Catalyzed Hairpin Assembly-Assisted DNA Dendrimer Enhanced Fluorescence Anisotropy for MicroRNA Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Apparatus
2.2. General Procedure
3. Results and Discussion
3.1. Working Principle
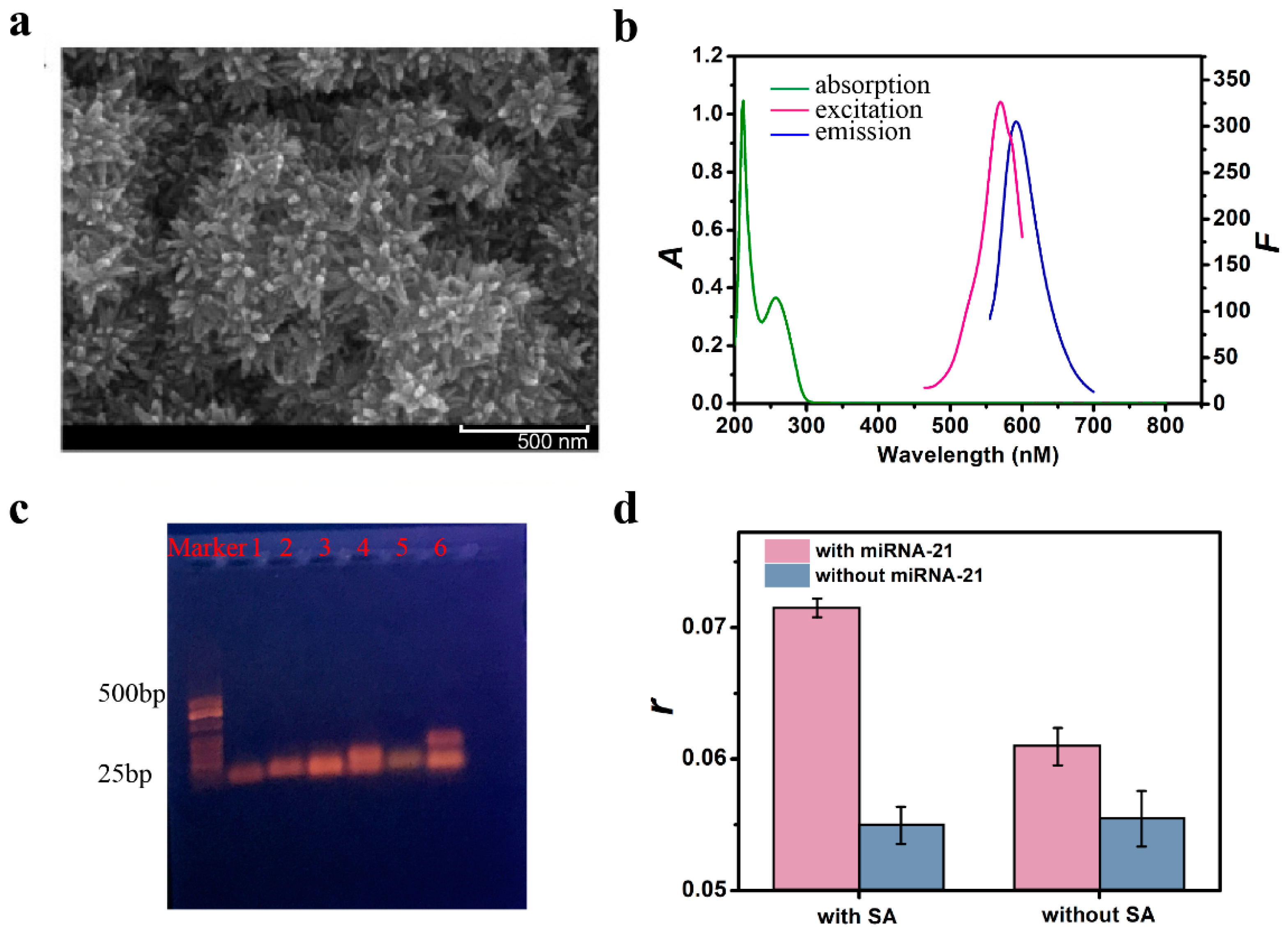
3.2. Feasibility Study
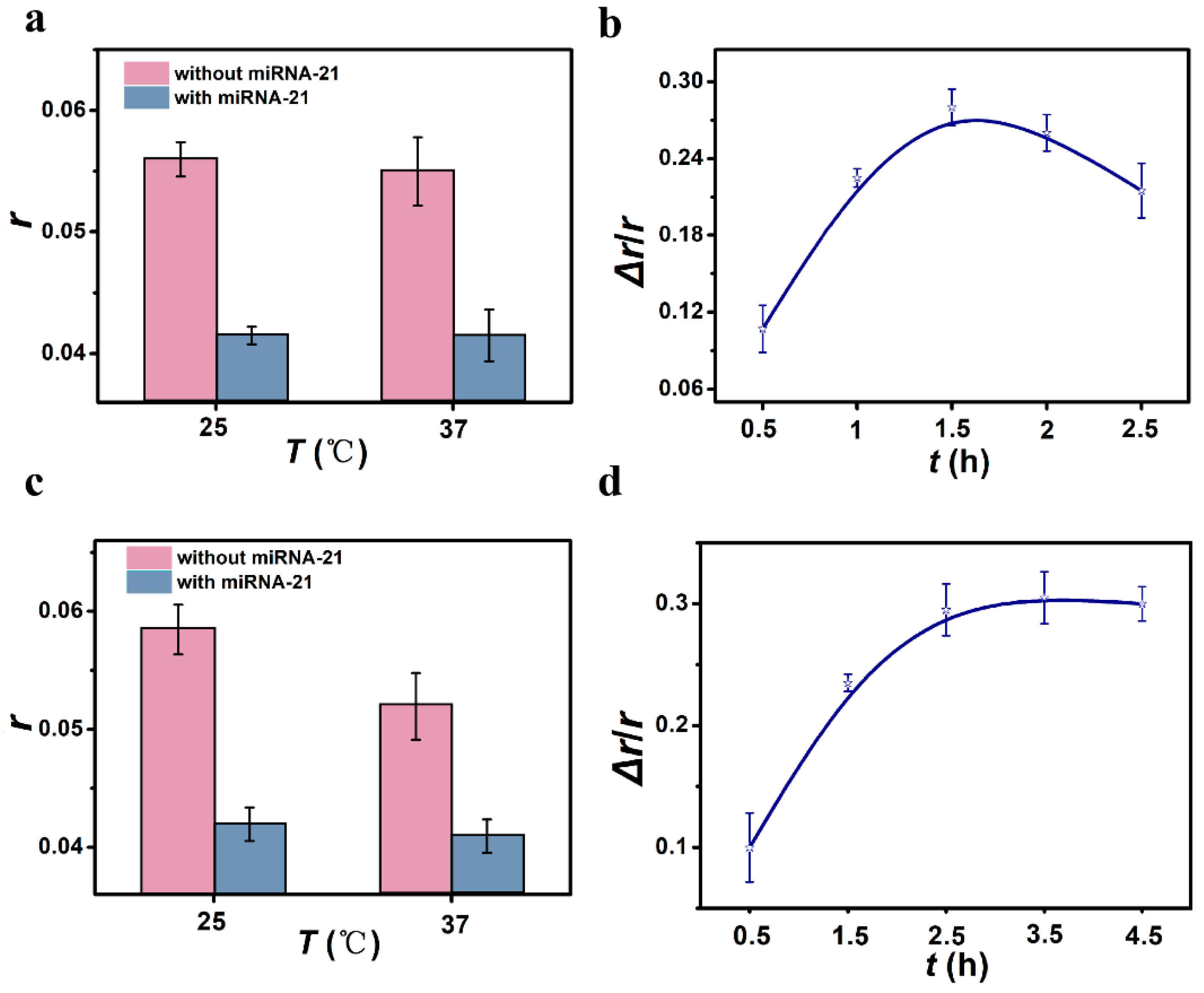
3.3. Optimal Conditions
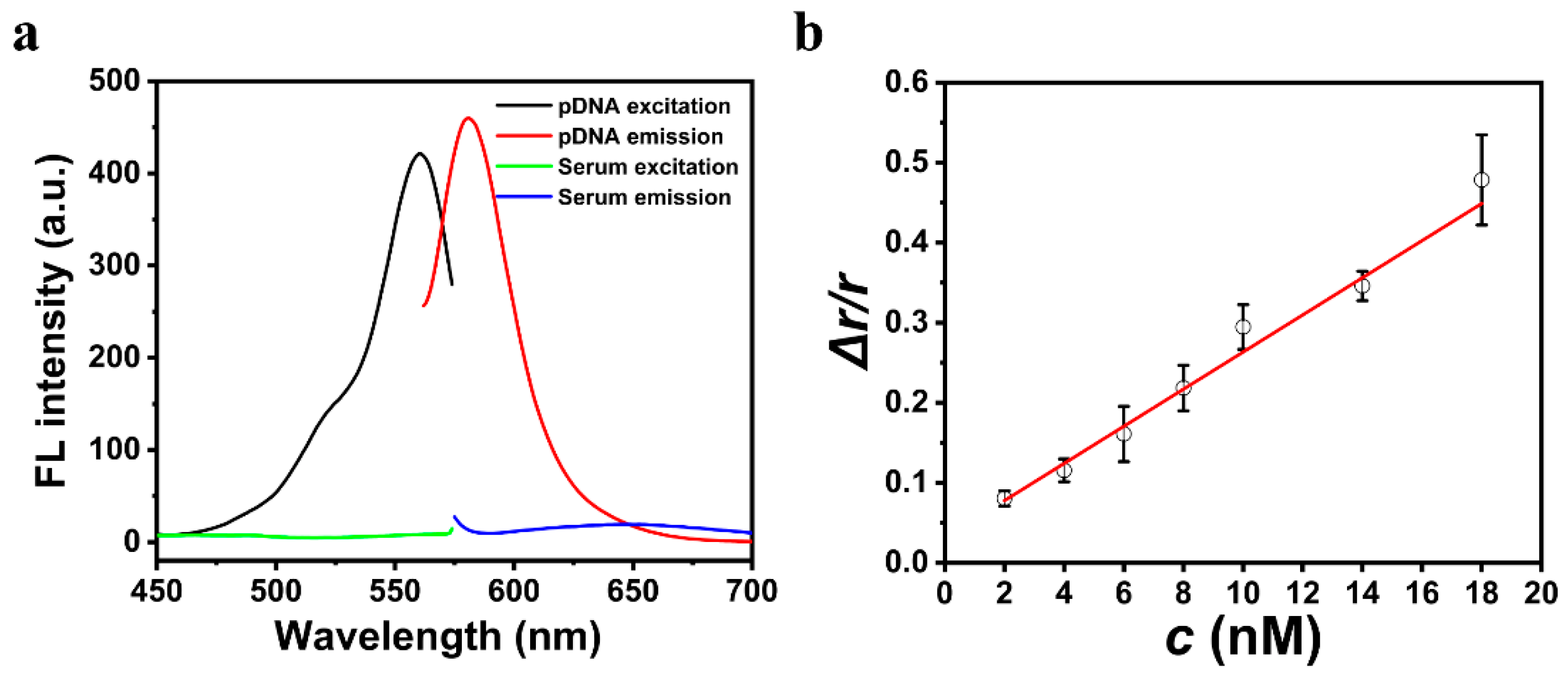
3.4. The Sensitivity and Specificity of the Strategy
3.5. Detection of miRNA-21 in Human Serum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jameson, D.M.; Ross, J.A. Fluorescence Polarization/Anisotropy in Diagnostics and Imaging. Chem. Rev. 2010, 110, 2685–2708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Li, Y.F.; Huang, C.Z.; Zhen, S.J. A novel graphene oxide amplified fluorescence anisotropy assay with improved accuracy and sensitivity. Chem. Commun. 2015, 51, 16080–16083. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, J.; Chen, X.; Qiu, H. Recent progress in nanomaterial-enhanced fluorescence polarization/anisotropy sensors. Chin. Chem. Lett. 2019, 30, 1575–1580. [Google Scholar] [CrossRef]
- Cui, L.; Zou, Y.; Lin, N.; Zhu, Z.; Jenkins, G.; Yang, C.J. Mass Amplifying Probe for Sensitive Fluorescence Anisotropy Detection of Small Molecules in Complex Biological Samples. Anal. Chem. 2012, 84, 5535–5541. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Yang, B.; Zhang, X.; Cui, L.; Meng, H.; Mei, L.; Wu, C.; Ren, S.; Tan, W. Enzymatic cleavage and mass amplification strategy for small molecule detection using aptamer-based fluorescence polarization biosensor. Anal. Chim. Acta 2015, 879, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, R.B.; Chen, X.; Zhou, N.; Cavagnero, S. Fluorescence Anisotropy Decays and Microscale-Volume Viscometry Reveal the Compaction of Ribosome-Bound Nascent Proteins. J. Phys. Chem. B 2021, 125, 6543–6558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.; Wang, H.; Le, X.C. Nucleic acid aptamers improving fluorescence anisotropy and fluorescence polarization assays for small molecules. Trends Analyt. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Chaudhary, A.; Schneitz, K. Using Steady-State Fluorescence Anisotropy to Study Protein Clustering. Methods Mol. Biol. 2022, 2457, 253–260. [Google Scholar] [PubMed]
- Li, Y.; Sun, Y.; Ye, J.; Pan, F.; Peng, B.; Li, H.; Zhang, M.; Xu, Y. Sensitive and selective detection of microRNA in complex biological samples based on protein-enhanced fluorescence anisotropy. Anal. Methods 2020, 12, 687–692. [Google Scholar] [CrossRef]
- Yoo, H.; Drummond, D.A. Using fluorescence anisotropy to monitor chaperone dispersal of RNA-binding protein condensates. STAR Protoc. 2022, 3, 101409. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, N.; Wang, H.; Zhao, Q. Fluorescence Anisotropy-Based Signal-Off and Signal-On Aptamer Assays Using Lissamine Rhodamine B as a Label for Ochratoxin, A. J. Agric. Food Chem. 2020, 68, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Xie, T.J.; Li, C.H.; Ye, Q.C.; Tian, L.L.; Li, Y.F.; Huang, C.Z.; Zhen, S.J. A crosslinked submicro-hydrogel formed by DNA circuit-driven protein aggregation amplified fluorescence anisotropy for biomolecules detection. Anal. Chim. Acta 2021, 1154, 338319. [Google Scholar] [CrossRef]
