Progress of Microfluidics Combined with SERS Technology in the Trace Detection of Harmful Substances
Abstract
:1. Introduction
2. Microfluidic and SERS
2.1. Microfluidics: An Excellent Detection Platform
2.2. SERS: Principles and Substrate
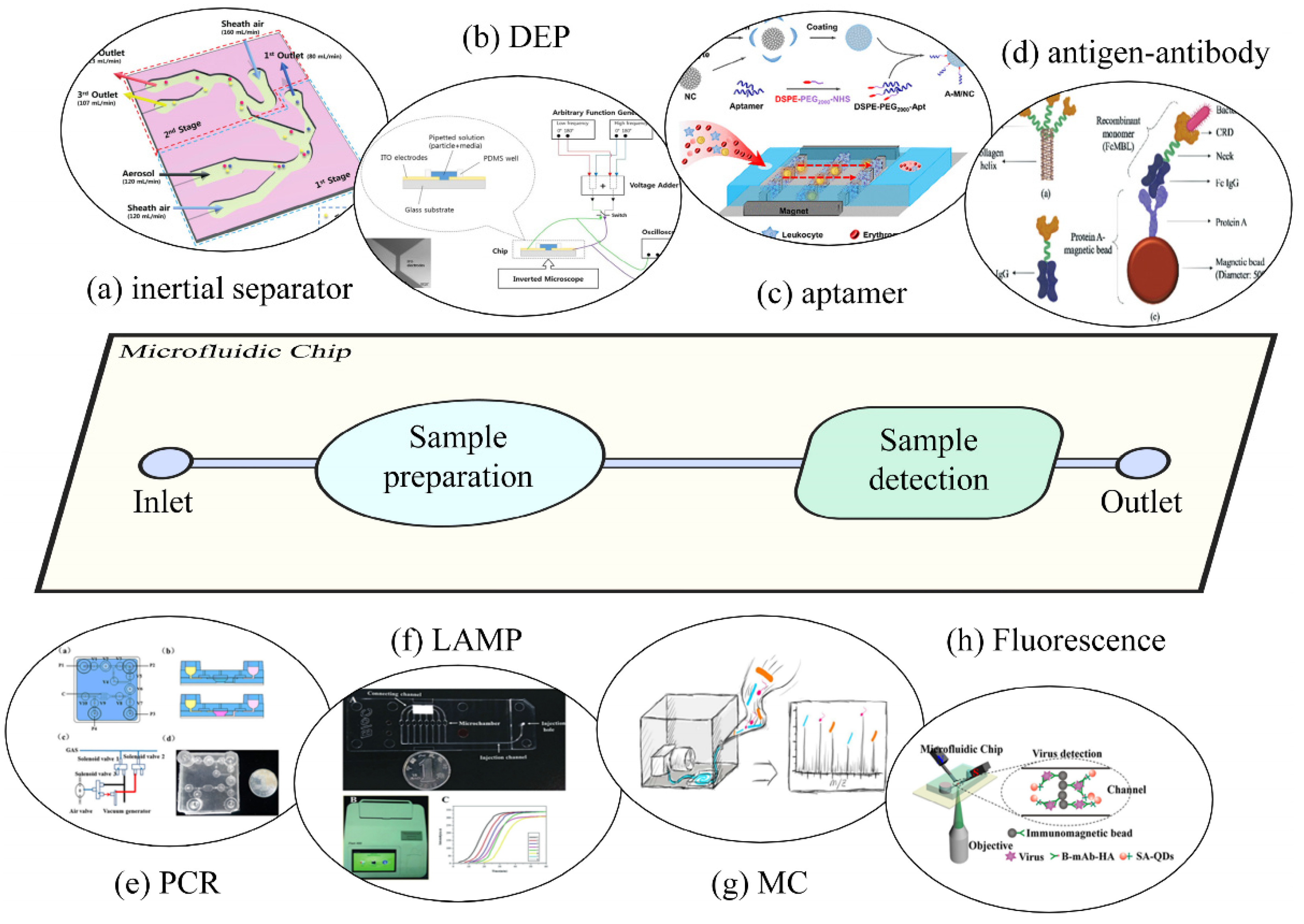
3. Microfluidics Combined with SERS
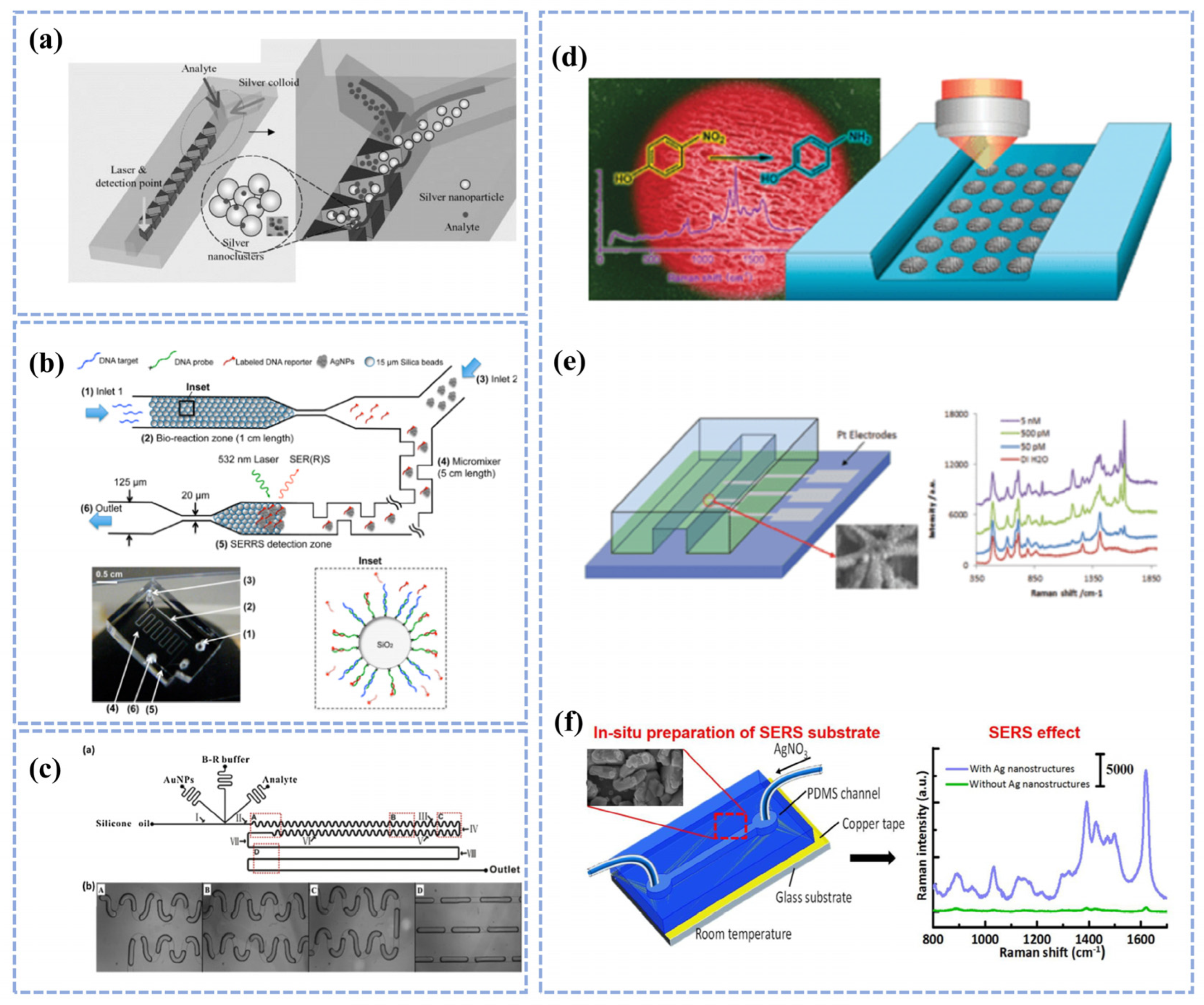
3.1. Key Factors for Combining Microfluidics with SERS
3.1.1. Mixing of Nanocolloids and Analytes in Microchannels
3.1.2. Synthesis and Integration of SERS Active Substrates in Microfluidic Chips
3.1.3. Target Trapping
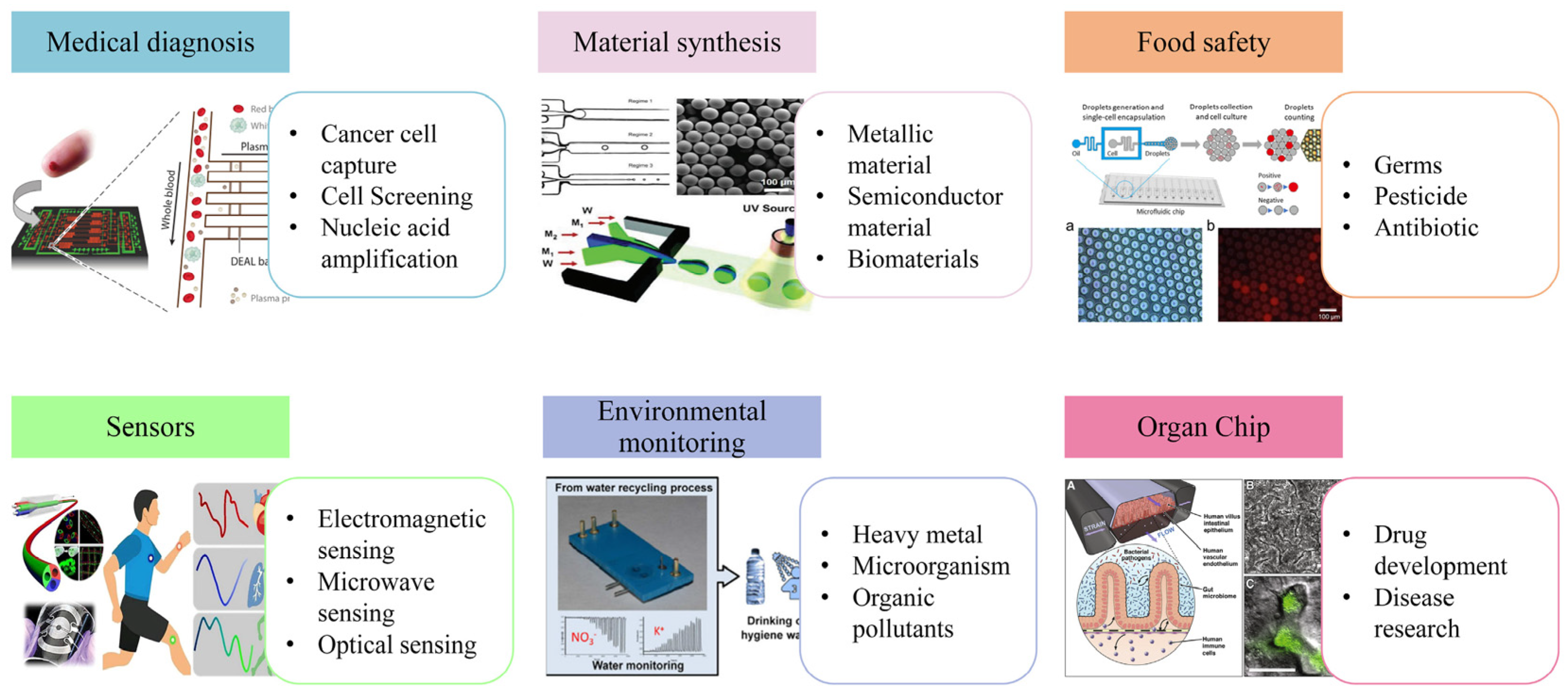
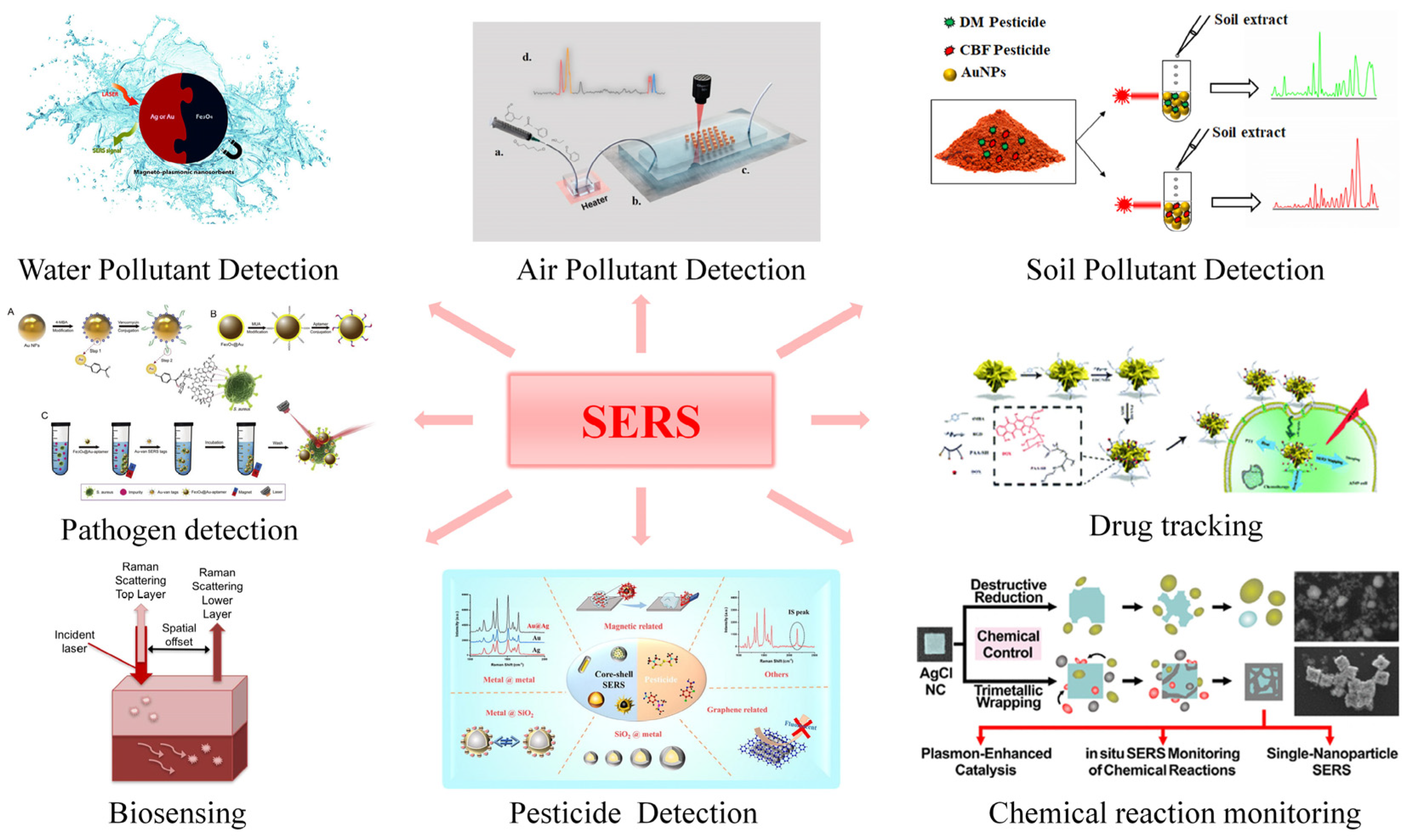
3.2. Application Prospect of the Microfluidic-SERS Detection System
3.3. Smart Algorithms and Results’ Visualization for SERS Data
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samsidar, A.; Siddiquee, S.; Shaarani, S.M. A review of extraction, analytical and advanced methods for determination of pesticides in environment and foodstuffs. Trends Food Sci. Technol. 2018, 71, 188–201. [Google Scholar] [CrossRef]
- Jin, B.; Xie, L.; Guo, Y.; Pang, G. Multi-residue detection of pesticides in juice and fruit wine: A review of extraction and detection methods. Food Res. Int. 2012, 46, 399–409. [Google Scholar] [CrossRef]
- Sulaiman, N.S.; Rovina, K.; Joseph, V.M. Classification, extraction and current analytical approaches for detection of pesticides in various food products. J. Consum. Prot. Food Saf. 2019, 14, 209–221. [Google Scholar] [CrossRef]
- Reyzer, M.L.; Brodbelt, J.S. Analysis of fire ant pesticides in water by solid-phase microextraction and gas chromatography/mass spectrometry or high-performance liquid chromatography/mass spectrometry. Anal. Chim. Acta 2001, 436, 11–20. [Google Scholar] [CrossRef]
- Yogarajah, N.; Tsai, S.S.H. Detection of trace arsenic in drinking water: Challenges and opportunities for microfluidics. Environ. Sci. Water Res. Technol. 2015, 1, 426–447. [Google Scholar] [CrossRef]
- Dong, J.; Ueda, H. ELISA-type assays of trace biomarkers using microfluidic methods. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol. 2017, 9, e1457. [Google Scholar] [CrossRef] [PubMed]
- Piorek, B.D.; Lee, S.J.; Moskovits, M.; Meinhart, C.D. Free-surface microfluidics/surface-enhanced Raman spectroscopy for real-time trace vapor detection of explosives. Anal. Chem. 2012, 84, 9700–9705. [Google Scholar] [CrossRef]
- Cui, F.; Rhee, M.; Singh, A.; Tripathi, A. Microfluidic Sample Preparation for Medical Diagnostics. Annu. Rev. Biomed. Eng. 2015, 17, 267–286. [Google Scholar] [CrossRef]
- Zhao, Q.; Cui, H.; Wang, Y.; Du, X. Microfluidic Platforms toward Rational Material Fabrication for Biomedical Applications. Small 2020, 16, e1903798. [Google Scholar] [CrossRef]
- Seo, M.; Nie, Z.; Xu, S.; Mok, M.; Lewis, P.C.; Graham, R.; Kumacheva, E. Continuous microfluidic reactors for polymer particles. Langmuir 2005, 21, 11614–11622. [Google Scholar] [CrossRef]
- Nie, Z.; Li, W.; Seo, M.; Xu, S.; Kumacheva, E. Janus and ternary particles generated by microfluidic synthesis: Design, synthesis, and self-assembly. J. Am. Chem. Soc. 2006, 128, 9408–9412. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Zuo, P.; Ye, B.C. A single cell droplet microfluidic system for quantitative determination of food-borne pathogens. Talanta 2020, 209, 120571. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yu, Y.; Cai, L.; Wang, Y.; Shi, K.; Shang, L.; Pan, J.; Zhao, Y. Microfluidics for flexible electronics. Mater. Today 2021, 44, 105–135. [Google Scholar] [CrossRef]
- Pol, R.; Céspedes, F.; Gabriel, D.; Baeza, M. Microfluidic lab-on-a-chip platforms for environmental monitoring. TrAC Trends Anal. Chem. 2017, 95, 62–68. [Google Scholar] [CrossRef]
- Calvo-Lopez, A.; Arasa-Puig, E.; Puyol, M.; Casalta, J.M.; Alonso-Chamarro, J. Biparametric potentiometric analytical microsystem for nitrate and potassium monitoring in water recycling processes for manned space missions. Anal. Chim. Acta 2013, 804, 190–196. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [Green Version]
- Baker, C.A.; Duong, C.T.; Grimley, A.; Roper, M.G. Recent advances in microfluidic detection systems. Bioanalysis 2009, 1, 967–975. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2020, 16, e1903916. [Google Scholar] [CrossRef]
- Patabadige, D.E.; Jia, S.; Sibbitts, J.; Sadeghi, J.; Sellens, K.; Culbertson, C.T. Micro Total Analysis Systems: Fundamental Advances and Applications. Anal. Chem. 2016, 88, 320–338. [Google Scholar] [CrossRef]
