). Additionally, within the CHD group, we have identified a subset of pregnant patients who are dealing with severe congestive heart failure, further underscoring the critical nature of early CHD detection during pregnancy. To gather data for analysis, we meticulously collected electrocardiogram (ECG) signals from these pregnant individuals over an extensive monitoring period, approximately spanning 20 h. Each data entry in our dataset is composed of a pair of ECG channels, recorded at a frequency of 250 Hertz. This comprehensive data collection process ensures that we have access to a wealth of information that can shed light on the cardiac health of pregnant women.
The significance of our study becomes apparent in several key aspects:
- Maternal and Fetal Health: Pregnancy imposes unique physiological demands on a woman’s cardiovascular system. Detecting CHD during pregnancy is not only about the mother’s health but also about ensuring optimal fetal development. The health of both mother and child is intertwined, making accurate detection a matter of utmost importance.
- Timely Intervention: Identifying CHD in pregnant women at an early stage allows for timely medical interventions and the implementation of tailored healthcare strategies. These interventions can potentially prevent complications that might arise during pregnancy or childbirth.
- Advanced Technology: Our study leverages advanced machine learning techniques, tailored specifically to the pregnant population. By training our model on this exclusive dataset of pregnant women, we can capture intricate patterns and relationships that are highly relevant to this demographic.
- Unique Dataset: A distinctive feature of our study is the exclusive focus on pregnant women’s datasets. This focus bridges a critical gap in research and ensures that our findings are directly applicable to the population of pregnant individuals.
In conclusion, our research is poised to make a substantial contribution to the field of prenatal care. The advanced machine learning model we have developed, trained on data exclusively from pregnant women, demonstrates its effectiveness in accurately predicting and categorizing CHD cases within this specific population. Our aim is to enhance prenatal care, safeguarding the health and well-being of both expectant mothers and their unborn children.
3.2. Materials
The ECG signals used in this study were obtained from publicly available databases, specifically the PhysioBank repository. The severity of congenital heart disease (CHD) indications was classified using the New York Heart Association (NYHA) measure. According to this classification system, Category 1 represents a mild condition with no discernible restriction on exercise, while Category 2 indicates a benign condition with slight movement restrictions. In Category 3, there is a moderate level of physical activity with significant limitations, and Category 4 signifies a severe impairment that completely restricts physical activity.
The ECG data related to CHD used in this study are classified under the Category 3 and 4 groups. Four distinct databases, namely Categories A, B, C, and D, were employed in this study. Categories A and B consist of complete ECG information that needs to be balanced, whereas Categories C and D contain equal amounts of ECG information. From the complete dataset, an average of thirty thousand ECG information points were randomly extracted for Categories C and D.
3.3. Preprocessing
Preprocessing of the ECG data in this research study involved several steps to ensure data quality and consistency. The databases used, namely Fantasia, ECG, and MIT-BIH Normal Sinus, had different sampling rates of 250 Hz and 128 Hz, respectively. To maintain uniformity, the signals obtained from the National Solar Radiation Database were upsampled to 250 Hz.
To facilitate further analysis, the ECG records were segmented into 2-s intervals without simultaneous R-peak identification. Each segment consisted of 500 samples, corresponding to a duration of 2 s. Before being inputted into the system, the ECG signals underwent regularization through Z-score standardization. This standardization process involved adjusting each data point to have a mean of zero and a standard deviation of one. Applying these preprocessing techniques ensures that the ECG data are standardized and ready for subsequent analysis. These steps help enhance the accuracy and reliability of the findings by addressing sampling rate discrepancies and normalizing the data for consistent comparison and evaluation.
3.4. LSTM Architecture
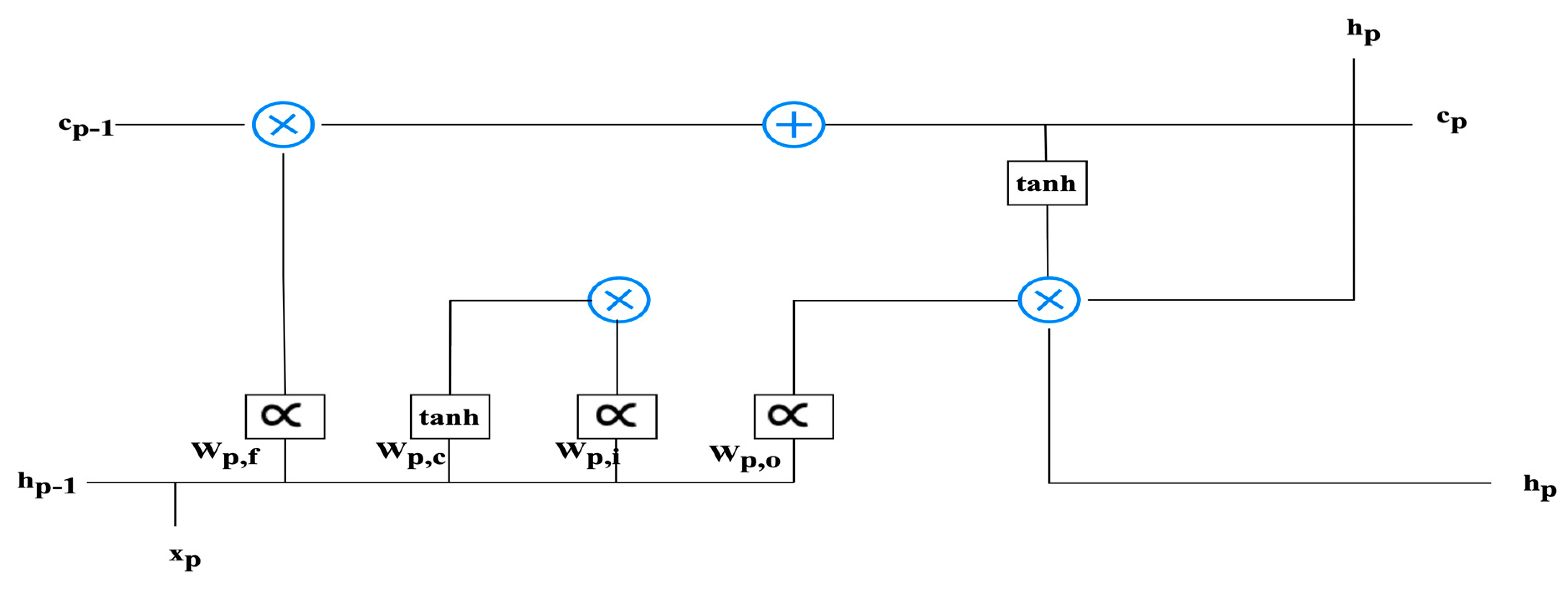
LSTM is a variant of the artificial neural network architecture that incorporates the dual memory systems, known as long-term and short-term memory. Time-series information is prevalent in various fields and is often addressed using LSTM as a solution. LSTM is a recurrent neural network architecture that is equipped with trainable memory cells. The LSTM cell consists of two distinct states: the long-term cell status (c_t) and the short-term cell status (h_t). These states represent the intermediate results of the cell at a given time period, denoted by t. LSTM cells possess three modifiable gates: the input gate, output gate, and forget gate. These gates control the flow of information into and out of the LSTM cell. One of the remarkable features of LSTM cells is their ability to retain information about specific values over multiple time periods. The gates serve as cellular components that regulate the flow of information within the cell. During training, the LSTM cells learn to determine which data are relevant to retain and which to discard. In summary, LSTM architecture is designed to handle time-series data by incorporating long-term and short-term memory components. The modifiable gates enable the LSTM cells to control the flow of information, allowing for the retention and utilization of relevant data during training.
Within the architecture of Long Short-Term Memory (LSTM) cells, they play a pivotal role in serving as conduits for the seamless transmission of data. These data transmissions are meticulously regulated by gates, which act as gatekeepers, determining the permissibility of information based on the current cellular context. Specifically, the neural network architecture governing the behavior of the forgetting gate within the LSTM cell is structured as a fundamental single-layer design. The structural configuration of the LSTM cell is visually depicted in Figure 3, providing an insightful overview of its inner workings. To elucidate the activation process of the forgetting gate, Equation (1) comes into play:
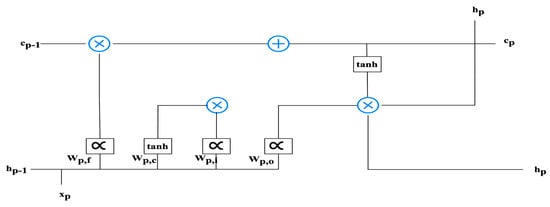
Figure 3.
The structure of the LSTM.
The notation represents the sequential input information, while denotes the result of the preceding LSTM block. signifies the memory of the previous LSTM block. The bias vector is represented by , the weight vectors are denoted by , and the activating function is . The activating functions that are most commonly utilized are tanh and sigmoid. The mathematical expression representing the sigmoid activating function is presented in Equation (2).
