Seeing the Woods for the Trees Again: Analyzing Evolutionary Diagrams in German and US University-Level Textbooks
Abstract
:1. Introduction
1.1. Historical Development of Evolutionary Trees
1.2. Importance of Evolutionary Trees
1.3. Difficulty of Reading Tree-Diagrams
1.4. Aim and Scope
2. Materials and Methods
2.1. Textbook Selection
2.2. Diagram Selection
2.3. Coding
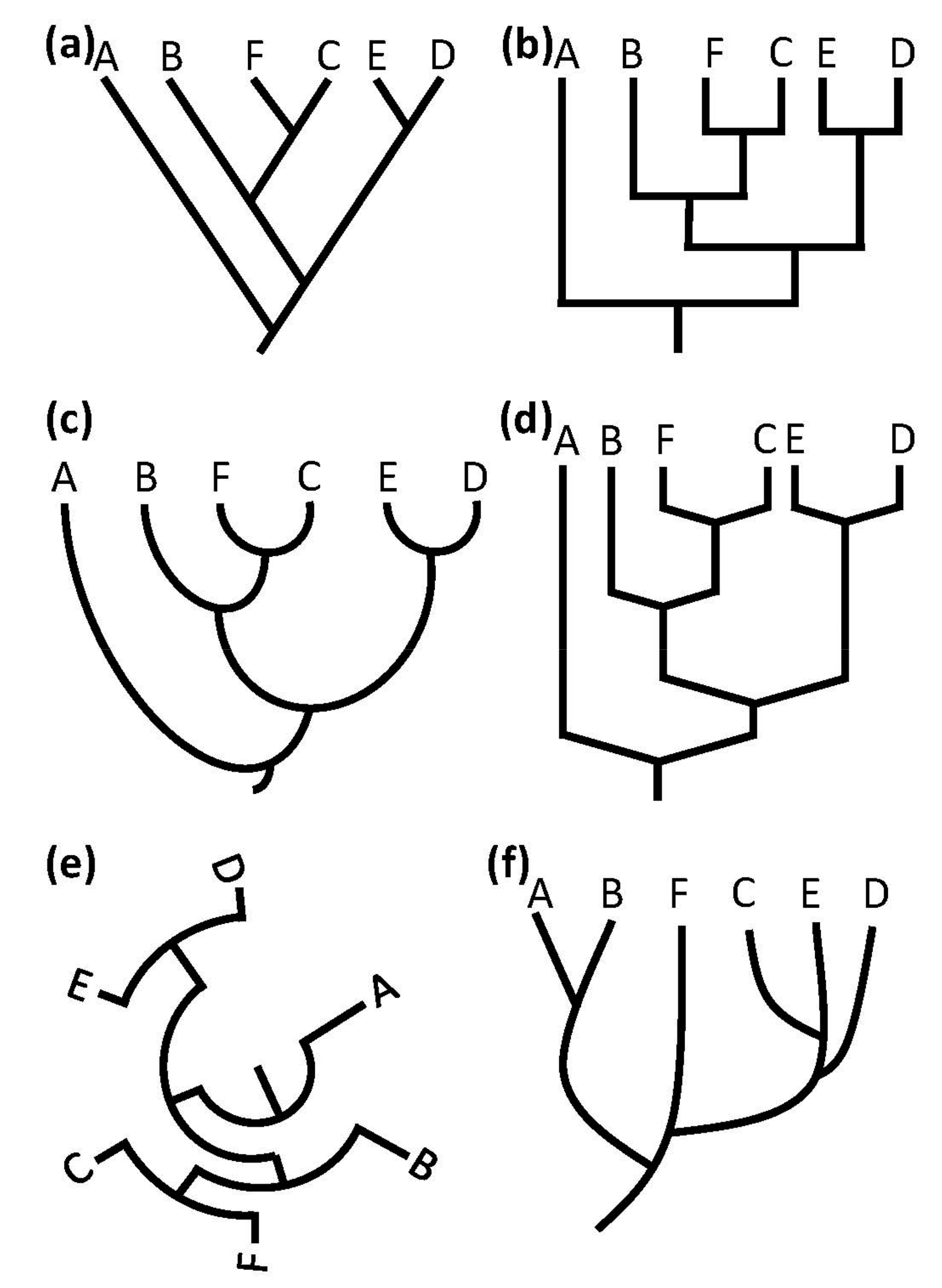
2.4. Tree Structure/Topology
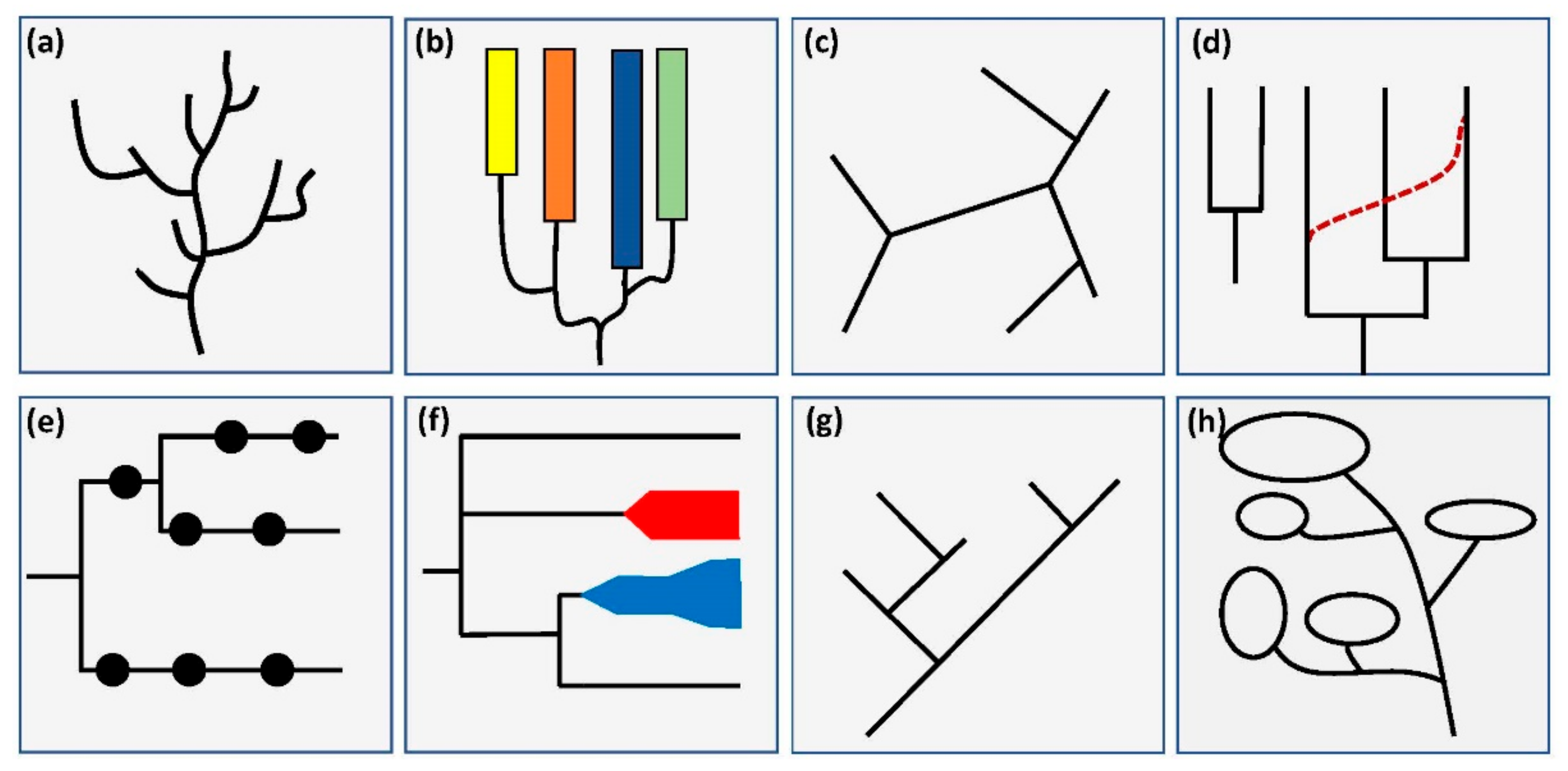
2.5. Branch Structure
- Non-dichotomous diagram—a node branches into at least three edges, at least at one node in the diagram. This characteristic can indicate a simplified version of a more sophisticated tree or unclear relationships between the species [3].
- Terminal nodes at different levels—terminal nodes are not presented along a line (or at the same radius in a circular diagram). This trait can either represent species that went extinct or can be an indication for the use of some form of metric, be it time, mutation rate, or some other scale. As there are multiple potential concepts which can be indicated by this trait, it can be ambiguous to interpret, especially to novice learners [3,41].
- Lateral gene transfer—in at least one point in the diagram, lines connect again after branching, or a lateral transfer of genes is presented in another way. Displays of lateral gene transfer violate the graph theoretical basis of tree diagrams but can visualize important events in the history of life. In many cases, lines indicating lateral gene transfers have a different meaning than other lines in a tree, which represent direct descent. Learners can potentially misinterpret lateral gene transfer as direct descent.
- Unrooted tree—the tree diagram is presented without a root. Unrooted trees do not show the way in which evolutionary processes develop. Therefore, branches and nodes do not represent clades but rather sets of lineages. However, the actual direction of development remains unclear [3]. This category was not included in Catley and Novick’s 2008 study, but we included it to explore potential different compositions of properties from what other tree types offer.