- Raman, C.V.; Krishnan, K.S. A New Type of Secondary Radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Liu, G.-k.; Zheng, H.; Lu, J.-l. Recent progress and perspective of trace antibiotics detection in aquatic environment by surface-enhanced Raman spectroscopy. Trends Environ. Anal. Chem. 2017, 16, 16–23. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Yang, B.; Jin, S.; Guo, S.; Park, Y.; Chen, L.; Zhao, B.; Jung, Y.M. Recent Development of SERS Technology: Semiconductor-Based Study. ACS Omega 2019, 4, 20101–20108. [Google Scholar] [CrossRef]
- Bantz, K.C.; Meyer, A.F.; Wittenberg, N.J.; Im, H.; Kurtulus, O.; Lee, S.H.; Lindquist, N.C.; Oh, S.H.; Haynes, C.L. Recent progress in SERS biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551–11567. [Google Scholar] [CrossRef]
- Yuan, Y.; Panwar, N.; Yap, S.H.K.; Wu, Q.; Zeng, S.; Xu, J.; Tjin, S.C.; Song, J.; Qu, J.; Yong, K.-T. SERS-based ultrasensitive sensing platform: An insight into design and practical applications. Coord. Chem. Rev. 2017, 337, 1–33. [Google Scholar] [CrossRef]
- Votteler, M.; Carvajal Berrio, D.A.; Pudlas, M.; Walles, H.; Stock, U.A.; Schenke-Layland, K. Raman spectroscopy for the non-contact and non-destructive monitoring of collagen damage within tissues. J. Biophotonics 2012, 5, 47–56. [Google Scholar] [CrossRef]
- Guo, J.; Zeng, F.; Guo, J.; Ma, X. Preparation and application of microfluidic SERS substrate: Challenges and future perspectives. J. Mater. Sci. Technol. 2020, 37, 96–103. [Google Scholar] [CrossRef]
- Lin, Z.; He, L. Recent advance in SERS techniques for food safety and quality analysis: A brief review. Curr. Opin. Food Sci. 2019, 28, 82–87. [Google Scholar] [CrossRef]
- Homola, J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Pinheiro, P.C.; Daniel-da-Silva, A.L.; Nogueira, H.I.S.; Trindade, T. Functionalized Inorganic Nanoparticles for Magnetic Separation and SERS Detection of Water Pollutants. Eur. J. Inorg. Chem. 2018, 2018, 3443–3461. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Z.; Liu, C.; Dai, X.; Zhou, W.; Jiang, K.; Zhang, T.; Yin, J.; Gao, J.; Yin, H.; et al. Continuous in situ portable SERS analysis of pollutants in water and air by a highly sensitive gold nanoparticle-decorated PVDF substrate. Anal. Bioanal. Chem. 2021, 413, 5469–5482. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiao, S.; Dong, T.; Nie, P. Gold Nanoparticles for Qualitative Detection of Deltamethrin and Carbofuran Residues in Soil by Surface Enhanced Raman Scattering (SERS). Int. J. Mol. Sci. 2019, 20, 1731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef]
- Song, C.; Dou, Y.; Yuwen, L.; Sun, Y.; Dong, C.; Li, F.; Yang, Y.; Wang, L. A gold nanoflower-based traceable drug delivery system for intracellular SERS imaging-guided targeted chemo-phototherapy. J. Mater. Chem B 2018, 6, 3030–3039. [Google Scholar] [CrossRef]
- Moore, T.J.; Moody, A.S.; Payne, T.D.; Sarabia, G.M.; Daniel, A.R.; Sharma, B. In Vitro and In Vivo SERS Biosensing for Disease Diagnosis. Biosens 2018, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.J.; Shin, H.; Oh, S.; Joo, J.H.; Choi, Y.; Lee, J.S. Wrapping AgCl Nanostructures with Trimetallic Nanomeshes for Plasmon-Enhanced Catalysis and in Situ SERS Monitoring of Chemical Reactions. ACS Appl. Mater. Interfaces 2020, 12, 2842–2853. [Google Scholar] [CrossRef]
- Kim, D.; Campos, A.R.; Datt, A.; Gao, Z.; Rycenga, M.; Burrows, N.D.; Greeneltch, N.G.; Mirkin, C.A.; Murphy, C.J.; Van Duyne, R.P.; et al. Microfluidic-SERS devices for one shot limit-of-detection. Analyst 2014, 139, 3227–3234. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Choo, J. Recent advances in surface-enhanced Raman scattering detection technology for microfluidic chips. Electrophoresis 2008, 29, 1815–1828. [Google Scholar] [CrossRef]
- Wang, C.; Yu, C. Analytical characterization using surface-enhanced Raman scattering (SERS) and microfluidic sampling. Nanotechnology 2015, 26, 092001. [Google Scholar] [CrossRef]
- Visaveliya, N.; Lenke, S.; Kohler, J.M. Composite Sensor Particles for Tuned SERS Sensing: Microfluidic Synthesis, Properties and Applications. ACS Appl. Mater. Interfaces 2015, 7, 10742–10754. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zong, S.; Zhang, Y.; Qian, Z.; Liu, Y.; Zhu, K.; Li, L.; Li, N.; Wang, Z.; Cui, Y. Array-Assisted SERS Microfluidic Chips for Highly Sensitive and Multiplex Gas Sensing. ACS Appl. Mater. Interfaces 2020, 12, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Pu, H.; Xiao, W.; Sun, D.-W. SERS-microfluidic systems: A potential platform for rapid analysis of food contaminants. Trends Food Sci. Technol. 2017, 70, 114–126. [Google Scholar] [CrossRef]