The input gate () is a component that utilizes the recently introduced memory’s hyperbolic tangent activating function and the preceding memory blocks. The operations are executed using Equations (3) and (4).
shows the sequential input information, shows the result of the preceding LSTM block. denotes the memory of the previous LSTM block, and denotes the memory of the LSTM block. The bias vector is shown by , , the weight vectors are shown by , and the activating function is . The LSTM block’s results designate the exit gate. The LSTM results are computed using Equations (5) and (6).
shows the sequential input information, shows the result of the preceding LSTM block. denotes the memory of the LSTM block, and the output layer is marked . The bias vector is shown by , the weight vectors are shown by , and the activating function is .
3.5. Attention Mechanisms
The utilization of attention systems is prevalent in diverse domains, including but not limited to natural language processing and image detection. The process in question resembles the visual system observed in the human brain. Its objective is to accentuate more crucial data while screening out irrelevant data not pertinent to the current task. Numerous empirical investigations have demonstrated the efficacy of utilizing this process to enhance the discernment of data, and as such, it has been incorporated into the suggested structure. The system of attention calculations is established in Equations (7)–(10).
The weight matrix is denoted as , while the bias vector is represented by . The symbol is employed in evaluating the resemblance of and the context vector . The concealed layer is denoted , the transpose function is denoted , and the computation result is denoted .
3.6. Training Procedure of CHD Diagnosis System
The training procedure comprises the following subsequent primary stages.
Step 1: The data input necessary to develop the proposed ML-CHDPM system is obtained.
Step 2: Data standardization is a technique utilized to enhance the efficacy of a framework in the presence of significant variations in the input information. The z-score approach is incorporated into the input normalization process, as outlined in Equation (10).
The symbols and denote the input information’s normalized value and standard deviation. and denote the input information and its corresponding average.
Step 3: Layers of the proposed model are linked with weights and biases to initialize the networks.
Step 4: The computational process within the CNN layer is systematically transmitted via the convolutional and pooling layers that are present within the CNN layer. Within this stratum, the process of obtaining features is achieved from the input information, and the resultant output values are ascertained.
Step 5: The computation of the hidden layer associated with the BiLSTM layer is contingent upon the output information generated by the CNN layer, which is utilized to ascertain its resultant value.
Step 6: The AM layer is employed to compute the output information of the BiLSTM layer to ascertain its output value.
Step 7: The computation of the model’s results is contingent upon the result value of the AM layer.
Step 8: The calculation of error involves determining the disparity between the genuine value of the data grouping and the established computation value generated by the result of the layer.
Step 9: Determine whether the categorization process has reached its termination condition. Within this particular context, it is deemed that the criteria for dismissal are achieved under three circumstances: firstly, when the rate of forecasting error falls below the designated threshold value; secondly, upon completion of a certain number of phases; and thirdly, when the weight is lower than the particular threshold value. The training process is deemed to be concluded upon fulfilling any previous circumstances.
Step 10: The training procedure reverts to Step 4 to resume the training procedure, following the update of biases and weights for each layer and the propagation of the computed error in the reverse direction.
The diagnosis algorithm for CHD detection is shown in Algorithm 1.
| Algorithm 1 CHD detection method |
| Input: The input training data Output: The trained model Step 1: Derive input data derivedData <- deriveInputData(trainingData) Step 2: Apply data standardization standardizedData <- applyDataStandardization(derivedData) Step 3: Initialize weights and biases initializeWeightsAndBiases() Step 4: Training loop while not terminationConditionMet() do: Step 4.1: Forward pass forwardPass(standardizedData) Step 4.2: Compute the hidden layer output hiddenOutput <- computeHiddenLayerOutput() Step 4.3: Apply the AM layer to compute the output output <- applyAMLayer(hiddenOutput) Step 4.4: Compute the final output finalOutput <- computeFinalOutput(output) Step 4.5: Compute the error error <- computeError(finalOutput, trainingData) Step 4.6: Check termination criteria if terminationCriteriaMet(error) then: exit the loop Step 4.7: Update weights and biases updateWeightsAndBiases() Step 5: Return the trained model |
The “Train Model” algorithm takes an input training dataset and outputs a trained model. The algorithm consists of several steps. Firstly, the input data are derived and then standardized for preprocessing. Weights and biases are initialized to set up the model. The training loop begins, where the forward pass is performed to compute the hidden layer output, followed by applying an activation function to generate the output. The final output is computed, and the error is calculated by comparing it with the desired training data. Termination criteria are checked, and if the error meets the criteria, the loop is exited. Otherwise, weights and biases are updated to optimize the model. This loop continues until the termination condition is satisfied. Finally, the trained model is returned as the output of the algorithm.
3.7. Classification Method of CHD
Accurately identifying and categorizing CHD pathology is contingent upon the training procedure and its successful culmination. The procedural details of the detection procedure are explicated as follows.
Step 1: The first step considers the input information necessary to identify and categorize CHD.
Step 2: The input information is standardized.
Step 3: The ML-CHDPM system that had undergone training is utilized to potentially process the standardized information as input to determine its corresponding output value.
Step 4: Restoring data normalization to its initial value is accomplished using Equation (11).
denotes the input information’s standard deviation. and denote the output result and the input information, respectively.
Step 5: The resultant value is obtained as the outcome of a categorization issue that distinguishes ECG commands into typical and CHD disease indications.
The process is shown below.
- Input Sequences:
The LSTM block receives sequential data representing the clinical and demographic features pertinent to CHD. The input sequence can be denoted as , where each reflects the input at a specific time step i.
- Input Embedding:
It is common practice to utilize an embedding layer before feeding the input sequence into the LSTM block. The layer in question facilitates the mapping of input values onto a continuous vector space, enabling the capture of connections between various input characteristics. The input sequence X is transformed into a novel illustration using an embedding layer. This new sequence illustration is denoted as E and comprises individual elements .
- LSTM Architecture:
The LSTM block comprises memory cells that retain information throughout the sequential inputs. The LSTM architecture should consist of three gates, namely the input, forget, and output gates, which regulate the data flow within each cell.
- Hidden State Initialization:
The LSTM block necessitates an initial concealed state and the beginning cell state at the onset of the sequences. Usually, these states start as vectors consisting of zeros or learned variables.
- LSTM Computation:
The LSTM block sequentially procedures the input sequence, iteratively modifying the concealed state and cell state at each period step.
- The computation of gates involves the input gate (), forget gate (), and output gate () for every time step x. These gates are calculated based on the present input () and the before hidden state ().
- Updating the memory cell () involves the integration of the input gate, forget gate, and the last memory cell (). The information storage process is governed by the input gate, which selects the relevant data to be retained, and the forget gate, which decides the data to be disregarded.
- The computation of the hidden state () is contingent upon the revised memory cell () and the output gate () in the process of hidden state update. The latent state conveys pertinent data from the input sequence to the following temporal intervals.
- The computation of the output at a given time step x () involves the application of a linear modification to the hidden states (), followed by the potential application of an activating function. Applying an activating function depends on the particular categorization task specifications.
- Final Time Step:
Upon completion of the input sequence preparation, the LSTM block generates a conclusive hidden state, denoted as , and a result corresponding to the final time step.
- Classification Layer:
The LSTM block’s result is subsequently inputted into the categorization layer, which comprises one or more densely connected layers. The layers acquire the ability to establish a correlation between the output of the LSTM and the intended categorization labels, thereby encapsulating intricate patterns within the dataset.
- Training and Optimization:
The labeled database of CHD instances is utilized for training the LSTM block in conjunction with the categorization layer. The LSTM block and categorization layer variables are optimized using gradient descent and backpropagation through duration to reduce a suitable loss coefficient.
- Inference:
Upon completing the training process of the LSTM block, it becomes viable to classify novel and unobserved data. The LSTM block is utilized to process the input sequence, and subsequently, the anticipated CHD categorization is obtained by passing the result through the categorization layer.
3.8. Training and Testing of the Model
The Xavier initialization technique is commonly employed to initialize the weights of an algorithm. The ML-CHDPM algorithm utilized in the present research was updated using backpropagation with a batch limit of 10. The cross-entropy functioning is utilized for the assessment of network loss. The ML-CHDPM architecture was trained with specific variables to optimize its diagnostic efficiency. These variables include lambda (L1 normalization) set to 0.2, a learning rate 3 × 10−4, and a momentum value of 0.3. The variables hinder the overfitting of the information through normalization, facilitate data convergence via learning rate, and modulate the pace of learning through movement.
The study utilizes the Leaky Rectifier Linear Unit (LeakyRelu) as the activating function for sections 1, 3, 5, 7, 9, and 10, as depicted in Equation (12). Additionally, layer L1 is carried out with the SoftMax operation, as presented in Equation (13).