- Branches vary in thickness—at least some branches in the tree are not represented by lines but show differing thicknesses. In many cases, these diagrams are insufficiently labeled and can represent the population size, number of species, geographical distribution, or other concepts. The use of this diagrammatic trait is regarded as questionable [41].
- Bar chart—the developmental lines of at least some taxa in a diagram are represented by a bar or rectangular box instead of a line. Bars displayed instead of lines can be interpreted as single, static entities instead of lineages undergoing continuous change. Thereby, learners’ misconceptions, such as the idea that evolutionary changes only happen at internal nodes [47], can be reinforced by interchanging lines with bars or boxes.
- Anagenesis—the graph shows different taxa along a linear developmental line, implying that one species turns into another. Diagrams following this idea can foster anagenetic evolutionary concepts. A diagram displaying this trait could be interpreted in a way that the bars represent unchanging entities instead of the lineage of one species.
- Unlinked groups—not all groups presented in the diagram are linked by the edges. This category was added to the system, as we found unlinked groups in early samples and wanted to investigate how widespread this property was.
- Direction of the tree—this category describes the direction in which a tree is presented and was coded by investigating where the root is located and the direction in which the tree develops. Five subcategories were used: root location left/right/top/bottom and no major direction (e.g., in circular or unrooted trees). As some research indicates that the orientation of a tree can influence reading difficulty [48], this category was added to the coding manual.