- Ochoa-Vazquez, G.; Kharisov, B.; Arizmendi-Morquecho, A.; Cario, A.; Aymonier, C.; Marre, S.; Lopez, I. Microfluidics and Surface-Enhanced Raman Spectroscopy: A Perfect Match for New Analytical Tools. IEEE Trans. Nanobiosci. 2019, 18, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, C.; Yu, Z.; Song, Y.; Zhang, X.; Ni, D.; Zhang, D.; Liang, P. Optimum synthesis of cactus-inspired SERS substrate with high roughness for paraquat detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 268, 120703. [Google Scholar] [CrossRef]
- Li, S.; Liang, P.; Chen, Q.; Sun, B.; Shang, Z.; Huang, J.; Zou, M.; Qi, X.; Wu, J. One-pot fabrication of Mo1-xWxS2 alloy nanosheets as SERS substrates with highly Raman enhancement effect and long-term stability. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 279, 121465. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Zhang, X.; Liang, P.; Chen, Q.; Qi, X.; Zou, M. Fabrication of magnetic Au/Fe3O4/MIL-101(Cr) (AF-MIL) as sensitive surface-enhanced Raman spectroscopy (SERS) platform for trace detection of antibiotics residue. Appl. Surf. Sci. 2022, 596, 153550. [Google Scholar] [CrossRef]
- Song, Y.; Chen, J.; Yang, X.; Zhang, D.; Zou, Y.; Ni, D.; Ye, J.; Yu, Z.; Chen, Q.; Jin, S.; et al. Fabrication of Fe3O4@Ag magnetic nanoparticles for highly active SERS enhancement and paraquat detection. Microchem. J. 2022, 173, 107019. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, Y.; Wang, Y.; Liang, P.; Zou, M.; Li, S.; Liu, J.; Qi, X.; Zhang, X.; Shang, Z.; et al. Measurement of trace bisphenol A in drinking water with combination of immunochromatographic detection technology and SERS method. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 267, 120519. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, D.; Zhao, H.; Sun, B.; Liang, P.; Ye, J.; Yu, Z.; Jin, S. Intelligent spectral algorithm for pigments visualization, classification and identification based on Raman spectra. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 250, 119390. [Google Scholar] [CrossRef]
- Hu, J.; Zou, Y.; Sun, B.; Yu, X.; Shang, Z.; Huang, J.; Jin, S.; Liang, P. Raman spectrum classification based on transfer learning by a convolutional neural network: Application to pesticide detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 265, 120366. [Google Scholar] [CrossRef]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B: Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Kovarik, M.L.; Ornoff, D.M.; Melvin, A.T.; Dobes, N.C.; Wang, Y.; Dickinson, A.J.; Gach, P.C.; Shah, P.K.; Allbritton, N.L. Micro total analysis systems: Fundamental advances and applications in the laboratory, clinic, and field. Anal. Chem. 2013, 85, 451–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novotny, J.; Foret, F. Fluid manipulation on the micro-scale: Basics of fluid behavior in microfluidics. J. Sep. Sci. 2017, 40, 383–394. [Google Scholar] [CrossRef]
- Hong, S.C.; Kang, J.S.; Lee, J.E.; Kim, S.S.; Jung, J.H. Continuous aerosol size separator using inertial microfluidics and its application to airborne bacteria and viruses. Lab Chip 2015, 15, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, L.; Nie, W.; Huang, L.; Zhang, J.; Li, F.; Xie, H.Y. Biomimetic Microfluidic System for Fast and Specific Detection of Circulating Tumor Cells. Anal. Chem. 2019, 91, 15726–15731. [Google Scholar] [CrossRef]
- Fang, Y.L.; Wang, C.H.; Chen, Y.S.; Chien, C.C.; Kuo, F.C.; You, H.L.; Lee, M.S.; Lee, G.B. An integrated microfluidic system for early detection of sepsis-inducing bacteria. Lab Chip 2021, 21, 113–121. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, R.; Li, Y.; Kong, W.; Guo, X.; Yang, Y.; Wu, F.; Liu, W.; Song, H.; Hao, R. Rapid detection of multiple respiratory viruses based on microfluidic isothermal amplification and a real-time colorimetric method. Lab Chip 2018, 18, 3507–3515. [Google Scholar] [CrossRef]
- Bian, X.; Lan, Y.; Wang, B.; Zhang, Y.S.; Liu, B.; Yang, P.; Zhang, W.; Qiao, L. Microfluidic Air Sampler for Highly Efficient Bacterial Aerosol Collection and Identification. Anal. Chem. 2016, 88, 11504–11512. [Google Scholar] [CrossRef]
- Zhang, R.Q.; Liu, S.L.; Zhao, W.; Zhang, W.P.; Yu, X.; Li, Y.; Li, A.J.; Pang, D.W.; Zhang, Z.L. A simple point-of-care microfluidic immunomagnetic fluorescence assay for pathogens. Anal. Chem. 2013, 85, 2645–2651. [Google Scholar] [CrossRef]