The function is represented by the notation. At the same time, indicates the probability of transportation over the complete set of classes, and x signifies the total number of types. The present study employs a categorized ten-fold cross-validation approach (). The ECGs of the four distinct groups have been partitioned into ten discrete sections. The model is trained using nine parts, while the remainder is reserved for testing. Each partitioned segment comprises a comparable proportion of the classroom target as the complete dataset. This study consists of ten repetitions.
3.9. Model Evaluation
To assess the effectiveness of the case segmentation approach, the Mask-RCNN results are subjected to six measurements for validation and evaluation. These indicators include categorization loss, segmentation losses, identification loss, Jaccard indicators, Dice coefficient resemblance for segmentation, and mean average accuracy for identifying objects.
The Jaccard resemblance coefficients, known as the DCS, are utilized to assess the resemblance and variation of sets of specimens. In this instance, the objective is to evaluate the efficacy of predictive pictures accompanied by comprehensive truth labeling. Equation (14) depicts the DCS.
The equation for calculating the expected outcomes, denoted by , in a given number of operates, M, is dependent on the corresponding truth label, . The DCS’s pixel index, which falls within the range of [0, 1], quantifies the likelihood of correspondence between the anticipated and actual images.
The quantitative evaluation of the Mask R-CNN approach, which was trained and verified, was conducted using mean average precision (mAP). The mAP is commonly used to evaluate object detection theories. The mAP values for the different groups were calculated and subsequently averaged. While the model could identify multiple objects, the definitive classification of said objects was occasionally ascertainable. Even with the accuracy of the anticipated classification for an object or examples, the output metric necessitates an evaluation of the model’s spatial localization performance within the image. The widely utilized mAP is represented by Equation (15).
The variable represents the aggregate of distinct classes, while denotes the overall count of pixels belonging to class x. The time is denoted .
This section outlines a comprehensive methodology for the timely identification and diagnosis of cardiovascular ailments by utilizing data derived from the IoMT. The implementation uses an LSTM design and Attention Mechanism to effectively capture the data’s temporal relationships and significant features. The precision of disease categorization is further improved by developing a diagnosis model utilizing ML-CHDPM. This section comprises a comprehensive assessment and juxtaposition of diverse models using the Heart Disease UCI and Cardiovascular Disease databases, thereby underscoring the efficacy of the proposed approach.
4. Simulation Analysis and Outcomes
The simulation analysis section provides a comprehensive overview of the experiments conducted to evaluate the effectiveness of the proposed strategy. The article describes the datasets used and the simulation measurements employed, highlighting the findings and conclusions attained. It demonstrates the efficacy of the proposed approach in identifying and assessing cardiovascular disease.
4.1. Datasets
Our research methodology is intricately woven around the utilization of diverse datasets, each carefully chosen to address specific facets of our investigative objectives. The Heart Disease UCI dataset [21,22,23,24] serves as a foundational element, standing out with its comprehensive array of 76 attributes that encompass a wide range of patient-related information. From this dataset, we identified a subset of attributes crucial to our analysis. To uphold the privacy of individuals, meticulous anonymization measures were implemented, replacing any personally identifiable information, such as patient identities and security numbers, with synthetic values. Our focus within this dataset is specifically directed towards the subset of attributes deemed functionally significant for our research objectives.
In contrast, the Cardiovascular Disease Dataset [25] emerges as a cornerstone in the execution of our proposed hybridized technique. This dataset comprises a voluminous repository of 70,000 individual patient records, each intricately characterized by 11 distinct features. Despite the initial dataset presenting an extensive pool of 210 factors, we conducted a rigorous curation process to select only the most relevant eight features for our analytical framework. Within this dataset, 93 individuals received a diagnosis of coronary artery disease, while 116 individuals were identified as free from cardiovascular disease. The diagnostic categorization is represented by a binary variable, where 0 indicates the absence of coronary artery disease, and 1 signifies its presence. The deliberate selection and curation of features within this dataset are pivotal, aligning meticulously with our research objectives and ensuring a nuanced exploration of factors integral to our study.
A crucial shift in our research direction involves an exclusive focus on 5-min ECG data sourced from pregnant women. This strategic refinement aligns our study more precisely with its overarching objectives, allowing for an in-depth analysis of short-term heart rate variability (HRV) patterns. This adjustment is particularly critical in our pursuit of refining early detection methods for cardiovascular conditions during pregnancy, underscoring the dynamic nature of our research and ensuring a targeted exploration of factors specifically relevant to our evolving goals. The inclusion of these datasets forms the bedrock of our proposed methodology, providing the necessary breadth and depth for the development and validation of our novel hybridized technique.
Beyond these primary datasets, we also leverage the Fantasia dataset—a pivotal component contributing to the diversity and richness of our analysis. Fantasia, a publicly available dataset, comprises a curated collection of electrocardiogram (ECG) recordings obtained from a diverse group of subjects. Notably, the dataset includes recordings from individuals with various cardiac conditions, offering a broad spectrum of cardiac signals that align with the scope of our research objectives.
The MIT-BIH Arrhythmia Database further enhances the breadth of our analysis. Widely recognized and utilized in cardiac research, this database includes annotated long-term ECG recordings from a variety of subjects, capturing a range of cardiac arrhythmias and conditions. The meticulous annotations within this dataset provide a valuable resource for the development and validation of algorithms aimed at detecting and classifying arrhythmias, aligning seamlessly with the goals of our study.
Together, these datasets contribute to the richness and diversity of our research, providing a robust foundation for the development, validation, and refinement of our proposed methodologies.
4.2. Parameter Settings and System Configuration
The Bi-LSTM system was configured with 8 epochs, a concealed layer size of 150, and a dropout rate of 0.2. The Adam optimization algorithm was employed for network optimization, using a fixed iteration count of 50 and a population size of 50. The proposed model for coronary heart disease (CHD) identification was implemented on a computational system equipped with an Intel i7 processor, 16 GB memory, and a 6 GB graphics card.
4.3. Metrics
The analysis utilizes the following metrics: true positive (Tpo), true negative (Tne), false positive (Fpo), and false negative (Fne). These metrics are employed in the analysis section to assess the accuracy of the predictions. Accuracy measures the count of correctly predicted outcomes for a given set of input specimens, calculated using Equation (16).
Precision is a reliable metric in scenarios where false positives are high. Its computation is shown in Equation (17):
The sensitivity or recall metric is determined by dividing the number of true positives by the total of true positives and false negatives. The recall is shown in Equation (18).
The measurement of specificity pertains to instances wherein the true state of the condition is absent, particularly in the context of negative attack categorization. The calculation is performed according to Equation (19).
The false positive rate (FPR) is a metric that quantifies the proportion of misclassified samples that were predicted as negative, despite belonging to the positive category. It is calculated using Equation (20).
The false negative rate (FNR) is a metric that quantifies the proportion of misclassified samples that belong to the positive class. It is calculated using Equation (21).
5. Experimental Results
In the results section of our study, we aim to provide a detailed exposition of how each component of our innovative framework contributes to enhancing the accuracy of congenital heart disease (CHD) detection. This analysis is essential for a comprehensive understanding of the role played by each element in achieving our research objectives.
Our framework integrates three key components: Convolutional Neural Networks (CNNs), Bi-directional Long Short-Term Memory (BiLSTM) networks, and Attention Mechanisms (AMs). Each of these components plays a pivotal role in fortifying the model’s ability to accurately identify CHD cases.
The CNN component, a foundational pillar of our methodology, excels at feature extraction by focusing on salient characteristics within the input data. In the results section, we delve into how CNNs effectively capture intricate patterns and subtle nuances within the data. This detailed feature engineering process significantly enhances the model’s diagnostic capabilities, ultimately leading to improved CHD detection. Complementing the CNN, our model incorporates BiLSTM networks, which are instrumental in capturing temporal dependencies within the data. In this section, we elucidate how BiLSTM networks conduct an in-depth analysis of preprocessed electrocardiogram (ECG) signals, a critical aspect of CHD diagnosis. We highlight how considering the temporal dynamics of the data empowers the model to adapt and make accurate predictions, especially when dealing with time-series data like ECG signals.
Furthermore, we emphasize the role of Attention Mechanisms (AMs) in refining predictive accuracy. In the results section, we elaborate on how AMs dynamically assign varying degrees of importance to past time series data and state information features. This strategic use of AMs enhances the model’s adaptive capabilities, contributing to more precise CHD predictions. By providing a detailed account of how each part of our framework contributes to the results, we offer a nuanced understanding of their individual and collective impact on CHD detection. This analysis underscores the significance of our innovative approach and its potential to transform the field of CHD diagnosis. In this section, the simulation analysis and results using the proposed method are compared with those of existing techniques.