2.6. Extent of Labeling
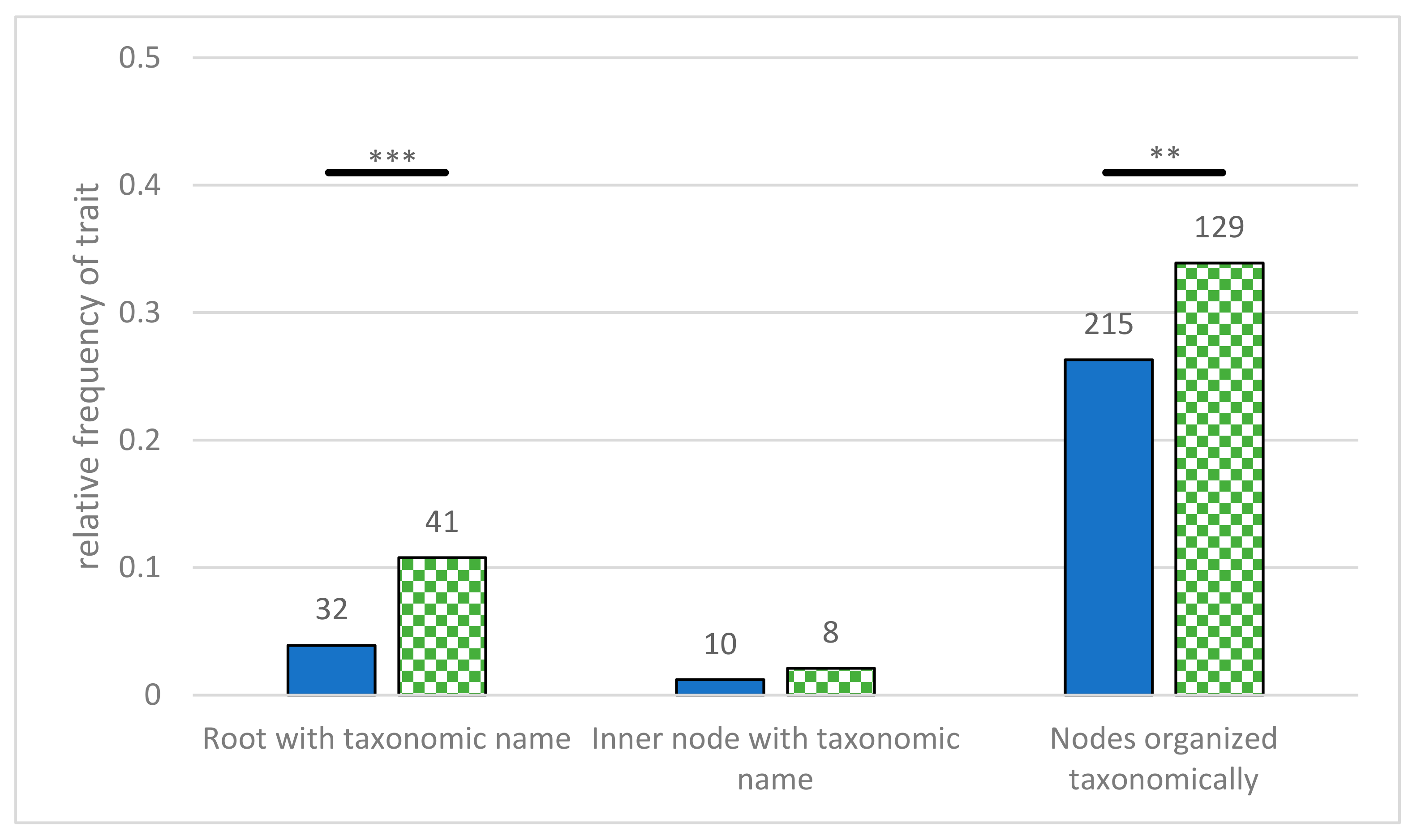
- Root labeled with taxonomic name—the root is labeled with the name of a taxonomic group. This can taxonomically group all presented organisms, but by using the name of a species, it can imply anagenetic ideas.
- Inner nodes labeled with taxa—non-terminal nodes were labeled with taxonomic names. These diagrams can easily be interpreted as one species turning into another, thus fostering anagenetic interpretations [41].
- Taxonomic grouping of nodes—different groups were represented as belonging to a taxonomic group. This can be achieved by coloring the branches, adding bars or brackets along the terminal nodes, or by labeling the inner nodes with taxonomic group names. As this is a common feature in evolutionary diagrams, and it could support reading and identification processes, we wanted to investigate its prevalence.
- Terminal nodes labeled—terminal nodes were labeled using text or graphics. We determined whether all or only some nodes were labeled. In early samples, we found trees in which nodes were only partially labeled, especially with regards to graphical labels. As the extent of labeling may influence reading difficulty, it was included in the coding manual.
2.7. Further Information
- Part of figure—the evolutionary tree is part of a larger figure: for example, in combination with pictures, graphs, and/or other diagrams of evolutionary relationships. The graphical representation labeling parts of the tree are ignored. Some authors argue that adding additional information, like other diagrams connected to the species and concepts represented in a tree, can support reading processes [49,50]. Therefore, this category was included in the present investigation. In the following, this category will also be referenced under the term MER (multiple external representation).
- Apomorphies—new evolutionary traits were labeled along the branches. This can be done in many forms; the most common are written or graphical labels along the branches. The inclusion of apomorphies in trees is known to facilitate reading processes [46]. Therefore, we investigated the prevalence of apomorphies in the evolutionary trees.