- Salman, A.; Carney, H.; Bateson, S.; Ali, Z. Shunting microfluidic PCR device for rapid bacterial detection. Talanta 2020, 207, 120303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Hu, A.; Cui, J.; Yang, K.; Zhu, X.; Liu, Y.; Deng, G.; Zhu, L. A Lab-on-a-Chip Device Integrated DNA Extraction and Solid Phase PCR Array for the Genotyping of High-Risk HPV in Clinical Samples. Micromachines 2019, 10, 537. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Mao, S.; Khan, M.; Li, W.; Zhang, Q.; Lin, J.M. Single-cell identification by microfluidic-based in situ extracting and online mass spectrometric analysis of phospholipids expression. Chem. Sci. 2019, 11, 253–256. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Chen, B.; Wang, H.; He, M.; Hu, B. Facile Chip-Based Array Monolithic Microextraction System Online Coupled with ICPMS for Fast Analysis of Trace Heavy Metals in Biological Samples. Anal. Chem. 2017, 89, 6878–6885. [Google Scholar] [CrossRef]
- Guo, P.L.; Tang, M.; Hong, S.L.; Yu, X.; Pang, D.W.; Zhang, Z.L. Combination of dynamic magnetophoretic separation and stationary magnetic trap for highly sensitive and selective detection of Salmonella typhimurium in complex matrix. Biosens. Bioelectron. 2015, 74, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wu, J.; Fang, X.; Kong, J. Washing-free centrifugal microchip fluorescence immunoassay for rapid and point-of-care detection of protein. Anal. Chim. Acta 2020, 1118, 18–25. [Google Scholar] [CrossRef]
- Nge, P.N.; Rogers, C.I.; Woolley, A.T. Advances in microfluidic materials, functions, integration, and applications. Chem. Rev. 2013, 113, 2550–2583. [Google Scholar] [CrossRef] [Green Version]
- Pang, S.; Yang, T.; He, L. Review of surface enhanced Raman spectroscopic (SERS) detection of synthetic chemical pesticides. TrAC Trends Anal. Chem. 2016, 85, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, D.J.; Barrow, B.; Schatz, G.C. Understanding the chemical contribution to the enhancement mechanism in SERS: Connection with Hammett parameters. J. Chem. Phys. 2020, 153, 124706. [Google Scholar] [CrossRef]
- Zhang, D.; Liang, P.; Chen, W.; Tang, Z.; Li, C.; Xiao, K.; Jin, S.; Ni, D.; Yu, Z. Rapid field trace detection of pesticide residue in food based on surface-enhanced Raman spectroscopy. Mikrochim. Acta 2021, 188, 370. [Google Scholar] [CrossRef]
- Wang, T.; Wang, S.; Cheng, Z.; Wei, J.; Yang, L.; Zhong, Z.; Hu, H.; Wang, Y.; Zhou, B.; Li, P. Emerging core–shell nanostructures for surface-enhanced Raman scattering (SERS) detection of pesticide residues. Chem. Eng. J. 2021, 424, 130323. [Google Scholar] [CrossRef]
- Fornasaro, S.; Cialla-May, D.; Sergo, V.; Bonifacio, A. The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents. Chemosensors 2022, 10, 128. [Google Scholar] [CrossRef]
- Wang, X.; Huang, S.-C.; Hu, S.; Yan, S.; Ren, B. Fundamental understanding and applications of plasmon-enhanced Raman spectroscopy. Nat. Rev. Phys. 2020, 2, 253–271. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguie, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef] [Green Version]
- González Fá, A.; Pignanelli, F.; López-Corral, I.; Faccio, R.; Juan, A.; Di Nezio, M.S. Detection of oxytetracycline in honey using SERS on silver nanoparticles. TrAC Trends Anal. Chem. 2019, 121, 115673. [Google Scholar] [CrossRef]
- Kamran, M.; Haroon, M.; Popoola, S.A.; Almohammedi, A.R.; Al-Saadi, A.A.; Saleh, T.A. Characterization of valeric acid using substrate of silver nanoparticles with SERS. J. Mol. Liq. 2019, 273, 536–542. [Google Scholar] [CrossRef]
- Hassan, M.M.; Zareef, M.; Jiao, T.; Liu, S.; Xu, Y.; Viswadevarayalu, A.; Li, H.; Chen, Q. Signal optimized rough silver nanoparticle for rapid SERS sensing of pesticide residues in tea. Food Chem. 2021, 338, 127796. [Google Scholar] [CrossRef]
- Dasary, S.S.; Singh, A.K.; Senapati, D.; Yu, H.; Ray, P.C. Gold nanoparticle based label-free SERS probe for ultrasensitive and selective detection of trinitrotoluene. J. Am. Chem. Soc. 2009, 131, 13806–13812. [Google Scholar] [CrossRef]
- Britto Hurtado, R.; Cortez-Valadez, M.; Ramírez-Rodríguez, L.P.; Larios-Rodriguez, E.; Alvarez, R.A.B.; Rocha-Rocha, O.; Delgado-Beleño, Y.; Martinez-Nuñez, C.E.; Arizpe-Chávez, H.; Hernández-Martínez, A.R.; et al. Instant synthesis of gold nanoparticles at room temperature and SERS applications. Phys. Lett. A 2016, 380, 2658–2663. [Google Scholar] [CrossRef]
- Al-Saadi, A.A.; Haroon, M.; Popoola, S.A.; Saleh, T.A. Sensitive SERS detection and characterization of procaine in aqueous media by reduced gold nanoparticles. Sens. Actuators B Chem. 2020, 304, 127057. [Google Scholar] [CrossRef]
