Development of a Versatile Strategy for Inkjet-Printed Molecularly Imprinted Polymer Microarrays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials, Chemicals, Devices
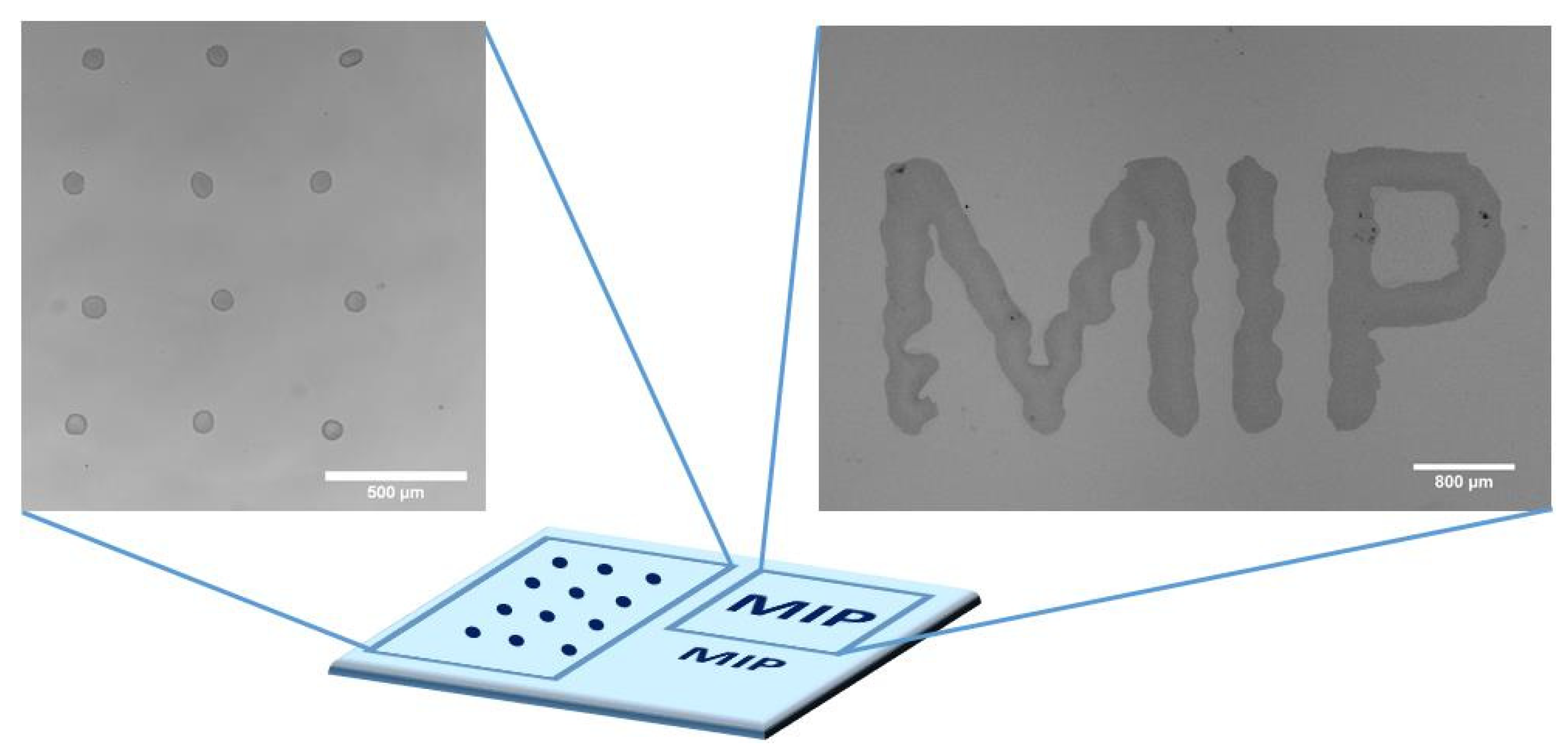
2.2. Design of Inkjet-Printed MIP Biochips
2.3. Sample and Substrate Preparation
2.3.1. Substrates
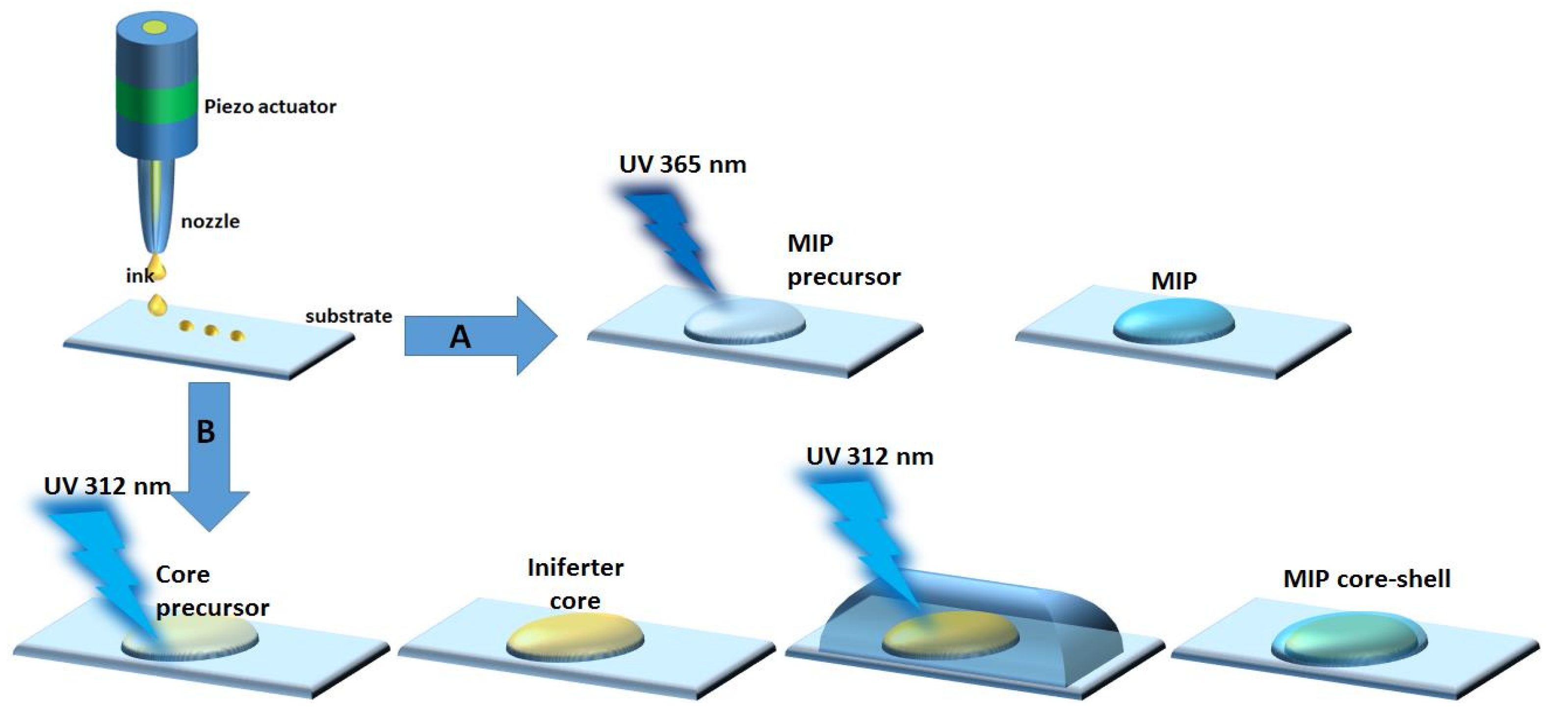
2.3.2. Inkjet Printing of Bulk MIPs
2.3.3. Inkjet Printing of Core–Shell MIPs
2.4. Evaluation of Fabricated Devices
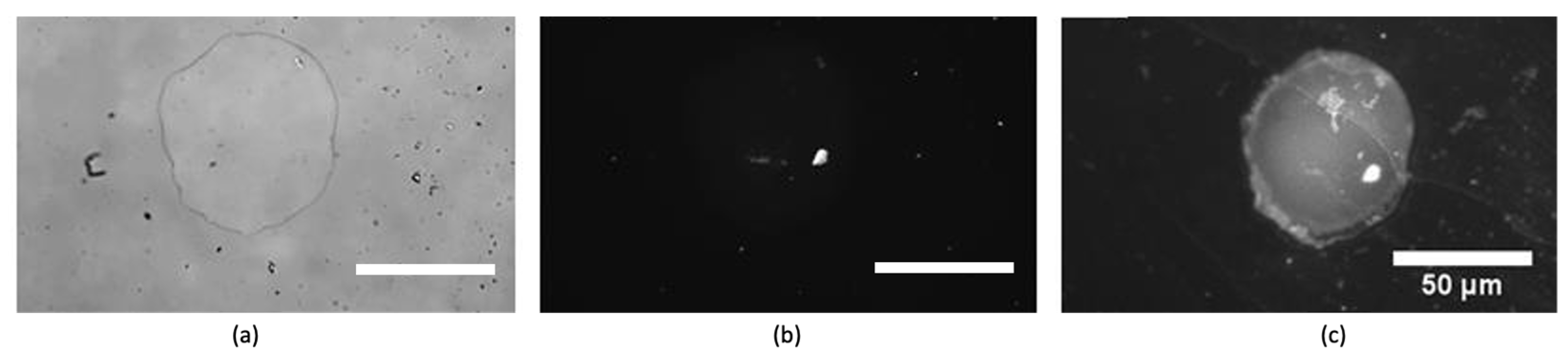
2.5. Binding Tests of the Core–Shell Structures
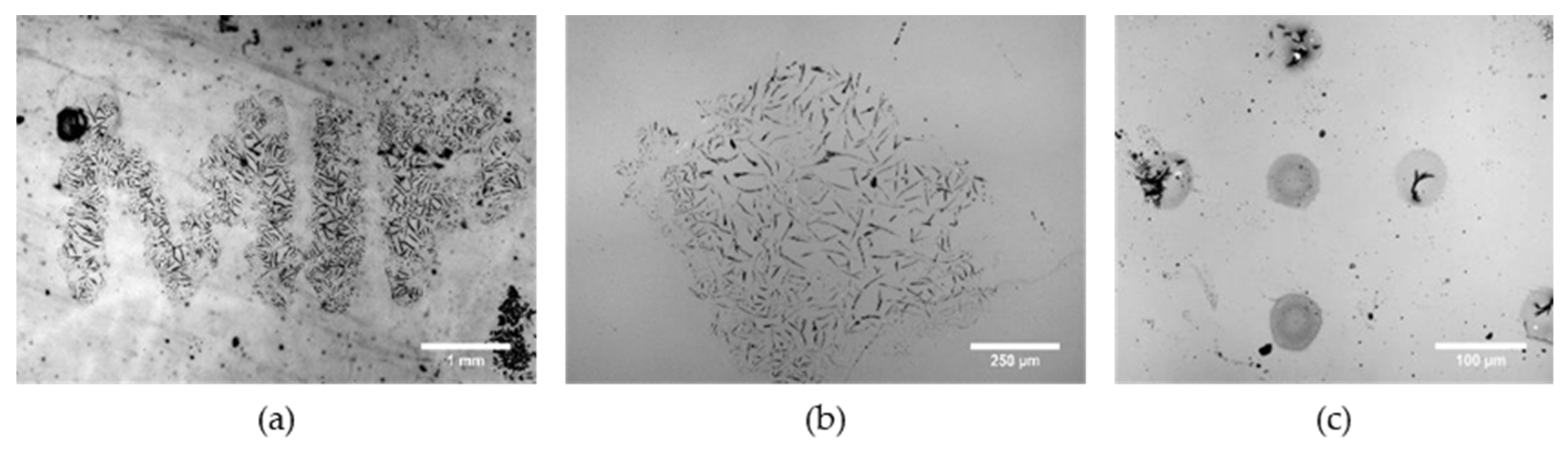
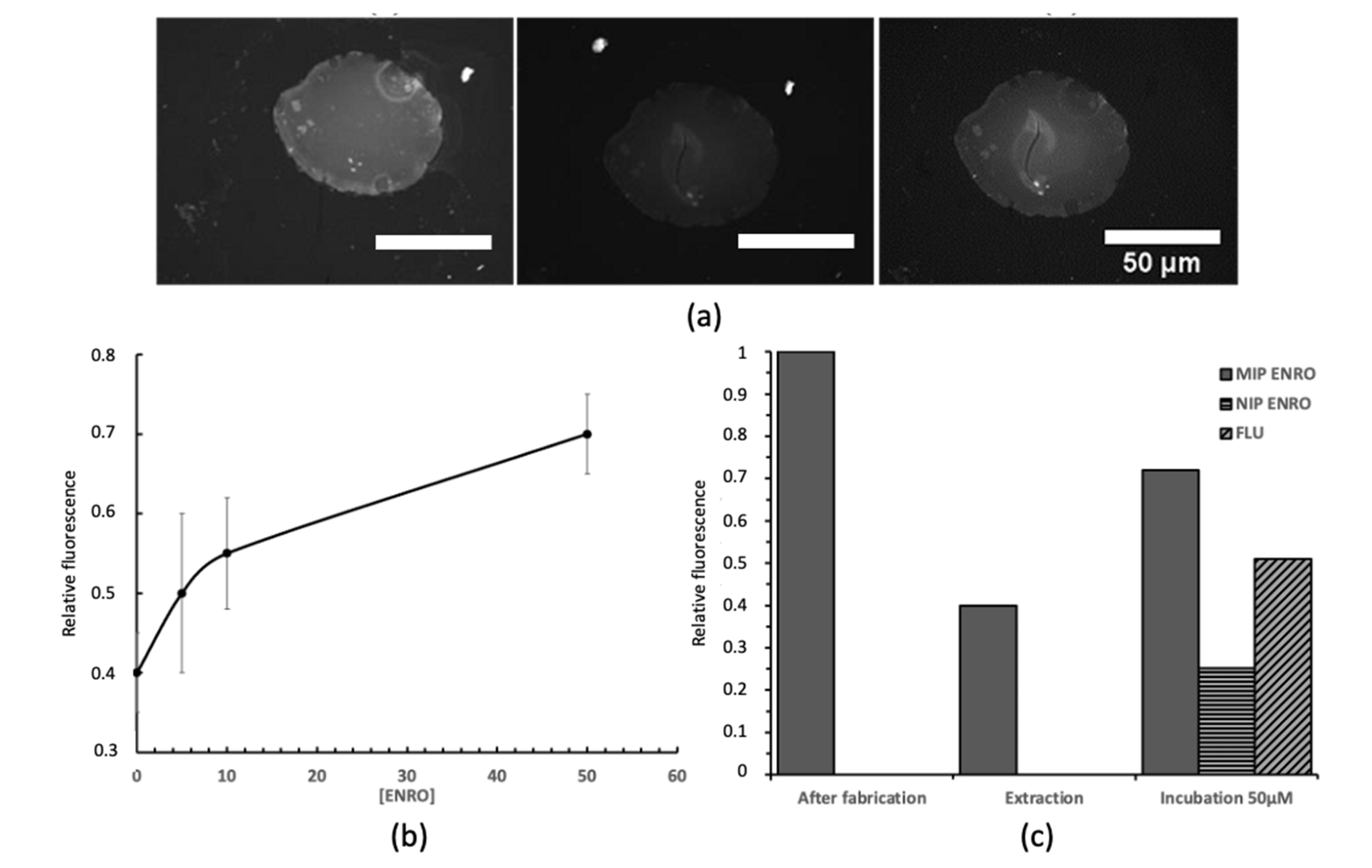
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Persidis, A. Biochips. Nat. Biotechnol. 1998, 16, 981–983. [Google Scholar] [CrossRef] [PubMed]
- Kamar, A.Z.; Shamzi, M.H. Desktop fabrication of lab-on-chip devices on flexible substrates: A brief review. Micromachines 2020, 11, 126. [Google Scholar]