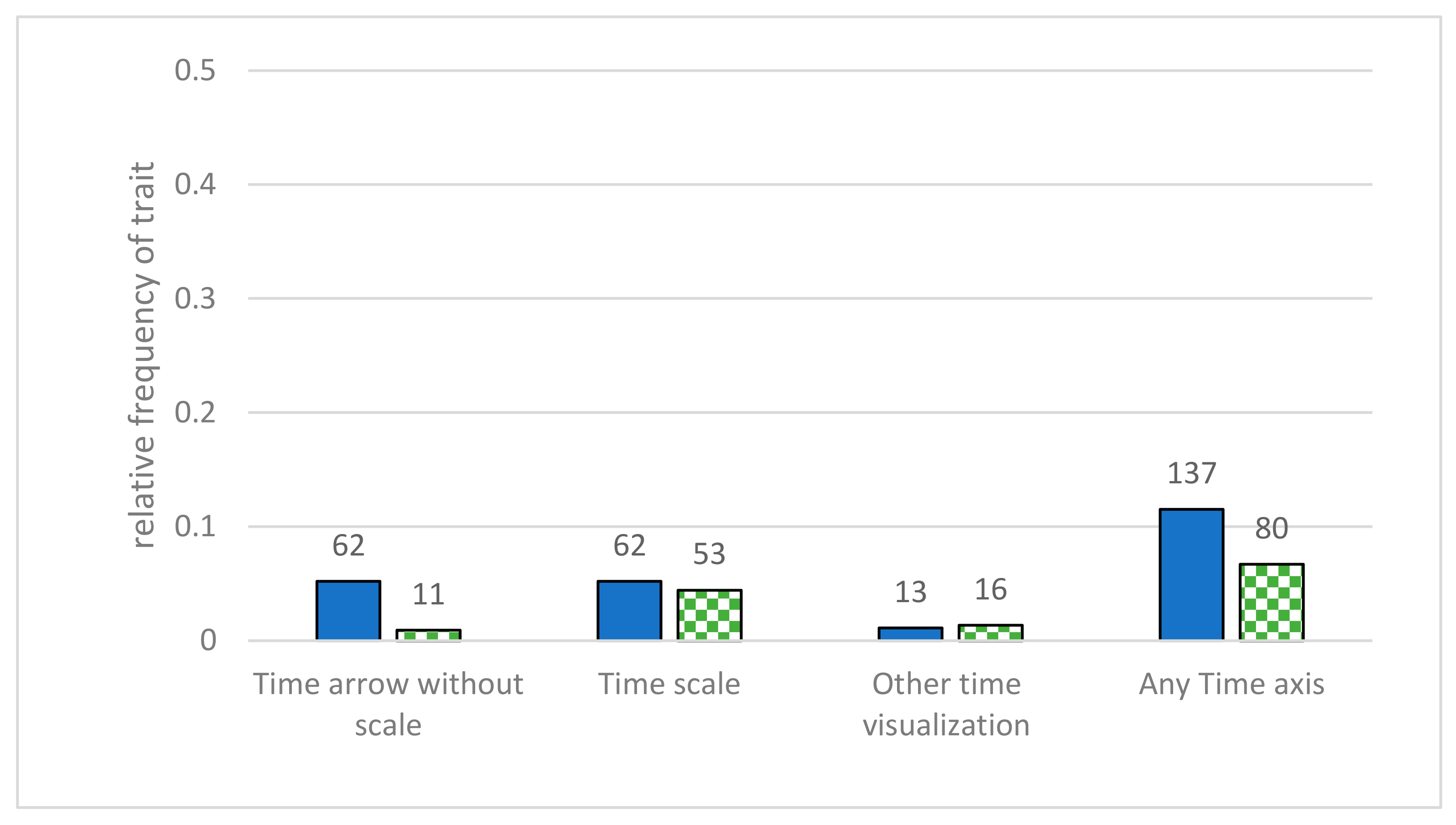
- Time scale—time and direction were represented in some way in the diagram. We distinguished between a time axis without scaling, a time or generation axis with a timescale, and other time representations (axis of mutation rate, singular points of time, or similar). This category was added to the coding manual, as students often misinterpret the direction of the flow of time [11,19], and adding an indication of the direction of time can reduce this risk [51,52]. In the study of Novick and Catley, they only investigated whether any indication of time is displayed in any given diagram [41]; in this study, this was extended by further investigating in which way time is displayed.
2.8. Presented Organisms
- Number of terminal nodes—the number of terminal nodes indicates the size of the presented tree. This was coded in steps of five nodes per level, with the highest level being more than 25 nodes. Because tree size may influence the difficulty of obtaining information, this category was included. Additionally, this could be beneficial in the design of educational materials and research instruments by informing educators of what tree sizes students have the most exposure to.
- Represented organisms—this indicates which groups of organisms are represented in the diagram. Because item-feature effects in the context of evolution are well known [53], we wanted to investigate which species are present in evolutionary trees. If at least one species from one of the predetermined groups was represented, this group was categorized, leading in many cases to multiple entries per tree. The groups investigated were animals (only when a node labeled animals is presented in the tree), non-mammalian animals, non-human mammals, (early) humans, plants, fungi, protozoa, genes, viruses, abstract/no context/no labeling, and non-biological context. In the previous study, special focus was put on diagrams only displaying hominid evolution [41]; in this study, investigating displayed organisms was done in more detail.
- Focal taxon—a certain group in the tree is placed in a special, emphasized position, leading to a focal taxon. This category was included in the investigation because focal taxa can support common misconceptions [1,52], and their prevalence in educational literature has not been investigated before. For example, humans are often presented in the far right, top, or bottom of a tree, giving novice learners the idea that they are “more developed” than other species. We coded this category separately for humans, mammals, and animals.