- Kim, K.; Ryoo, H.; Shin, K.S. Adsorption and aggregation characteristics of silver nanoparticles onto a poly(4-vinylpyridine) film: A comparison with gold nanoparticles. Langmuir 2010, 26, 10827–10832. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Moyano, D.F.; Parnsubsakul, A.; Papadopoulos, A.; Wang, L.S.; Landis, R.F.; Das, R.; Rotello, V.M. Ultrastable and Biofunctionalizable Gold Nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 14096–14101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, N.; Pu, H.; Sun, D.W. Core size optimized silver coated gold nanoparticles for rapid screening of tricyclazole and thiram residues in pear extracts using SERS. Food Chem. 2021, 350, 129025. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, T.; Pu, H.; Sun, D.W. Fabrication of silver-coated gold nanoparticles to simultaneously detect multi-class insecticide residues in peach with SERS technique. Talanta 2019, 196, 537–545. [Google Scholar] [CrossRef]
- Sankar, S.S.; Sangeetha, K.; Karthick, K.; Anantharaj, S.; Ede, S.R.; Kundu, S. Pt nanoparticle tethered DNA assemblies for enhanced catalysis and SERS applications. New. J. Chem. 2018, 42, 15784–15792. [Google Scholar] [CrossRef]
- Fu, J.H.; Zhong, Z.; Xie, D.; Guo, Y.J.; Kong, D.X.; Zhao, Z.X.; Zhao, Z.X.; Li, M. SERS-Active MIL-100(Fe) Sensory Array for Ultrasensitive and Multiplex Detection of VOCs. Angew. Chem. Int. Ed. Engl. 2020, 59, 20489–20498. [Google Scholar] [CrossRef]
- Zhao, X.; Deng, M.; Rao, G.; Yan, Y.; Wu, C.; Jiao, Y.; Deng, A.; Yan, C.; Huang, J.; Wu, S.; et al. High-Performance SERS Substrate Based on Hierarchical 3D Cu Nanocrystals with Efficient Morphology Control. Small 2018, 14, e1802477. [Google Scholar] [CrossRef]
- Mustafa, D.E.; Yang, T.; Xuan, Z.; Chen, S.; Tu, H.; Zhang, A. Surface Plasmon Coupling Effect of Gold Nanoparticles with Different Shape and Size on Conventional Surface Plasmon Resonance Signal. Plasmonics 2010, 5, 221–231. [Google Scholar] [CrossRef]
- Mahmoud, M.A.; El-Sayed, M.A. Substrate effect on the plasmonic sensing ability of hollow nanoparticles of different shapes. J. Phys. Chem. B 2013, 117, 4468–4477. [Google Scholar] [CrossRef]
- McLellan, J.M.; Li, Z.Y.; Siekkinen, A.R.; Xia, Y. The SERS activity of a supported Ag nanocube strongly depends on its orientation relative to laser polarization. Nano Lett. 2007, 7, 1013–1017. [Google Scholar] [CrossRef]
- Fang, J.; Yi, Y.; Ding, B.; Song, X. A route to increase the enhancement factor of surface enhanced Raman scattering (SERS) via a high density Ag flower-like pattern. Appl. Phys. Lett. 2008, 92, 131115. [Google Scholar] [CrossRef]
- Alsammarraie, F.K.; Lin, M. Using Standing Gold Nanorod Arrays as Surface-Enhanced Raman Spectroscopy (SERS) Substrates for Detection of Carbaryl Residues in Fruit Juice and Milk. J. Agric. Food Chem. 2017, 65, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Krajczewski, J.; Kedziora, M.; Kolataj, K.; Kudelski, A. Improved synthesis of concave cubic gold nanoparticles and their applications for Raman analysis of surfaces. RSC Adv. 2019, 9, 18609–18618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Zhao, J.; Wang, M.; Zhu, S. Snowflake-like gold nanoparticles as SERS substrates for the sensitive detection of organophosphorus pesticide residues. Food Control. 2020, 108, 106835. [Google Scholar] [CrossRef]
- Kamholz, A.E.; Weigl, B.H.; Finlayson, B.A.; Yager, P. Quantitative analysis of molecular interaction in a microfluidic channel: The T-sensor. Anal. Chem. 1999, 71, 5340–5347. [Google Scholar] [CrossRef]
- Mengeaud, V.; Josserand, J.; Girault, H.H. Mixing processes in a zigzag microchannel: Finite element simulations and optical study. Anal. Chem. 2002, 74, 4279–4286. [Google Scholar] [CrossRef]
- Stroock, A.D.; Dertinger, S.K.; Ajdari, A.; Mezic, I.; Stone, H.A.; Whitesides, G.M. Chaotic mixer for microchannels. Science 2002, 295, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Yasui, T.; Omoto, Y.; Osato, K.; Kaji, N.; Suzuki, N.; Naito, T.; Okamoto, Y.; Tokeshi, M.; Shamoto, E.; Baba, Y. Confocal microscopic evaluation of mixing performance for three-dimensional microfluidic mixer. Anal. Sci. 2012, 28, 57–59. [Google Scholar] [CrossRef] [Green Version]