- Fruncillo, S.; Su, X.; Liu, H.; Wong, L.S. Lithographic processes for the scalable fabrication of micro-and nanostructures for biochips and biosensors. ACS Sens. 2021, 6, 2002–2024. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Kang, B.-H.; Jeong, K.-H. Based biochip assays and recent developments: A review. BioChip J. 2018, 12, 1–10. [Google Scholar] [CrossRef]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of biochip technology: A review from lab-on-a-chip to organ-on-a-chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]
- London, J.W.; Diliën, H.; Singla, P.; Peeters, M.; Cleij, T.J.; Van Grinsven, B.; Eersels, K. MIPs for commercial application in low-cost sensors and assays–An overview of the current status quo. Sens. Actuators B Chem. 2020, 325, 128973. [Google Scholar]
- Refaat, D.; Aggour, M.G.; Farghali, A.A.; Mahajan, R.; Wiklander, J.G.; Nicholls, I.A.; Piletsky, S.A. Strategies for molecular imprinting and the evolution of MIP nanoparticles as plastic antibodies—Synthesis and applications. Int. J. Mol. Sci. 2019, 20, 6304. [Google Scholar] [CrossRef]
- Bokeloh, F.; Ayela, C.; Haupt, K. Micro and Nanofabrication Methods of Molecularly Imprinted Polymers. In Molecularly Imprinted Polymers for Analytical Chemistry Application; Royal Society of Chemistry: London, UK, 2018; Chapter 6. [Google Scholar]
- Haupt, K.; Linares, A.V.; Bompart, M.; Bui, B.T. Molecularly imprinting. Top. Curr. Chem. 2012, 325, 1–28. [Google Scholar]
- Paruli, E., III; Soppera, O.; Haupt, K.; Gonzato, C. Photopolymerization and Photostructuring of Molecularly Imprinted Polymers. ACS Appl. Polym. Mater. 2021, 3, 4769–4790. [Google Scholar] [CrossRef]
- Lalo, H.; Ayela, C.; Dague, E.; Vieu, C.; Haupt, K. Nanopatterning molecularly imprinted polymers by soft lithography: A hierarchical approach. Lab Chip 2010, 10, 1316–1318. [Google Scholar] [CrossRef]
- Belmont, A.-S.; Sokuler, M.; Haupt, K.; Heber, L.A. Direct writing of molecularly imprinted microstructures using a nanofountain pen. Appl. Phys. Lett. 2007, 90, 193101. [Google Scholar] [CrossRef]