- Xiao, X.; Tao, J.; Zhang, H.Z.; Huang, C.Z.; Zhen, S.J. Exonuclease III-assisted graphene oxide amplified fluorescence anisotropy strategy for ricin detection. Biosens. Bioelectron. 2016, 85, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, N.; Zhang, L.; Zhao, J.; Zhao, S. A T7 exonuclease assisted dual-cycle signal amplification assay of miRNA using nanospheres-enhanced fluorescence polarization. Talanta 2019, 202, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Jourdain, M.; Grandjean, A.; Titz, A.; Jung, G. Beta-Boronic Acid-Substituted Bodipy Dyes for Fluorescence Anisotropy Analysis of Carbohydrate Binding. Anal. Chem. 2022, 94, 6112–6119. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Guo, C.; Zhai, J.; Xie, X. Fluorescence Anisotropy as a Self-Referencing Readout for Ion-Selective Sensing and Imaging Using Homo-FRET between Chromoionophores. Anal. Chem. 2022, 94, 9793–9800. [Google Scholar] [CrossRef]
- Jain, P.; Aida, T.; Motosuke, M. Fluorescence Anisotropy as a Temperature-Sensing Molecular Probe Using Fluorescein. Micromachines 2021, 12, 1109. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.-W.; Xu, Z.; Li, H.-B.; Hu, X.-Y.; Guo, D.-S. Study on assembling compactness of amphiphilic calixarenes by fluorescence anisotropy. Supramol. Chem. 2021, 33, 527–533. [Google Scholar] [CrossRef]
- Ye, H.; Duan, N.; Gu, H.; Wang, H.; Wang, Z. Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers. Mikrochim. Acta 2019, 186, 173. [Google Scholar] [CrossRef]