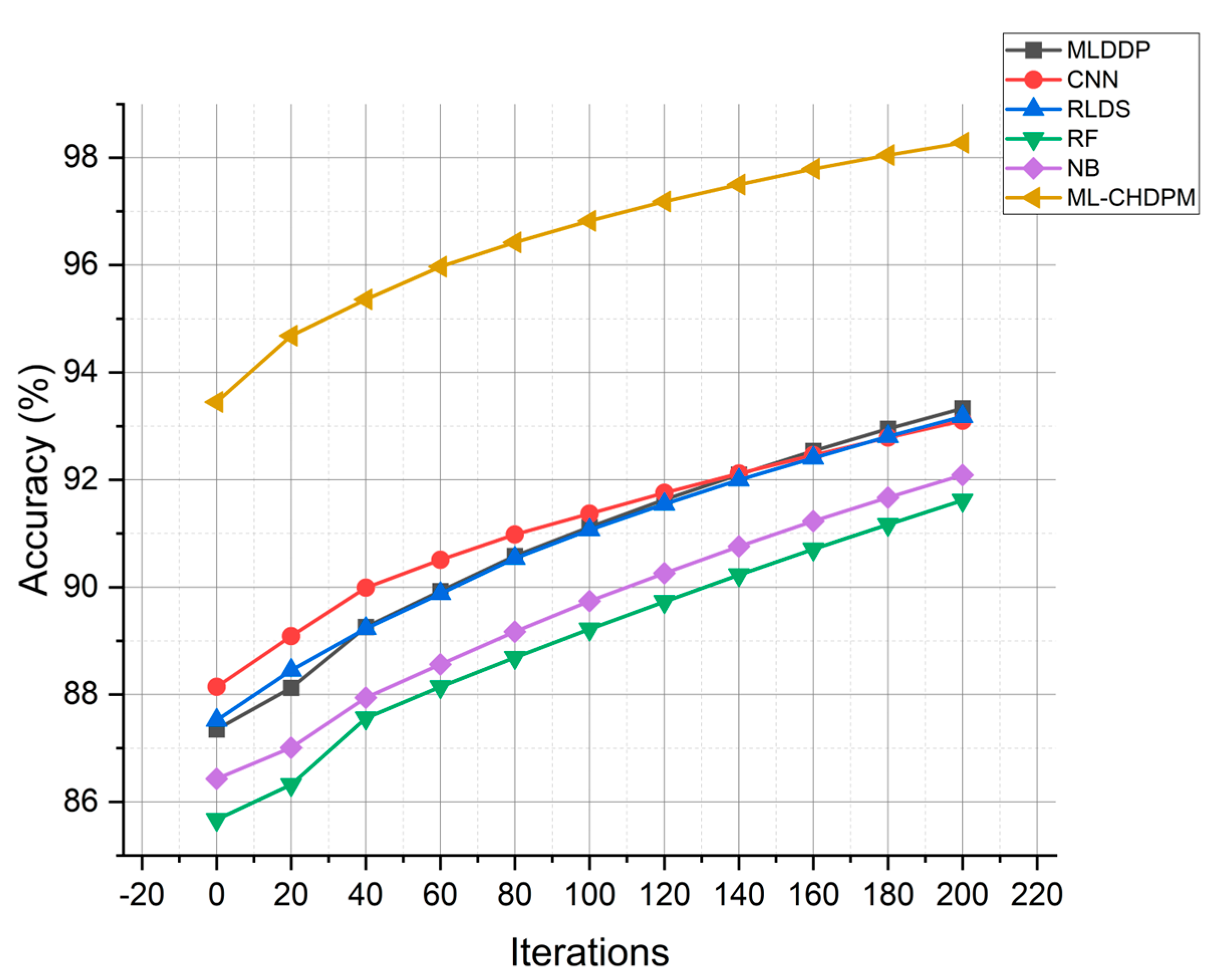
The accuracy results of various approaches, including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM, across multiple iterations from 0 to 200, are visually depicted in Figure 4. The mean accuracy values for these different methods are as follows: MLDDP (91.07%), CNN (91.39%), RLDS (91.23%), RF (89.04%), NB (89.19%), and our novel ML-CHDPM method introduced in this study (96.51%). It is evident that the ML-CHDPM outperforms alternative methodologies, demonstrating its ability to achieve significantly higher levels of accuracy. This observed improvement can be attributed to the successful integration of the LSTM framework, the Attention Mechanism, and the tailored training regimen within the ML-CHDPM diagnostic model. These techniques empower our approach to effectively extract and leverage essential characteristics from the input data, resulting in enhanced precision of predictions and an exceptional overall efficacy in diagnosing cardiovascular ailments.
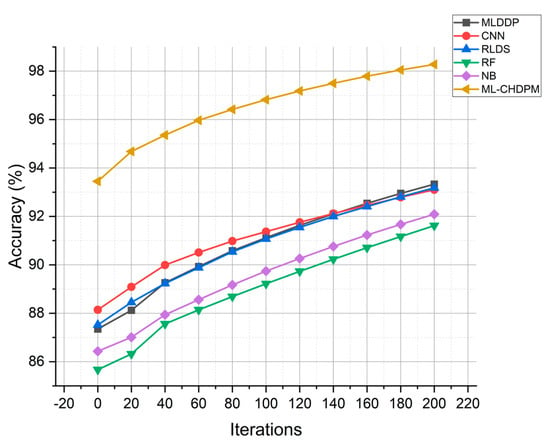
Figure 4.
Accuracy evaluation for CHD classification.
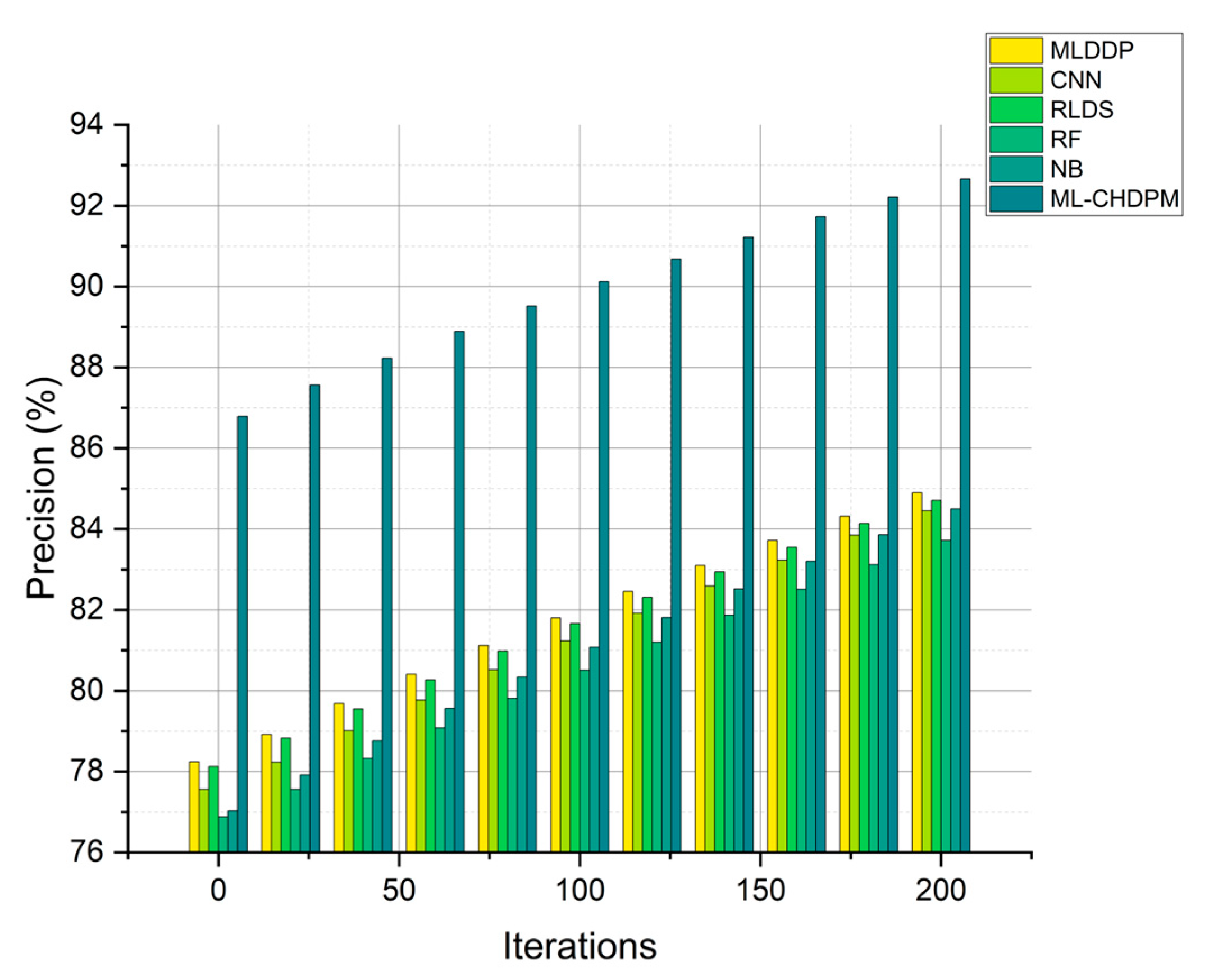
Precision results for multiple methods, including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM, across a range of iterations from 0 to 200, are graphically presented in Figure 5. These precision values for each method were meticulously calculated, yielding the following results: MLDDP at 81.56%, CNN at 81.21%, RLDS at 82.38%, RF at 80.31%, NB at 80.97%, and our innovative ML-CHDPM at 89.14%. It is noteworthy that the ML-CHDPM model demonstrates a significant enhancement in precision when compared to alternative approaches. Consistently outperforming its counterparts, our model achieves an average precision rate of 89.14%. This exceptional precision improvement can be attributed to the strategic incorporation of the LSTM architecture, the Attention Mechanism, and the tailored training process within the ML-CHDPM diagnostic system. These techniques empower our approach to adeptly acquire and leverage informative characteristics from the input data, resulting in more accurate predictions and heightened precision in diagnosing cardiovascular ailments.