3. Results
3.1. Tree Style or Topology
3.2. Branch Structure
3.3. Extent of Labeling
3.4. Additional Information in the Diagrams
3.5. Presented Organisms
4. Discussion
4.1. Tree Structure/Topology
4.2. Branch Structure
4.3. Extent of Labeling
4.4. Further Information
4.5. Presented Organisms
4.6. Applications and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Authors | Name | Year | Edition | Country | Number of Diagrams |
|---|---|---|---|---|---|
| US Sample | |||||
| Audesirk; Audesirk; Byers | Biology-Life on Earth with Physiology * | 2017 | 11 | US | 16 |
| Bergstrom; Dugatkin | Evolution | 2016 | 2 | US | 219 |
| Brooker et al. | Principles of Biology | 2018 | 2 | US | 49 |
| Freemen et al. | Biological Science * | 2017 | 6 | US | 126 |
| Hoefnagels; Taylor | Biology-The Essentials | 2015 | 2 | US | 78 |
| Mader; Windelspecht | Biology * | 2015 | 4 | US | 21 |
| Raven et al. | Biology * | 2017 | 11 | US | 98 |
| Sadava; Hillis; Heller | Life-The Science of Biology * | 2017 | 11 | US | 101 |
| Simon; Dickey; Hogan; Reece | Campbell Essential Biology with Physiology * | 2016 | 5 | US | 33 |
| Starr; Dickey; Hogan; Reece | Biology: The Unity and Diversity of Life * | 2016 | 14 | US | 20 |
| Taylor et al. | Biology-Concepts & Connections * | 2018 | 9 | US | 31 |
| German sample | |||||
| Campbell et al. | Campbell Biologie † | 2016 | 10 | Ger | 99 |
| Kleesattel | Die Evolution | 2011 | 1 | Ger | 15 |
| Kutschera | Evolutionsbiologie | 2015 | 4 | Ger | 45 |
| Munk | Grundstudium Biologie | 2000 | 1 | Ger | 19 |
| Munk; Brose | Ökologie-Evolution | 2009 | 1 | Ger | 23 |
| Oschmann | Evolution der Erde | 2016 | 1 | Ger | 18 |
| Richard et al. | Biologie im Überblick | 2013 | 1 | Ger | 3 |
| Sadava et al. | Biologie † | 2011 | 9 | Ger | 79 |
| Storch; Welsch; Wink | Evolutionsbiologie | 2013 | 3 | Ger | 69 |
| Tomiuk; Loeschke | Grundlagen der Evolutionsbiologie und Formalen Genetik | 2016 | 1 | Ger | 6 |
| Zrzavý et al. | Evolution-Ein Lese-Lehrbuch | 2013 | 2 | Ger | 29 |
| Total: | 1197 | ||||
Appendix B
| Category | Kappa | Frequency in Dual Coding Sample |
|---|---|---|
| Tree Topology | 0.91 | 250 |
| non-dichotomous | 0.82 | 55 |
| Terminal nodes on different levels | 0.85 | 36 |
| Lateral gene transfer | 0.91 | 6 |
| Unrooted tree | 0.75 | 4 |
| Branches vary in thickness | 0.66 | 6 |
| Bar chart | 1.00 | 2 |
| Anagenesis | 0.82 | 7 |
| Unlinked groups | 0.50 | 3 |
| Direction of the tree | 0.92 | 250 |
| Inner nodes labeled with taxa | 0.57 | 4 |
| Taxonomic grouping of nodes | 0.78 | 83 |
| Terminal nodes labeled graphically completely | 0.77 | 215 |
| Terminal nodes labeled graphically partially | 0.64 | 13 |
| Terminal nodes labeled/written completely | 0.82 | 49 |
| Terminal nodes labeled/written partially | 0.85 | 25 |
| Part of Figure/MER | 0.88 | 104 |
| Apomorphies | 0.80 | 79 |
| Time axis without scale | 0.74 | 12 |
| Time axis | 0.90 | 22 |
| Other kind of time representation | 0.72 | 5,00 |
| Number of terminal nodes | 0.93 | 250 |
| Animals | 0.80 | 17 |
| non-mammalian animal | 0.87 | 81 |
| non-human mammal | 0.96 | 58 |
| (early) human | 0.94 | 27 |
| Plant | 0.90 | 58 |
| Fungi | 0.92 | 29 |
| Gene | 0.66 | 6 |
| Single celled organisms | 0.83 | 31 |
| Virus | 1.00 | 3 |
| Abstract/no context | 0.90 | 42 |
| Non-biological | 1.00 | 1 |
| Other | 0.50 | 3 |
| Limited on humans | 0.79 | 12 |
| limited on horses | 1.00 | 2 |
| Animals focused | 0.66 | 8 |
| Mammals focused | 0.80 | 17 |
| Humans focused | 0.83 | 13 |
References
- Meisel, R.P. Teaching Tree-Thinking to Undergraduate Biology Students. Evolution 2010, 3, 621–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehm, R.H.; Kampourakis, K. History and Philosophy of Science and the Teaching of Macroevolution. In International Handbook of Research in History, Philosophy and Science Teaching; Matthews, M.R., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 401–421. [Google Scholar]
- Baum, D.A.; Smith, S.D. Tree Thinking: An Introduction to Phylogenetic Biology; Roberts and Company Publishers: Greenwood Village, CO, USA, 2013. [Google Scholar]
- Baum, D.A.; Smith, S.D.; Donovan, S.S.S. Evolution. The tree-thinking challenge. Science 2005, 310, 979–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catley, K.M.; Novick, L.R.; Funk, D. The Promise and Challenges of Introducing Tree Thinking into Evolution Education. In Evolution Challenges: Integrating Research and Practice in Teaching and Learning about Evolution; Rosengren, K.S., Brem, S.K., Evans, E.M., Eds.; Oxford University Press: Oxford, UK, 2012; pp. 93–101. [Google Scholar]