- Ye, H.; Lu, Q.; Duan, N.; Wang, Z. GO-amplified fluorescence polarization assay for high-sensitivity detection of aflatoxin B-1 with low dosage aptamer probe. Anal. Bioanal. Chem. 2019, 411, 1107–1115. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, H. Fluorescence Anisotropy Reduction of An Allosteric G-Rich Oligonucleotide for Specific Silver Ion and Cysteine Detection Based on the G-Ag+-G Base Pair. Anal. Chem. 2019, 91, 14538–14544. [Google Scholar] [CrossRef] [PubMed]
- Shokri, E.; Hosseini, M.; Sadeghan, A.A.; Bahmani, A.; Nasiri, N.; Hosseinkhani, S. Virus-directed synthesis of emitting copper nanoclusters as an approach to simple tracer preparation for the detection of Citrus Tristeza Virus through the fluorescence anisotropy immunoassay. Sens. Actuators B Chem. 2020, 321, 128634. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, S.; Chen, Z.-F.; Liu, Y.-C.; Liang, H. Ultrasensitive endonuclease activity and inhibition detection using gold nanoparticle-enhanced fluorescence polarization. Chem. Commun. 2011, 47, 4763–4765. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Q. Aptamer Structure Switch Fluorescence Anisotropy Assay for Small Molecules Using Streptavidin as an Effective Signal Amplifier Based on Proximity Effect. Anal. Chem. 2019, 91, 7379–7384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Xiao, X.; Li, C.H.; Men, C.; Ye, Q.C.; Lv, W.Y.; Li, Y.F.; Huang, C.Z.; Zhen, S.J. DNA nanosheet as an excellent fluorescence anisotropy amplification platform for accurate and sensitive biosensing. Talanta 2020, 211, 120730. [Google Scholar] [CrossRef]