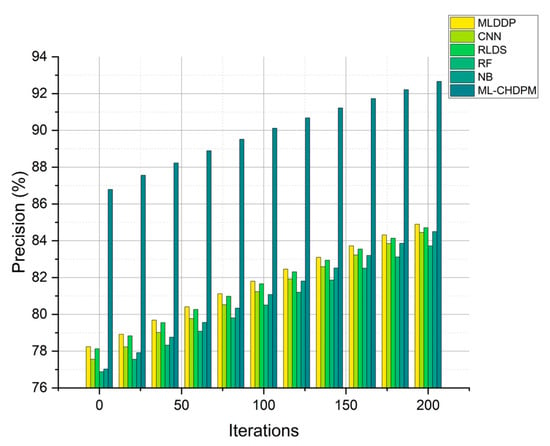
Figure 5.
Precision evaluation for CHD classification.
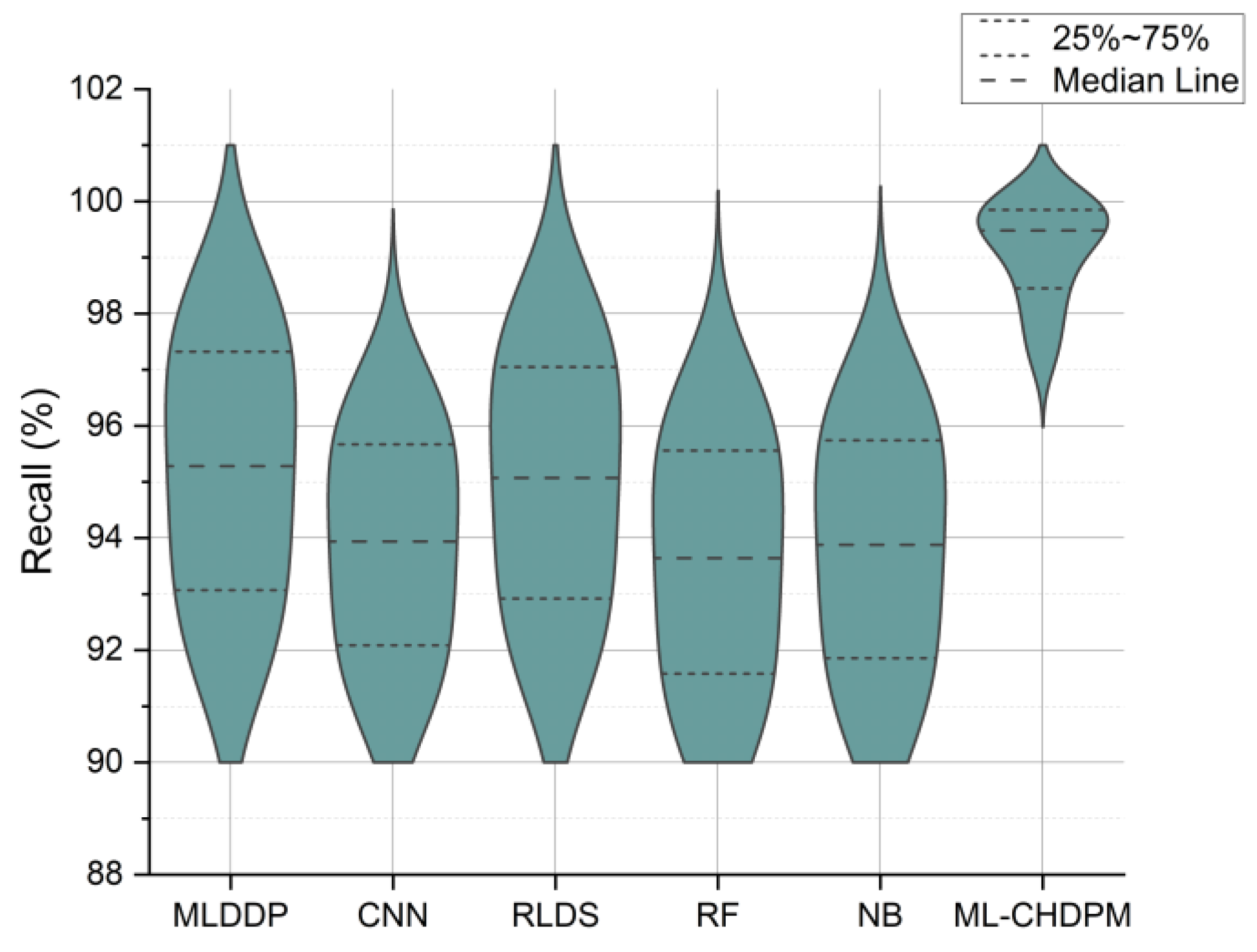
Figure 6 presents the recall results expressed as percentages across a range of iterations from 0 to 200, evaluating various methods, including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM. The mean recall rates for these methods are as follows: MLDDP (94.27%), CNN (93.25%), RLDS (94.02%), RF (92.56%), NB (92.97%), and the innovative ML-CHDPM (99.19%). Remarkably, the ML-CHDPM model showcases a significant enhancement in recall compared to alternative methodologies. It consistently achieves the maximum recall rate, averaging at an impressive 99.19%. These exceptional results can be attributed to the proficient utilization of the LSTM framework, the strategic incorporation of the Attention Mechanism, and the tailored training regimen within the ML-CHDPM diagnostic model. These techniques enable our methodology to adeptly grasp and leverage pertinent data from the input, resulting in enhanced prognostic precision and heightened identification of cardiovascular ailments.
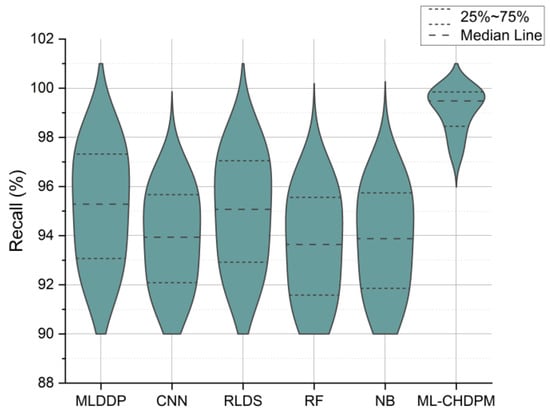
Figure 6.
Recall evaluation for CHD classification.