- Yates, T.L.; Mills, J.N.; Parmenter, C.A.; Ksiazek, T.G.; Parmenter, R.R.; Vande Castle, J.R.; Calisher, C.H.; Nichol, S.T.; Abbott, K.D.; Young, J.C.; et al. The Ecology and Evolutionary History of an Emergent Disease: Hantavirus Pulmonary Syndrome. BioScience 2002, 52, 989. [Google Scholar] [CrossRef] [Green Version]
- Searls, D.B. Pharmacophylogenomics: Genes, evolution and drug targets. Nat. Rev. Drug Discov. 2003, 2, 613–623. [Google Scholar] [CrossRef]
- Blacquiere, L.D.; Hoese, W.J. A valid assessment of students’ skill in determining relationships on evolutionary trees. Evol. Educ. Outreach 2016, 9, 979. [Google Scholar] [CrossRef]
- Dees, J.; Momsen, J.L.; Niemi, J.; Montplaisir, L. Student interpretations of phylogenetic trees in an introductory biology course. CBE Life Sci. Educ. 2014, 13, 666–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halverson, K.L. Improving Tree-Thinking One Learnable Skill at a Time. Evol. Educ. Outreach 2011, 4, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Meir, E.; Perry, J.; Herron, J.C.; Kingsolver, J. College Students’ Misconceptions About Evolutionary Trees. Am. Biol. Teach. 2007, 69, e71–e76. [Google Scholar] [CrossRef]
- Omland, K.E.; Cook, L.G.; Crisp, M.D. Tree thinking for all biology: The problem with reading phylogenies as ladders of progress. Bioessays 2008, 30, 854–867. [Google Scholar] [CrossRef]
- Omland, K.E. Interpretation of Phylogenetic Trees. In The Princeton Guide to Evolution; Losos, J.B., Ed.; Princeton University Press: Princeton, NJ, USA; Woodstock, UK, 2014; pp. 51–59. [Google Scholar]
- Bokor, J.R.; Landis, J.B.; Crippen, K.J. High school students’ learning and perceptions of phylogenetics of flowering plants. CBE Life Sci. Educ. 2014, 13, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Catley, K.M.; Phillips, B.C.; Novick, L.R. Snakes and Eels and Dogs!: Oh, My! Evaluating High School Students’ Tree-Thinking Skills: An Entry Point to Understanding Evolution. Res. Sci. Educ. 2013, 43, 2327–2348. [Google Scholar] [CrossRef]
- Pietsch, T.W. Trees of Life: A Visual History of Evolution; Johns Hopkins University Press: Baltimore, MD, USA, 2012. [Google Scholar]
- Gontier, N. Depicting the Tree of Life: The Philosophical and Historical Roots of Evolutionary Tree Diagrams. Evol. Educ. Outreach 2011, 4, 515–538. [Google Scholar] [CrossRef] [Green Version]
- Mogie, M. Historical baggage in biology: The case of ‘higher’ and ‘lower’ species. Bioessays 2000, 22, 868–869. [Google Scholar] [CrossRef]
- Gregory, T.R. Understanding Evolutionary Trees. Evol. Educ. Outreach 2008, 1, 121–137. [Google Scholar] [CrossRef] [Green Version]
- Kummer, T.A.; Whipple, C.J.; Jensen, J.L. Prevalence and Persistence of Misconceptions in Tree Thinking. J. Microbiol. Biol. Educ. 2016, 17, 389–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matuk, C. Images of Evolution. J. Biocommun. 2007, 33, E54–E61. [Google Scholar]
- Archibald, J.D. Aristotle’s Ladder, Darwin’s Tree: The Evolution of Visual Metaphors for Biological Order; Columbia University Press: New York, NY, USA, 2014. [Google Scholar]
- Kutschera, U. From the scala naturae to the symbiogenetic and dynamic tree of life. Biol. Direct. 2011, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Hennig, W. Grundzüge Einer Theorie der Phylogenetischen Systematik; Deutscher Zentralverlag: Berlin, Germany, 1950. [Google Scholar]
- Morrison, D.A. Is the tree of life the best metaphor, model, or heuristic for phylogenetics? Syst. Biol. 2014, 63, 628–638. [Google Scholar] [CrossRef] [Green Version]
- Bapteste, E.; van Iersel, L.; Janke, A.; Kelchner, S.; Kelk, S.; McInerney, J.O.; Morrison, D.A.; Nakhleh, L.; Steel, M.; Stougie, L.; et al. Networks: Expanding evolutionary thinking. Trends Genet. 2013, 29, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Intraspecific gene genealogies: Trees grafting into networks. Trends Ecol. Evol. 2001, 16, 37–45. [Google Scholar] [CrossRef]
- Dobzhansky, T. Nothing in Biology Makes Sense except in the Light of Evolution. Am. Biol. Teach. 1973, 35, 125–129. [Google Scholar] [CrossRef]
- Gould, G.C.; MacFadden, B.J. Chapter 17: Gigantism, Dwarfism, and Cope’s Rule: “Nothing in Evolution Makes Sense without a Phylogeny”. Bull. Am. Mus. Nat. Hist. 2004, 285, 219–237. [Google Scholar] [CrossRef]
- Young, A.; White, B.; Skurtu, T. Teaching undergraduate students to draw phylogenetic trees: Performance measures and partial successes. Evol. Educ. Outreach 2013, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Novick, L.R.; Catley, K.M. Fostering 21st-Century Evolutionary Reasoning: Teaching Tree Thinking to Introductory Biology Students. CBE Life Sci. Educ. 2016, 15. [Google Scholar] [CrossRef] [Green Version]