- Adamo, C.B.; Junger, A.S.; Bressan, L.P.; da Silva, J.A.F.; Poppi, R.J.; de Jesus, D.P. Fast and straightforward in-situ synthesis of gold nanoparticles on a thread-based microfluidic device for application in surface-enhanced Raman scattering detection. Microchem. J. 2020, 156, 104985. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, M.; Wang, Y.; Xu, Z. Droplet-based microfluidic synthesis of (Au nanorod@Ag)–polyaniline Janus nanoparticles and their application as a surface-enhanced Raman scattering nanosensor for mercury detection. Anal. Methods 2019, 11, 3966–3973. [Google Scholar] [CrossRef]
- Lawanstiend, D.; Gatemala, H.; Nootchanat, S.; Eakasit, S.; Wongravee, K.; Srisa-Art, M. Microfluidic approach for in situ synthesis of nanoporous silver microstructures as on-chip SERS substrates. Sens. Actuators B Chem. 2018, 270, 466–474. [Google Scholar] [CrossRef]
- Xu, B.B.; Zhang, R.; Liu, X.Q.; Wang, H.; Zhang, Y.L.; Jiang, H.B.; Wang, L.; Ma, Z.C.; Ku, J.F.; Xiao, F.S.; et al. On-chip fabrication of silver microflower arrays as a catalytic microreactor for allowing in situ SERS monitoring. Chem. Commun. (Camb) 2012, 48, 1680–1682. [Google Scholar] [CrossRef]
- Parisi, J.; Su, L.; Lei, Y. In situ synthesis of silver nanoparticle decorated vertical nanowalls in a microfluidic device for ultrasensitive in-channel SERS sensing. Lab Chip 2013, 13, 1501–1508. [Google Scholar] [CrossRef]
- Yan, S.; Chu, F.; Zhang, H.; Yuan, Y.; Huang, Y.; Liu, A.; Wang, S.; Li, W.; Li, S.; Wen, W. Rapid, one-step preparation of SERS substrate in microfluidic channel for detection of molecules and heavy metal ions. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 220, 117113. [Google Scholar] [CrossRef]
- Cheng, I.F.; Chang, H.C.; Chen, T.Y.; Hu, C.; Yang, F.L. Rapid (<5 min) identification of pathogen in human blood by electrokinetic concentration and surface-enhanced Raman spectroscopy. Sci. Rep. 2013, 3, 2365. [Google Scholar] [CrossRef] [Green Version]
- Krafft, B.; Tycova, A.; Urban, R.D.; Dusny, C.; Belder, D. Microfluidic device for concentration and SERS-based detection of bacteria in drinking water. Electrophoresis 2021, 42, 86–94. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, L.; Li, L.; Zong, S.; Wang, Z.; Cui, Y. Simultaneous and highly sensitive detection of multiple breast cancer biomarkers in real samples using a SERS microfluidic chip. Talanta 2018, 188, 507–515. [Google Scholar] [CrossRef]
- Yazdi, S.H.; Giles, K.L.; White, I.M. Multiplexed detection of DNA sequences using a competitive displacement assay in a microfluidic SERRS-based device. Anal. Chem. 2013, 85, 10605–10611. [Google Scholar] [CrossRef]
- Zhang, W.-S.; Wang, Y.-N.; Wang, Y.; Xu, Z.-R. Highly reproducible and fast detection of 6-thioguanine in human serum using a droplet-based microfluidic SERS system. Sens. Actuators B: Chem. 2019, 283, 532–537. [Google Scholar] [CrossRef]
- Ma, W.; Liu, L.; Zhang, X.; Liu, X.; Xu, Y.; Li, S.; Zeng, M. A microfluidic-based SERS biosensor with multifunctional nanosurface immobilized nanoparticles for sensitive detection of MicroRNA. Anal. Chim. Acta 2022, 1221, 340139. [Google Scholar] [CrossRef]
- Lafuente, M.; Pellejero, I.; Clemente, A.; Urbiztondo, M.A.; Mallada, R.; Reinoso, S.; Pina, M.P.; Gandia, L.M. In Situ Synthesis of SERS-Active Au@POM Nanostructures in a Microfluidic Device for Real-Time Detection of Water Pollutants. ACS Appl. Mater Interfaces 2020, 12, 36458–36467. [Google Scholar] [CrossRef]
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.; Lim, W.T.; Han, J.; Bhagat, A.A.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [Green Version]
- Shiriny, A.; Bayareh, M. Inertial focusing of CTCs in a novel spiral microchannel. Chem. Eng. Sci. 2021, 229, 116102. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.; Chaudhuri, P.K.; Tan, D.S.; Lim, W.T.; Lee, S.C.; Chen, P.C.; et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 2014, 14, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.; Kumar, G.; Papautsky, I. Inertial microfluidics for continuous particle separation in spiral microchannels. Lab Chip 2009, 9, 2973–2980. [Google Scholar] [CrossRef] [Green Version]
- Choi, W.; Min, Y.W.; Lee, K.Y.; Jun, S.; Lee, H.G. Dielectrophoresis-based microwire biosensor for rapid detection of Escherichia coli K-12 in ground beef. Lwt 2020, 132, 109230. [Google Scholar] [CrossRef]
- Yang, L.; Banada, P.P.; Chatni, M.R.; Seop Lim, K.; Bhunia, A.K.; Ladisch, M.; Bashir, R. A multifunctional micro-fluidic system for dielectrophoretic concentration coupled with immuno-capture of low numbers of Listeria monocytogenes. Lab Chip 2006, 6, 896–905. [Google Scholar] [CrossRef]
- Hashimoto, M.; Kaji, H.; Nishizawa, M. Selective capture of a specific cell type from mixed leucocytes in an electrode-integrated microfluidic device. Biosens. Bioelectron. 2009, 24, 2892–2897. [Google Scholar] [CrossRef]