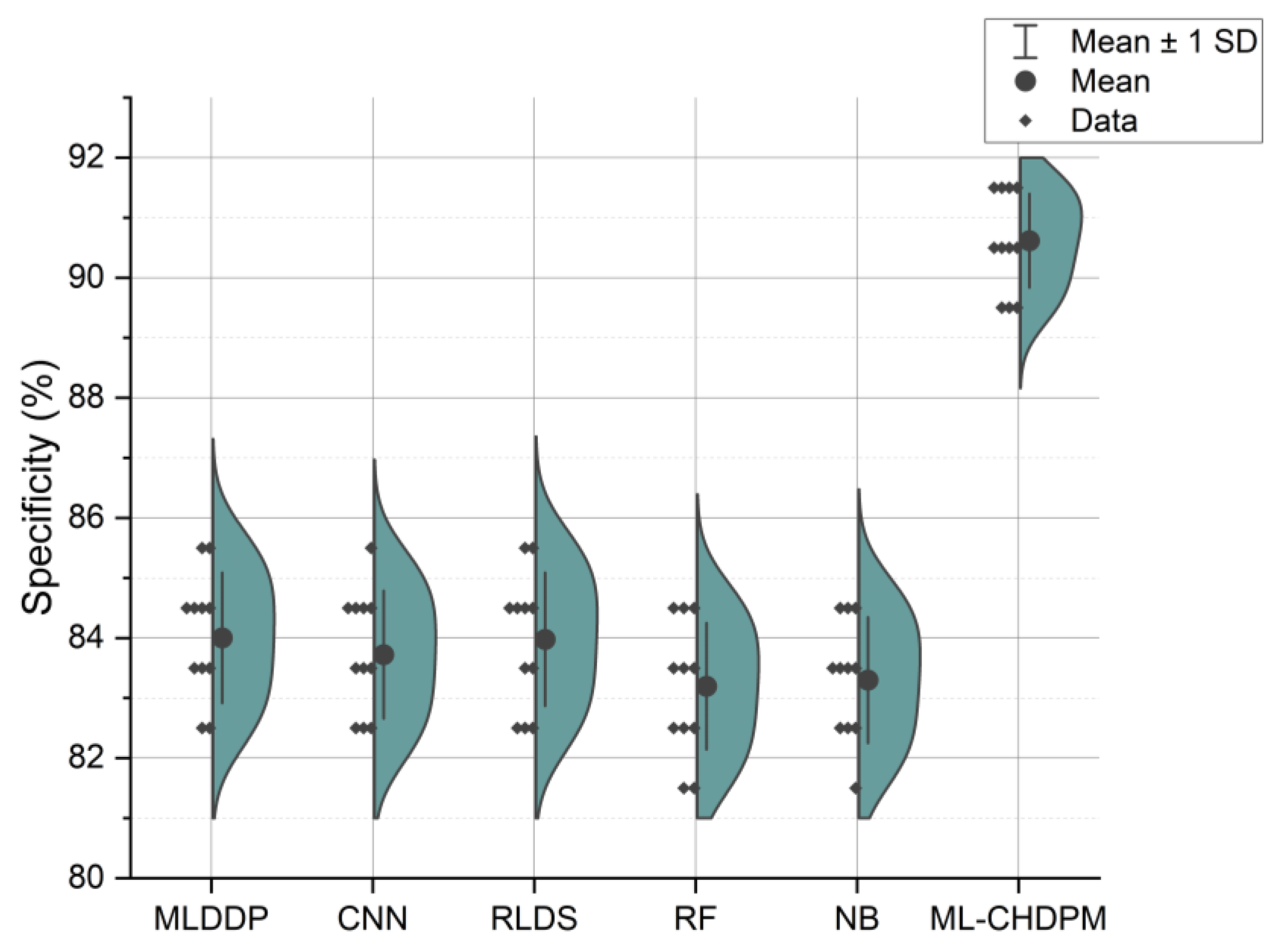
Figure 7 illustrates the specificity results presented in percentage values across a range of iterations from 0 to 200, employing various methods including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM. The mean specificity values for each of these methods are as follows: MLDDP (83.85%), CNN (83.16%), RLDS (83.93%), RF (82.05%), NB (82.34%), and our innovative ML-CHDPM method introduced in this study (90.11%). Notably, the ML-CHDPM model showcases a substantial enhancement in specificity when compared to alternative approaches. It consistently achieves the highest level of specificity, with an average of 90.11%. This improved efficacy can be attributed to the implementation of sophisticated methodologies, including the LSTM framework, the strategic incorporation of the Attention Mechanism, and the tailored training regimen within the ML-CHDPM diagnostic model. These techniques enable our method to efficiently acquire and utilize pertinent characteristics from the input data, resulting in enhanced prognostic precision and superior identification of cardiovascular ailments.
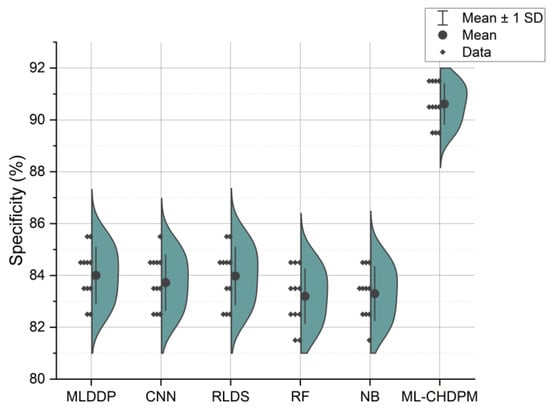
Figure 7.
Specificity evaluation for CHD classification.
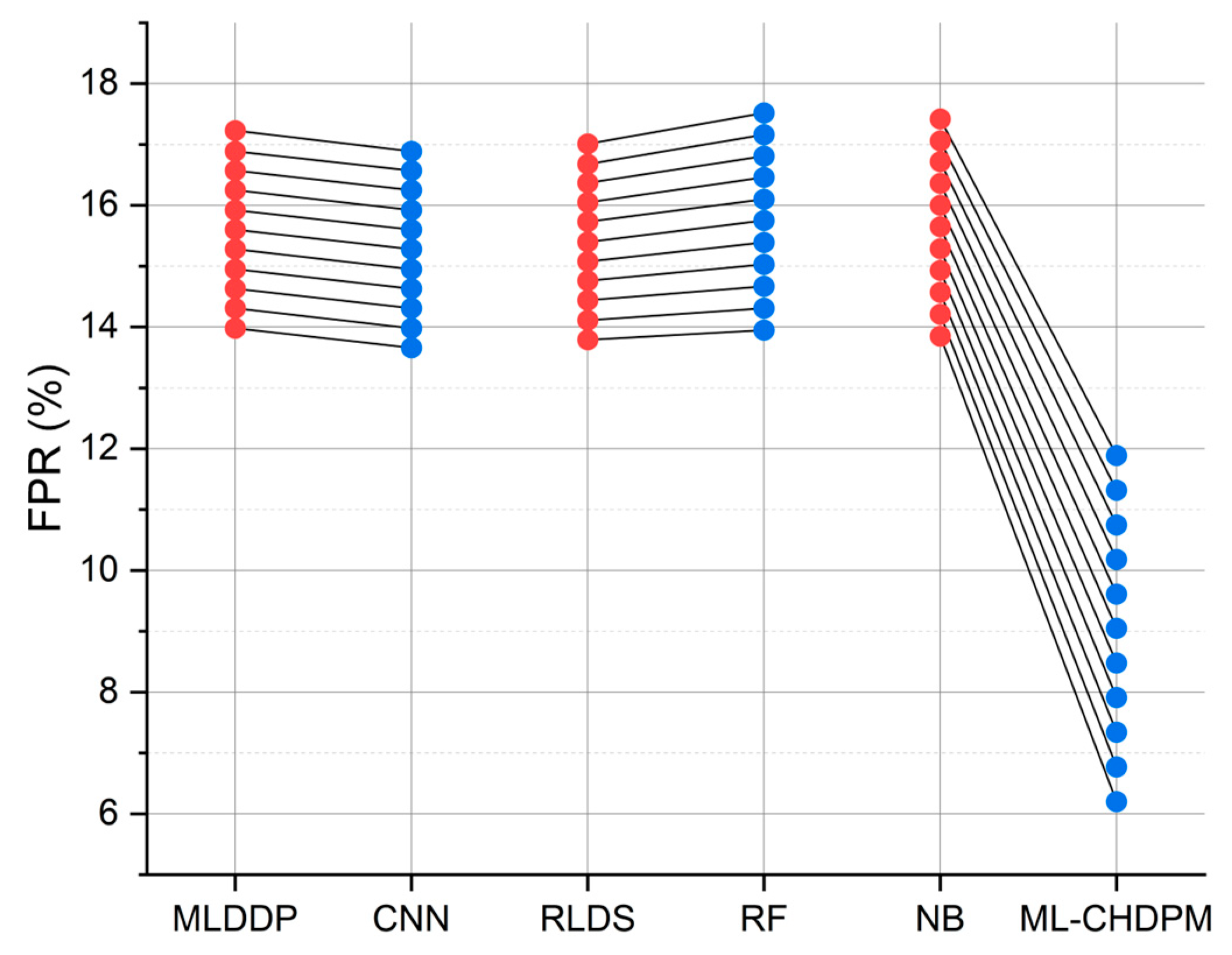
Figure 8 provides a comprehensive view of the false positive rate (FPR) outcomes achieved by various techniques, including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM, across different iterations ranging from 0 to 200. The mean FPR for each method is calculated as follows: MLDDP (15.69%), CNN (15.36%), RLDS (15.49%), RF (15.97%), NB (15.87%), and our innovative ML-CHDPM method introduced in this study (9.08%). Remarkably, the ML-CHDPM model showcases a significant reduction in the false positive rate compared to alternative methodologies. It consistently achieves the lowest rate of false positives, averaging at just 9.08%. This enhanced performance can be attributed to the incorporation of sophisticated methods, including the utilization of LSTM, the strategic integration of the Attention Mechanism, and the tailored training procedure within the ML-CHDPM diagnostic model. These methodologies enable our approach to effectively identify and minimize erroneous positive predictions, ultimately resulting in improved precision and reliability in the diagnosis of cardiovascular ailments.