- Novick, L.R.; Pickering, J.; MacDonald, T.; Diamond, J.; Ainsworth, S.; Aquino, A.E.; Catley, K.M.; Dodick, J.; Evans, E.M.; Matuk, C.; et al. Depicting the tree of life in museums: Guiding principles from psychological research. Evol. Educ. Outreach 2014, 7, 669. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, T.; Wiley, E.O. Communicating Phylogeny: Evolutionary Tree Diagrams in Museums. Evol. Educ. Outreach 2012, 5, 14–28. [Google Scholar] [CrossRef] [Green Version]
- Du Plessis, L.; McCrone, J.T.; Zarebski, A.E.; Peacock, S.; Pybus, O. COG-UK Consortium. Establishment & lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021. [Google Scholar] [CrossRef]
- Morel, B.; Barbera, P.; Czech, L.; Bettisworth, B.; Hübner, L.; Lutteropp, S.; Serdari, D.; Kostaki, E.; Mamais, I.; Kozlov, A.M.; et al. Phylogenetic Analysis of SARS-CoV-2 Data Is Difficult. Mol. Biol. Evol. 2021, 38, 1777–1791. [Google Scholar] [CrossRef]
- Kaustuv, R.; Hunt, G.; Jablonski, D. Phylogenetic conservatism of extinctions in marine bivalves. Science 2009, 325, 733–737. [Google Scholar] [CrossRef] [Green Version]
- Scaduto, D.I.; Brown, J.M.; Haaland, W.C.; Zwickl, D.J.; Hillis, D.M.; Metzker, M.L. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc. Natl. Acad. Sci. USA 2010, 107, 21242–21247. [Google Scholar] [CrossRef] [Green Version]
- Stern, F.; Kampourakis, K.; Huneault, C.; Silveira, P.; Müller, A. Undergraduate Biology Students’ Teleological and Essentialist Misconceptions. Educ. Sci. 2018, 8, 135. [Google Scholar] [CrossRef] [Green Version]
- Schramm, T.; Schmiemann, P. Teleological pitfalls in reading evolutionary trees and ways to avoid them. Evol. Educ. Outreach 2019, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Valverde, G.A.; Bianchi, L.J.; Wolfe, R.G.; Schmidt, W.H.; Houang, R.T. According to the Book: Using TIMSS to Investigate the Translation of Policy into Practice Through the World of Textbooks; Springer: Dordrecht, The Netherlands, 2002. [Google Scholar]
- Catley, K.M.; Novick, L.R. Seeing the Wood for the Trees: An Analysis of Evolutionary Diagrams in Biology Textbooks. BioScience 2008, 58, 976. [Google Scholar] [CrossRef] [Green Version]
- Nelson, G.; Platnick, N.I. Three-Taxon Statement: A More Precise Use of Parsimony? Cladistics 1991, 7, 351–366. [Google Scholar] [CrossRef]
- Crisp, M.D.; Cook, L.G. Do early branching lineages signify ancestral traits? Trends Ecol. Evol. 2005, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Novick, L.R.; Catley, K.M. Understanding phylogenies in biology: The influence of a Gestalt Perceptual Principle. J. Exp. Psychol. Appl. 2007, 13, 197–223. [Google Scholar] [CrossRef]
- Morabito, N.P.; Catley, K.M.; Novick, L.R. Reasoning about evolutionary history: Post-secondary students’ knowledge of most recent common ancestry and homoplasy. J. Biol. Educ. 2010, 44, 166–174. [Google Scholar] [CrossRef]
- Novick, L.R.; Catley, K.M.; Funk, D. Characters Are Key: The Effect of Synapomorphies on Cladogram Comprehension. Evol. Educ. Outreach 2010, 3, 539–547. [Google Scholar] [CrossRef] [Green Version]
- Kummer, T.A.; Whipple, C.J.; Bybee, S.M.; Adams, B.J.; Jensen, J.L. Development of an Evolutionary Tree Concept Inventory. J. Microbiol. Biol. Educ. 2019, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, B.C.; Novick, L.R.; Catley, K.M.; Funk, D. Interactive Effects of Diagrammatic Format and Teleological Beliefs on Tree Thinking.; Catrambone, R., Ohlsson, S., Eds.; Curran Associates Inc.: Red Hook, NY, USA, 2011. [Google Scholar]
- Mead, L.S. Transforming Our Thinking about Transitional Forms. Evo. Edu. Outreach 2009, 2, 310–314. [Google Scholar] [CrossRef] [Green Version]
- Padian, K. Trickle-down evolution: An approach to getting major evolutionary adaptive changes into textbooks and curricula. Integr. Comp. Biol. 2008, 48, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Baum, D.A.; Offner, S. Phylogenics & Tree-Thinking. Am. Biol. Teach. 2008, 70, 222–229. [Google Scholar] [CrossRef]
- Novick, L.R.; Catley, K.M. When Relationships Depicted Diagrammatically Conflict With Prior Knowledge: An Investigation of Students’ Interpretations of Evolutionary Trees. Sci. Ed. 2014, 98, 269–304. [Google Scholar] [CrossRef]
- Nehm, R.H.; Ha, M. Item feature effects in evolution assessment. J. Res. Sci. Teach. 2011, 48, 237–256. [Google Scholar] [CrossRef]
- Kim, H.Y. Statistical notes for clinical researchers: Chi-squared test and Fisher’s exact test. Restor. Dent. Endod. 2017, 42, 152–155. [Google Scholar] [CrossRef] [PubMed]
- MAXQDA 2020; VERBI Software: Berlin, Germany, 2019.