- Han, C.H.; Woo, S.Y.; Bhardwaj, J.; Sharma, A.; Jang, J. Rapid and selective concentration of bacteria, viruses, and proteins using alternating current signal superimposition on two coplanar electrodes. Sci. Rep. 2018, 8, 14942. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Torres, K.Y.; McLamore, E.S.; Arnold, D.P. A High-Throughput Microfluidic Magnetic Separation (microFMS) Platform for Water Quality Monitoring. Micromachines 2019, 11, 16. [Google Scholar] [CrossRef]
- Armbrecht, L.; Rutschmann, O.; Szczerba, B.M.; Nikoloff, J.; Aceto, N.; Dittrich, P.S. Quantification of Protein Secretion from Circulating Tumor Cells in Microfluidic Chambers. Adv. Sci. (Weinh) 2020, 7, 1903237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zhang, Z.; Liu, H.; Zhang, Z.; Lin, C.; Wang, B. Hybrid magnetic and deformability based isolation of circulating tumor cells using microfluidics. AIP Adv. 2019, 9, 025023. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, S.; Xun, M.; Li, M.; Kong, X.; Shao, R.; Zheng, T.; Pan, D.; Li, J.; Li, Q. Rapid and label-free optical assay of S-layer protein with high sensitivity using TiO2-coated porous silicon-based microfluidic biosensor. Sens. Actuators B: Chem. 2020, 321, 128524. [Google Scholar] [CrossRef]
- Rodoplu, D.; Chang, C.S.; Kao, C.Y.; Hsu, C.H. A simple magnetic-assisted microfluidic method for rapid detection and phenotypic characterization of ultralow concentrations of bacteria. Talanta 2021, 230, 122291. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, X.; Bai, M.; Zhang, J.; Yu, H.; Chen, F.; Zhao, Y. A microfluidic surface-enhanced Raman scattering (SERS) sensor for microRNA in extracellular vesicles with nucleic acid-tyramine cascade amplification. Chin. Chem. Lett. 2022, 33, 2101–2104. [Google Scholar] [CrossRef]
- Saha, A.; Jana, N.R. Paper-based microfluidic approach for surface-enhanced raman spectroscopy and highly reproducible detection of proteins beyond picomolar concentration. ACS Appl. Mater. Interfaces 2015, 7, 996–1003. [Google Scholar] [CrossRef]
- Jadhav, S.A.; Biji, P.; Panthalingal, M.K.; Murali Krishna, C.; Rajkumar, S.; Joshi, D.S.; Sundaram, N. Development of integrated microfluidic platform coupled with Surface-enhanced Raman Spectroscopy for diagnosis of COVID-19. Med. Hypotheses 2021, 146, 110356. [Google Scholar] [CrossRef]
- Wang, J.; Kao, Y.C.; Zhou, Q.; Wuethrich, A.; Stark, M.S.; Schaider, H.; Soyer, H.P.; Lin, L.L.; Trau, M. An Integrated Microfluidic-SERS Platform Enables Sensitive Phenotyping of Serum Extracellular Vesicles in Early Stage Melanomas. Adv. Funct. Mater. 2021, 32, 2010296. [Google Scholar] [CrossRef]
- Lin, H.Y.; Huang, C.H.; Hsieh, W.H.; Liu, L.H.; Lin, Y.C.; Chu, C.C.; Wang, S.T.; Kuo, I.T.; Chau, L.K.; Yang, C.Y. On-line SERS detection of single bacterium using novel SERS nanoprobes and a microfluidic dielectrophoresis device. Small 2014, 10, 4700–4710. [Google Scholar] [CrossRef]
- He, X.; Zhou, X.; Liu, Y.; Wang, X. Ultrasensitive, recyclable and portable microfluidic surface-enhanced raman scattering (SERS) biosensor for uranyl ions detection. Sens. Actuators B: Chem. 2020, 311, 127676. [Google Scholar] [CrossRef]
- Rodríguez-Lorenzo, L.; Garrido-Maestu, A.; Bhunia, A.K.; Espiña, B.; Prado, M.; Diéguez, L.; Abalde-Cela, S. Gold Nanostars for the Detection of Foodborne Pathogens via Surface-Enhanced Raman Scattering Combined with Microfluidics. ACS Appl. Nano Mater. 2019, 2, 6081–6086. [Google Scholar] [CrossRef]
- Lin, X.; Lin, S.; Liu, Y.; Zhao, H.; Liu, B.; Wang, L. Lab-on-paper surface-enhanced Raman spectroscopy platform based on self-assembled Au@Ag nanocube monolayer for on-site detection of thiram in soil. J. Raman Spectrosc. 2019, 50, 916–925. [Google Scholar] [CrossRef]
- Asgari, S.; Wu, G.; Aghvami, S.A.; Zhang, Y.; Lin, M. Optimisation using the finite element method of a filter-based microfluidic SERS sensor for detection of multiple pesticides in strawberry. Food Addit. Contam. Part. A Chem. Anal. Control. Expo. Risk Assess. 2021, 38, 646–658. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, G.; Guan, X.L.; Zhao, L. Rapid preparation of surface-enhanced Raman substrate in microfluidic channel for trace detection of amoxicillin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 235, 118262. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Q.; Tao, W.; Yu, B.; Du, Y. Quantitative analysis of thymine with surface-enhanced Raman spectroscopy and partial least squares (PLS) regression. Anal. Bioanal. Chem. 2010, 398, 1827–1832. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Weng, S.; Yang, L.; Liu, J. Detection and direct readout of drugs in human urine using dynamic surface-enhanced Raman spectroscopy and support vector machines. Anal. Chem. 2015, 87, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Dies, H.; Raveendran, J.; Escobedo, C.; Docoslis, A. Rapid identification and quantification of illicit drugs on nanodendritic surface-enhanced Raman scattering substrates. Sens. Actuators B Chem. 2018, 257, 382–388. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, H.; Wang, Y.; Qian, H.; Zhu, Y.; Dong, B.; Xu, F.; Chen, N.; Liu, S.; Pan, J.; et al. Deep convolutional neural networks combine Raman spectral signature of serum for prostate cancer bone metastases screening. Nanomedicine 2020, 29, 102245. [Google Scholar] [CrossRef]
- Das, S.K.; Bhattacharya, T.S.; Ghosh, M.; Chowdhury, J. Probing blood plasma samples for the detection of diabetes using SERS aided by PCA and LDA multivariate data analyses. New. J. Chem. 2021, 45, 2670–2682. [Google Scholar] [CrossRef]
- Zhu, J.; Sharma, A.S.; Xu, J.; Xu, Y.; Jiao, T.; Ouyang, Q.; Li, H.; Chen, Q. Rapid on-site identification of pesticide residues in tea by one-dimensional convolutional neural network coupled with surface-enhanced Raman scattering. Spectrochim Acta A Mol. Biomol. Spectrosc. 2021, 246, 118994. [Google Scholar] [CrossRef] [PubMed]
- Lussier, F.; Missirlis, D.; Spatz, J.P.; Masson, J.F. Machine-Learning-Driven Surface-Enhanced Raman Scattering Optophysiology Reveals Multiplexed Metabolite Gradients Near Cells. ACS Nano 2019, 13, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
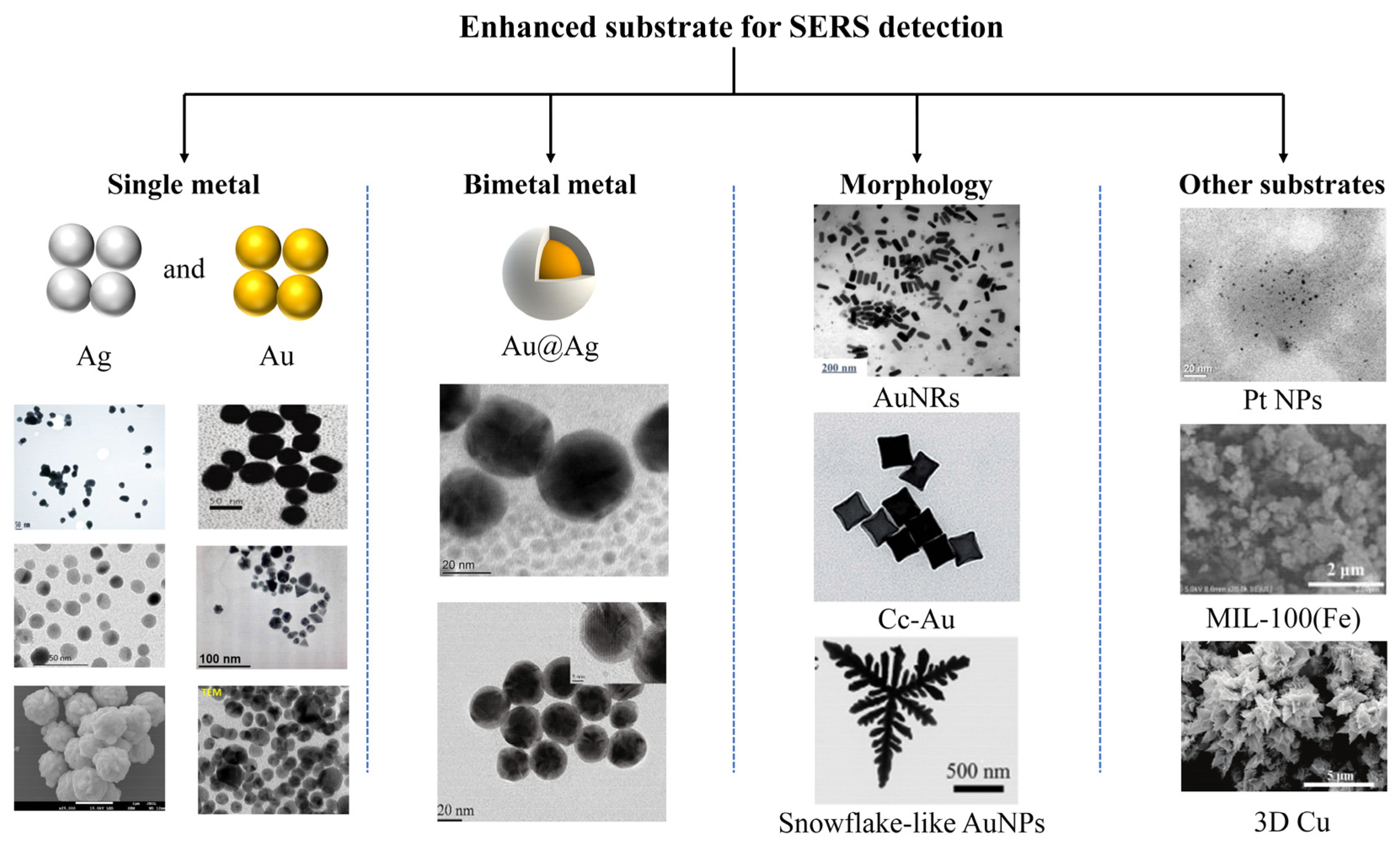

| Type | Substrates | Detector | LOD | Ref |
|---|---|---|---|---|
| Single metal | AgNPs | Oxytetracycline | 5 ppb | [75] |
| Valeric acid | 10 × 10−1 M | [76] | ||
| Methomyl | 5.58 × 10−4 µg/mL | [77] | ||
| Acetamiprid-(AC) | 1.88 × 10−4 µg/mL | |||
| 2,4-dichlorophenoxyacetic acid-(2,4-D) | 4.72 × 10−3 µg/mL | |||
| AuNPs | 2,4,6-Trinitrotoluene | 2 pM | [78] | |
| Chabazite | - | [79] | ||
| Procaine | 10−10 M | [80] | ||
| Bimetal metal | Au@AgNPs | Tricyclazole | 0.005 ppm | [83] |
| Thiram | 0.003 ppm | |||
| Thiacloprid | 0.1 mg/kg | [84] | ||
| Profenofos | 0.01 mg/kg | |||
| Oxamyl | 0.01 mg/kg | |||
| Morphology | AuNRs | Carbaryl | 391 ppb | [92] |
| cc-Au | p-MBA | - | [93] | |
| Snowflake | R6G | 3 × 10−9 M | [94] | |
| Organophosphorus Pesticides | 10−8 M | |||
| Other substrates | PtNPs | Methylene blue | 10−5 M | [85] |
| MIL-100(Fe) | Toluene | 2.5 ppm | [86] | |
| 3D Cu | 4-MBA | 10−5 M | [87] |
| Type | Year | Channel Structure | Result | Ref |
|---|---|---|---|---|
| Mix | 1999 | T-shaped | Diffusion mixing | [95] |
| 2002 | Z-shaped | Mixing at high Reynolds numbers | [96] | |
| 2002 | Staggered herringbone mixer (SHM) | Mixing at low Reynolds numbers | [97] | |
| 2008 | Alligator-toothed | Higher mixing efficiency | [39] | |
| 2012 | Baker’s transformation (MBT) | 70-fold faster than straight channel | [98] | |
| Synthetic Nanoparticles | 2020 | Cotton microfluidic device | AuNPs | [99] |
| 2019 | Droplet microfluidics | (AuNR@Ag)-PANI JNP | [100] | |
| 2018 | In situ | np-AgMS | [101] | |
| Substrate integration | 2012 | SMAs | High catalytic activity and SERS enhancement | [102] |
| 2013 | AgNPs’ nanowalls | LOD: 50 pM | [103] | |
| 2019 | Ag nanostructures | LOD: 10−7 M | [104] | |
| Target trapping | 2013 | DEP | Target trapping within 1 min | [105] |
| 2021 | Filtration and electrokinetic flow | Bacterial concentration | [106] | |
| 2018 | Antibody/antigen | LOD: 0.0001 U/mL | [107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, S.; Yao, F.; Bao, F.; Ge, Y.; Zou, M.; Liang, P.; Chen, Q. Progress of Microfluidics Combined with SERS Technology in the Trace Detection of Harmful Substances. Chemosensors 2022, 10, 449. https://doi.org/10.3390/chemosensors10110449
Chen J, Li S, Yao F, Bao F, Ge Y, Zou M, Liang P, Chen Q. Progress of Microfluidics Combined with SERS Technology in the Trace Detection of Harmful Substances. Chemosensors. 2022; 10(11):449. https://doi.org/10.3390/chemosensors10110449
Chicago/Turabian StyleChen, Junjie, Suyang Li, Fuqi Yao, Fubing Bao, Yuqing Ge, Minqiang Zou, Pei Liang, and Qiang Chen. 2022. "Progress of Microfluidics Combined with SERS Technology in the Trace Detection of Harmful Substances" Chemosensors 10, no. 11: 449. https://doi.org/10.3390/chemosensors10110449