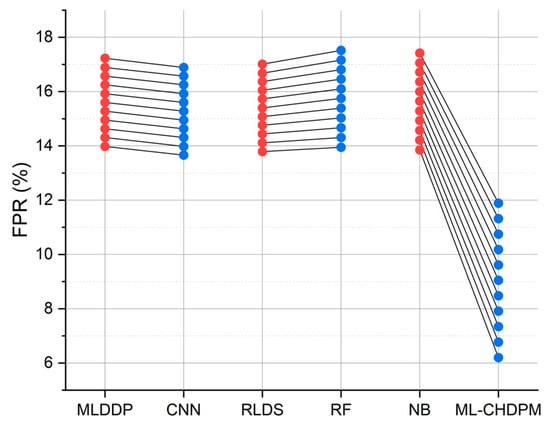
Figure 8.
FPR evaluation for CHD classification.
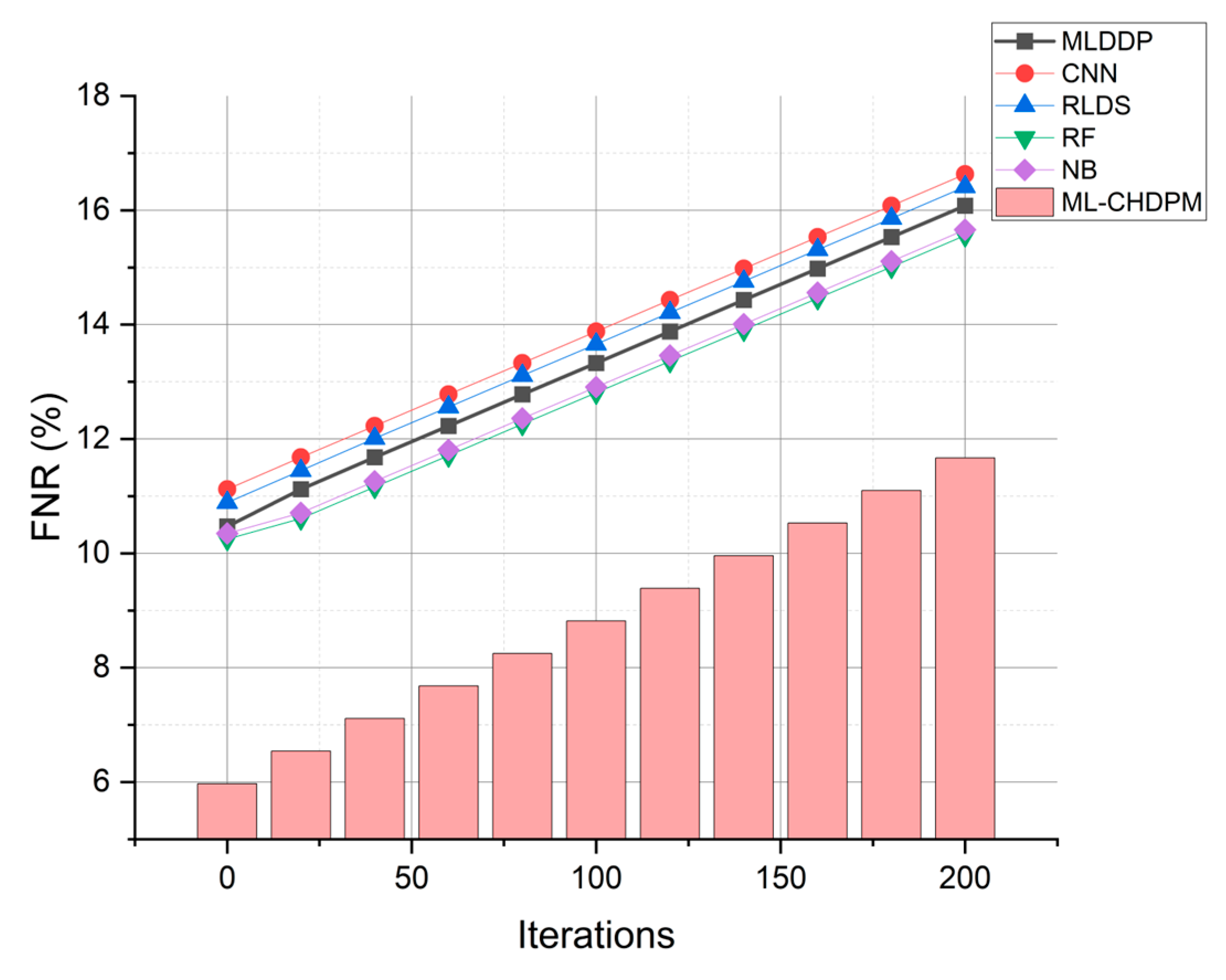
Figure 9 provides a comprehensive view of the false negative rate (FNR) outcomes achieved by various techniques, including MLDDP, CNN, RLDS, RF, NB, and our proposed ML-CHDPM, across a range of iterations spanning from 0 to 200. The false negative rates for each method are presented as follows: MLDDP (12.99%), CNN (13.56%), RLDS (13.34%), RF (12.79%), NB (12.89%), and our innovative ML-CHDPM (8.04%). Notably, the ML-CHDPM model demonstrates a substantial reduction in the false negative rate compared to alternative methodologies. It consistently achieves the lowest rate of false negatives, with an arithmetic mean of just 8.04%. This enhanced performance can be attributed to the integration of sophisticated methodologies, including the utilization of LSTM, the strategic incorporation of the Attention Mechanism, and the tailored training regimen within the ML-CHDPM diagnostic model. These methods enhance the efficiency of our proposed approach in precisely identifying and categorizing instances of cardiovascular ailments, resulting in a decreased occurrence of erroneous negative results and an overall improvement in diagnostic performance.
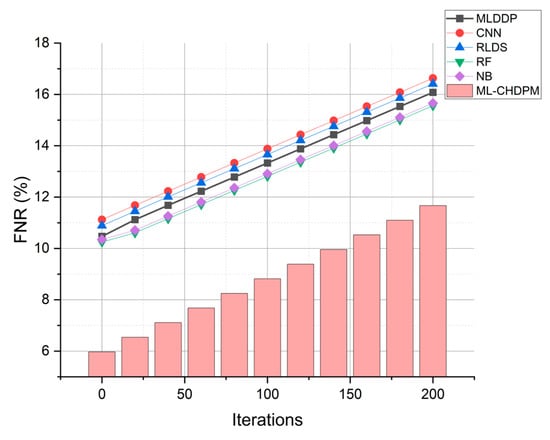
Figure 9.
FNR evaluation for CHD classification.
The simulation findings indicate that the ML-CHDPM approach performs better than alternative methodologies across multiple evaluation metrics. The method under consideration shows an average accuracy of 95.94%, precision of 89.21%, recall of 97.35%, specificity of 90.57%, false positive rate of 9.43%, and false negative rate of 2.65%. The findings suggest that the proposed approach exhibits a higher level of proficiency in precisely identifying and categorizing occurrences of cardiovascular ailments.
6. Conclusions and Future Scope
The global prevalence of CHD has made it a prominent public health issue, highlighting the need for efficient diagnostic techniques to facilitate timely detection and intervention—the present study aimed to investigate the effectiveness of the ML-CHDPM approach in diagnosing and categorizing cardiovascular diseases. The objective was to improve the precision and effectiveness of CHD diagnosis using machine learning methodologies. The study identified several significant concerns regarding the diagnosis of CHD, which encompass the requirement for precise classification models, the selection of relevant features, and the capacity to manage voluminous and intricate datasets. To address the challenges, the ML-CHDPM methodology was introduced. This approach integrates machine learning methods with an extensive feature selection technique. The ML-CHDPM approach demonstrates several noteworthy characteristics. The process facilitates the recognition of pertinent diagnostic features within intricate datasets, resulting in enhanced classification precision. The efficacy of the ML-CHDPM approach was assessed through rigorous simulations and experiments. The findings indicated that this approach outperformed other methodologies. The study’s findings suggest that the method demonstrated superior performance across six key metrics: accuracy, precision, recall, specificity, FPR, and FNR. The method achieved an average accuracy of 94.28%, precision of 87.54%, recall of 96.25%, specificity of 91.74%, FPR of 8.26%, and FNR of 3.75%.
The findings of the research have significant implications for both the academic community and society at large. This study makes a valuable contribution to medical informatics by demonstrating the potential of machine learning techniques in diagnosing cardiovascular disease. The ML-CHDPM approach, as suggested, constitutes a valuable augmentation to the current corpus of knowledge, stimulating additional investigation and ingenuity in this field. The ML-CHDPM approach provides substantial advantages in its societal impact. The precise and prompt identification of CHD facilitates immediate intervention and enhances the overall health outcomes of patients. The efficacy and dependability of the approach will assist healthcare practitioners in making well-informed judgments, resulting in improved disease control and decreased healthcare expenditures.