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novick, L.R.; Schreiber, E.G.; Catley, K.M. Deconstructing evolution education: The relationship between micro- and macroevolution. J. Res. Sci. Teach. 2014, 51, 759–788. [Google Scholar] [CrossRef]
- Kattmann, U. Learning biology by means of anthropomorphic conceptions. In Biology in Context: Learning and Teaching for the Twenty-First Century; Hammann, M., Reiss, M., Boulter, C., Eds.; Institute of Education Press: London, UK, 2008; pp. 7–17. [Google Scholar]
- Sandvik, H. Anthropocentricisms in cladograms. Biol. Philos. 2009, 24, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nee, S. The great chain of being. Nature 2005, 435, 429. [Google Scholar] [CrossRef]
- Halverson, K.L.; Friedrichsen, P. Learning Tree Thinking: Developing a New Framework of Representational Competence. In Multiple Representations in Biological Education; Treagust, D., Tsui, C.Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 185–201. [Google Scholar]
- Schramm, T.; Jose, A.; Schmiemann, P. Modeling and Measuring Tree-Reading Skills in Undergraduate and Graduate Students. CBE Life Sci. Educ. 2021, 20, ar32. [Google Scholar] [CrossRef]
- Gibson, J.P.; Cooper, J.T. Botanical Phylo-Cards: A Tree-Thinking Game to Teach Plant Evolution. Am. Biol. Teach. 2017, 79, 241–244. [Google Scholar] [CrossRef]
- Goldsmith, D.W. The Great Clade Race. Am. Biol. Teach. 2003, 65, 679–682. [Google Scholar] [CrossRef]
- Halverson, K.L. Using Pipe Cleaners to Bring the Tree of Life to Life. Am. Biol. Teach. 2010, 72, 223–224. [Google Scholar] [CrossRef]







| Main Categories | Coding Categories | Inclusion in Catley and Novick’s 2008 Study |
|---|---|---|
| Tree structure/Topology | ||
| Tree Diagram | ||
| Rectangular tree | Changed | |
| Diagonal tree | Changed | |
| Circular tree | Changed | |
| Tree of Life | Similar | |
| Other tree | Similar | |
| Non-tree diagram | New | |
| Branch structure | ||
| Non-dichotomous diagram | Similar | |
| Terminal nodes on different levels | Similar | |
| Lateral gene transfer | Similar | |
| Unrooted tree | New | |
| Branches vary in thickness | Similar | |
| Bar chart | Similar | |
| Anagenesis | Similar | |
| Unlinked groups | New | |
| Direction of the tree | New | |
| Extent of labeling | ||
| Inner nodes labeled with taxa | Similar | |
| Taxonomic grouping of nodes | New | |
| Terminal nodes labeled | New | |
| Further Information | ||
| Part of figure/MER | New | |
| Apomorphies | New | |
| Time scale | Changed | |
| Task of representation | ||
| Figure | New | |
| Advanced organizer | New | |
| Task | New | |
| Other | New | |
| Presented organisms | ||
| Number of terminal nodes | New | |
| Represented organisms | New | |
| Limited on special group | Changed | |
| Focal taxon | New | |
| US | ||||
| graphical labels | ||||
| all nodes | some nodes | none | ||
| written labels | all nodes | 25% | 8.9% | 42.9% |
| 209 | 73 | 351 | ||
| some nodes | 0.2% | 1.8% | 4.9% | |
| 2 | 15 | 40 | ||
| none | 3.4% | 0.3% | 11.7% | |
| 28 | 3 | 96 | ||
| Germany | ||||
| graphical labels | ||||
| all nodes | some nodes | none | ||
| written labels | all nodes | 22.9% | 5% | 56.8% |
| 87 | 19 | 216 | ||
| some nodes | 0.2% | 3.2% | 4.5% | |
| 1 | 12 | 17 | ||
| none | 2.6% | 0.5% | 4.2% | |
| 10 | 2 | 16 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schramm, T.; Jose, A.; Schmiemann, P. Seeing the Woods for the Trees Again: Analyzing Evolutionary Diagrams in German and US University-Level Textbooks. Educ. Sci. 2021, 11, 367. https://doi.org/10.3390/educsci11080367
Schramm T, Jose A, Schmiemann P. Seeing the Woods for the Trees Again: Analyzing Evolutionary Diagrams in German and US University-Level Textbooks. Education Sciences. 2021; 11(8):367. https://doi.org/10.3390/educsci11080367
Chicago/Turabian StyleSchramm, Thilo, Anika Jose, and Philipp Schmiemann. 2021. "Seeing the Woods for the Trees Again: Analyzing Evolutionary Diagrams in German and US University-Level Textbooks" Education Sciences 11, no. 8: 367. https://doi.org/10.3390/educsci11080367
APA StyleSchramm, T., Jose, A., & Schmiemann, P. (2021). Seeing the Woods for the Trees Again: Analyzing Evolutionary Diagrams in German and US University-Level Textbooks. Education Sciences, 11(8), 367. https://doi.org/10.3390/educsci11080367