- Ayela, C.; Dubourg, G.; Pellet, C.; Haupt, K. All-Organic Microelectromechanical Systems Integrating Specific Molecular Recognition–A New Generation of Chemical Sensors. Adv. Mater. 2014, 26, 5876–5879. [Google Scholar] [CrossRef]
- Arrabito, G.; Gulli, D.; Alfano, C.; Pignataro, B. “Writing biochips”: High-resolution droplet-to-droplet manufacturing of analytical platforms. Analyst 2022, 147, 1294–1312. [Google Scholar] [CrossRef]
- Li, X.; Liu, B.; Pei, B.; Chen, J.; Zhou, D.; Peng, J.; Zhang, X.; Jia, W.; Xu, T. Inkjet bioprinting of biomaterials. Chem. Rev. 2020, 120, 10793–10833. [Google Scholar] [CrossRef]
- Caro, E.; Marcé, R.M.; Cormack, P.A.G.; Sherrington, D.C.; Borrull, F. Novel enrofloxacin imprinted polymer applied to the solid-phase extraction of fluorinated quinolones from urine and tissue samples. Anal. Chim. Acta 2006, 562, 145–151. [Google Scholar] [CrossRef]
- Okerman, L.; Noppe, H.; Cornet, V.; De Zutter, L. Microbiological detection of 10 quinolone antibiotic residues and its application to artificially contaminated poultry samples. Food Addit. Contam. 2007, 24, 252–257. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, S.; Liang, Y.; Wang, X.; Zhuang, X.; Tian, C.; Luan, F.; Chen, L. Selective detection of enrofloxacin in biological and environmental samples using a molecularly imprinted electrochemiluminescence sensor based on functionalized copper nanoclusters. Talanta 2022, 236, 122835. [Google Scholar] [CrossRef]
- Wang, W.; Wang, R.; Liao, M.; Kidd, M.T. Rapid detection of enrofloxacin using a localized surface plasmon resonance sensor based on polydopamine molecular imprinted recognition polymer. J. Food Meas. Charact. 2021, 15, 3376–3386. [Google Scholar] [CrossRef]
- Liu, X.; Ren, J.; Su, L.; Gao, X.; Tang, Y.; Ma, T.; Zhu, L.; Li, J. Novel hybrid probe based on double recognition of aptamer-molecularly imprinted polymer grafted on upconversion nanoparticles for enrofloxacin sensing. Biosens. Bioelectron. 2017, 87, 203–208. [Google Scholar] [CrossRef]
- Sigma-Aldrich. Available online: www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/1/m6514pis.pdf (accessed on 1 April 2021).