Even with the encouraging outcomes, it is crucial to recognize the constraints of this study. The research was centered on a particular dataset, which constrained how the results could be applied to other contexts. Additional verification through the utilization of more extensive and heterogeneous datasets is imperative. The practical and ethical aspects necessitate careful deliberation for deploying the suggested approach in real-life clinical environments. It is recommended that further research be conducted to perform comparative analyses with other advanced machine learning algorithms to provide additional validation for the superior performance of the ML-CHDPM technique. Interpretability methods could be used to augment the model’s transparency and dependability by providing valuable insights into its decision-making process. It is necessary to conduct actual clinical trials to assess the efficacy and practicality of the approach in real healthcare environments.
Author Contributions
Formal analysis, A.K.J.S. and M.A. (Mohammed AlKhathami); Resources, D.A.; Writing—original draft, P.P. and R.C.P.; Writing—review & editing, R.C.P.; Funding acquisition, M.A. (Musleh Alsulami). All authors have read and agreed to the published version of the manuscript.
Funding
Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number: IFP22UQU4290525DSR237.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Laggerbauer, B.; Engelhardt, S. MicroRNAs as therapeutic targets in cardiovascular disease. J. Clin. Investig. 2022, 132, e159179. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.W.; Raghavan, S.; Marquez-Luna, C.; Luzum, J.A.; Damrauer, S.M.; Ashley, E.A.; O’Donnell, C.J.; Willer, C.J.; Natarajan, P. Polygenic risk scores for cardiovascular disease: A scientific statement from the American Heart Association. Circulation 2022, 146, e93–e118. [Google Scholar] [CrossRef] [PubMed]
- Sudheer, K.R.; Mohammad Koya, P.K.; Prakash, A.J.; Prakash, A.M.; Manoj Kumar, R.; Shyni, S.; Jagadeesan, C.K.; Jaikrishan, G.; Das, B. Evaluation of risk due to chronic low dose ionizing radiation exposure on the birth prevalence of congenital heart diseases (CHD) among the newborns from high-level natural radiation areas of Kerala coast, India. Genes Environ. 2022, 44, 1. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Ye, S.Q.; Xu, Z.H.; Penaloza, J.S.; Aljuhani, M.; Vetter, T.; Zhao, M.T.; McBride, K.L. Generation and characterization of a human induced pluripotent stem cell (iPSC) line from a patient with congenital heart disease (CHD). Stem Cell Res. 2022, 65, 102958. [Google Scholar] [CrossRef]
- Xu, W.; Yu, K.; Ye, J.; Li, H.; Chen, J.; Yin, F.; Xu, J.; Zhu, J.; Li, D.; Shu, Q. Automatic pediatric congenital heart disease classification based on heart sound signal. Artif. Intell. Med. 2022, 126, 102257. [Google Scholar] [CrossRef]
- Wong, J.; Kohari, K.; Bahtiyar, M.O.; Copel, J. Impact of prenatally diagnosed congenital heart defects on outcomes and management. J. Clin. Ultrasound 2022, 50, 646–654. [Google Scholar] [CrossRef]
- Bottelli, L.; Franzè, V.; Tuo, G.; Buffelli, F.; Paladini, D. Prenatal detection of congenital heart disease at 12–13 gestational weeks: Detailed analysis of false-negative cases. Ultrasound Obstet. Gynecol. 2023, 61, 577–586. [Google Scholar] [CrossRef]
- Gearhart, A.; Dwork, N.; Jone, P.N. Artificial intelligence in echocardiography to diagnose congenital heart disease and fetal echocardiography. Intell.-Based Med. 2022, 6, 100082. [Google Scholar] [CrossRef]
- Truong, V.T.; Nguyen, B.P.; Nguyen-Vo, T.H.; Mazur, W.; Chung, E.S.; Palmer, C.; Tretter, J.T.; Alsaied, T.; Pham, V.T.; Do, H.Q.; et al. Application of machine learning in screening for congenital heart diseases using fetal echocardiography. Int. J. Cardiovasc. Imaging 2022, 38, 1007–1015. [Google Scholar] [CrossRef]
- Ge, B.; Yang, H.; Ma, P.; Guo, T.; Pan, J.; Wang, W. Detection of pulmonary hypertension associated with congenital heart disease based on time-frequency domain and deep learning features. Biomed. Signal Process. Control 2023, 81, 104316. [Google Scholar]
- Suvorov, V.; Loboda, O.; Balakina, M.; Kulczycki, I. A New Three-Dimensional (3D) Printing Prepress Algorithm for Simulation of Planned Surgery for Congenital Heart Disease. Congenit. Heart Dis. 2023, 18, 491–505. [Google Scholar] [CrossRef]
- Ng, W.W.; Liang, H.; Peng, Q.; Zhong, C.; Dong, X.; Huang, Z.; Zhong, P.; Li, C.; Xu, M.; Sun, Y.; et al. An automatic framework for perioperative risks classification from retinal images of complex congenital heart disease patients. Int. J. Mach. Learn. Cybern. 2022, 13, 471–483. [Google Scholar] [CrossRef]
- Kobel, M.; Kalden, P.; Michaelis, A.; Markel, F.; Mensch, S.; Weidenbach, M.; Riede, F.T.; Löffelbein, F.; Bollmann, A.; Shamloo, A.S.; et al. Accuracy of the Apple Watch iECG in children with and without congenital heart disease. Pediatr. Cardiol. 2022, 43, 191–196. [Google Scholar] [CrossRef] [PubMed]
- van Genuchten, W.J.; Helbing, W.A.; Ten Harkel, A.D.; Fejzic, Z.; Md, I.M.K.; Slieker, M.G.; van der Ven, J.P.; Boersma, E.; Takken, T.; Bartelds, B. Exercise capacity in a cohort of children with congenital heart disease. Eur. J. Pediatr. 2023, 182, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, D.; Renumadhavi, C.H. An efficient multilayer deep detection perceptron (MLDDP) methodology for detecting testicular anomalies with or without congenital heart disease (TACHD). J. Supercomput. 2022, 78, 4057–4072. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Yang, Z.; Quan, J.; Liu, L.; Tian, J. Deep learning-based computer-aided heart sound analysis in children with left-to-right shunt congenital heart disease. Int. J. Cardiol. 2022, 348, 58–64. [Google Scholar] [CrossRef]
- Solvin, H.B.; Diab, S.G.; Elle, O.J.; Holmstrøm, H.; Brun, H. Real-Time Remote-Mentored Echocardiography in Management of Newborns with Critical Congenital Heart Defects. Congenit. Heart Dis. 2023, 18, 551–559. [Google Scholar] [CrossRef]
- Steeden, J.A.; Muthurangu, V.; Secinaro, A. Artificial Intelligence-Based Evaluation of Congenital Heart Disease. In Artificial Intelligence in Cardiothoracic Imaging; Springer International Publishing: Cham, Switzerland, 2022; pp. 365–376. [Google Scholar]
- Alici-Karaca, D.; Akay, B.; Yay, A.; Suna, P.; Nalbantoglu, O.U.; Karaboga, D.; Basturk, A.; Balcioglu, E.; Baran, M. A new lightweight convolutional neural network for radiation-induced liver disease classification. Biomed. Signal Process. Control 2022, 73, 103463. [Google Scholar] [CrossRef]
- Qiao, S.; Pang, S.; Luo, G.; Pan, S.; Yu, Z.; Chen, T.; Lv, Z. RLDS: An explainable residual learning diagnosis system for fetal congenital heart disease. Future Gener. Comput. Syst. 2022, 128, 205–218. [Google Scholar] [CrossRef]
- Satish, S.; Sridevi, S. CNN and XGBoost Based Hybrid Model in Classification of Fetal Ultrasound Scan Planes Images in Detection of Congenital Heart Defects. In IoT Based Control Networks and Intelligent Systems: Proceedings of 3rd ICICNIS 2022; Springer Nature: Singapore, 2022; pp. 41–54. [Google Scholar]
- Bai, J.; Fu, J.; Du, S.; Chen, X.; Zhang, C. Prediction of Heart Failure in Children with Congenital Heart Disease Based on Multichannel LSTM. Mob. Inf. Syst. 2022, 2022, 5901445. [Google Scholar] [CrossRef]
- Chung, L.M.; Hariharan, G.; Varma, S. Safety of stimulant medications for attention deficit hyperactivity disorder in pediatric congenital heart disease. J. Paediatr. Child Health 2023, 59, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.kaggle.com/datasets/redwankarimsony/heart-disease-data (accessed on 23 July 2022).
- Available online: https://ieee-dataport.org/documents/cardiovascular-disease-dataset (accessed on 17 August 2021).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).