- McCloy, R.A.; Rogers, S.; Caldon, C.E.; Lorca, T.; Castro, A.; Burgess, A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014, 13, 1400–1412. [Google Scholar] [CrossRef]
- Calvert, P. Inkjet printing for materials and devices. Chem. Mater. 2001, 13, 3299–3305. [Google Scholar] [CrossRef]
- Yang, H.; He, Y.; Tuck, C.; Wildman, R.; Ashcroft, I.; Dickens, P.; Hague, R. High viscosity jetting system for 3D reactive inkjet printing. In Proceedings of the 24th Annual International Solid Freeform Fabrication Symposium: An Additive Manufacturing Conference, Austin, TX, USA, 12 August 2013. [Google Scholar]
- Vandevelde, F.; Leichle, T.; Ayela, C.; Bergaud, C.; Nicu, L.; Haupt, K. Direct patterning of molecularly imprinted microdot arrays for sensors and biochips. Langmuir 2007, 23, 6490–6493. [Google Scholar] [CrossRef]
- Beyazit, S.; Bui, B.T.; Haupt, K.; Gonzato, C. Molecularly imprinted polymer nanomaterials and nanocomposites by controlled/living radical polymerization. Prog. Polym. Sci. 2016, 62, 1–21. [Google Scholar] [CrossRef]
- Sellergren, B.; Rückert, B.; Hall, A.J. Layer-by-layer grafting of molecularly imprinted polymers via iniferter modified supports. Adv. Mater. 2002, 14, 1204–1208. [Google Scholar] [CrossRef]
- Barrios, C.A.; Zhenhe, C.; Navarro-Villoslada, F.; López-Romero, D.; Moreno-Bondi, M.C. Molecularly imprinted polymer diffraction grating as label-free optical bio (mimetic) sensor. Biosens. Bioelectron. 2011, 26, 2801–2804. [Google Scholar] [CrossRef]
- Marchyk, N.; Maximilien, J.; Beyazit, S.; Haupt, K.; Bui, B.T. One-pot synthesis of iniferter-bound polystyrene core nanoparticles for the controlled grafting of multilayer shells. Nanoscale 2014, 6, 2872–2878. [Google Scholar] [CrossRef]
- Ton, X.-A.; Acha, V.; Haupt, K.; Bui, B.T. Direct fluorimetric sensing of UV-excited analytes in biological and environmental samples using molecularly imprinted polymer nanoparticles and fluorescence polarization. Biosens. Bioelectron. 2012, 36, 22–28. [Google Scholar] [CrossRef]
- Mipdatabase. Available online: www.mipdatabase.com (accessed on 1 April 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bokeloh, F.; Gibson, K.; Haupt, K.; Ayela, C. Development of a Versatile Strategy for Inkjet-Printed Molecularly Imprinted Polymer Microarrays. Chemosensors 2022, 10, 396. https://doi.org/10.3390/chemosensors10100396
Bokeloh F, Gibson K, Haupt K, Ayela C. Development of a Versatile Strategy for Inkjet-Printed Molecularly Imprinted Polymer Microarrays. Chemosensors. 2022; 10(10):396. https://doi.org/10.3390/chemosensors10100396
Chicago/Turabian StyleBokeloh, Frank, Kasia Gibson, Karsten Haupt, and Cédric Ayela. 2022. "Development of a Versatile Strategy for Inkjet-Printed Molecularly Imprinted Polymer Microarrays" Chemosensors 10, no. 10: 396. https://doi.org/10.3390/chemosensors10100396