- Brom, T.; Reddavide, F.V.; Heiden, S.; Thompson, M.; Zhang, Y. Influence of the geometry of fluorescently labelled DNA constructs on fluorescence anisotropy assay. Biochem. Biophys. Res. Commun. 2020, 533, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wan, B.; Du, W.; Hu, H.; Zeng, L.; Duan, X.; Liu, J.; Wei, Z.; Tang, L.; Peng, Y. A ligation-triggered and protein-assisted fluorescence anisotropy amplification platform for sensitive and selective detection of small molecules in a biological matrix. Rsc Adv. 2020, 10, 21789–21794. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, Q. A simple fluorescence anisotropy assay for detection of bisphenol A using fluorescently labeled aptamer. J. Environ. Sci. 2020, 97, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Bricker, W.P. Molecular interactions in DNA-scaffolded energy transfer systems. Biophys. J. 2022, 121, 65. [Google Scholar] [CrossRef]
- Xu, L.; Zhou, B.; Song, Y.; Cai, X.; Lu, W. Electron-Transfer Study and Single Nucleotide Discrimination of a DNA Sequence on a Polymer Gold Electrode (PGE) by Differential Pulse Voltammetry (DPV). Anal. Lett. 2022, 55, 1919–1931. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Z.; Liu, W.; Gao, F.; Guo, J.; Song, B.; Lee, L.P.; Zhang, F. The Selective Function of Quantum Biological Electron Transfer between DNA Bases and Metal Ions in DNA Replication. J. Phys. Chem. Lett. 2022, 13, 7779–7787. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Metternich, J.B.; Tiwari, P.; Benzenberg, L.R.; Harrison, J.A.; Liu, Q.; Zenobi, R. Structural Studies of a Stapled Peptide with Native Ion Mobility- Mass Spectrometry and Transition Metal Ion Fo?rster Resonance Energy Transfer in the Gas Phase. J. Am. Chem. Soc. 2022, 144, 14441–14445. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Li, M.; Peng, K.; Ye, T.; Wu, X.; Yuan, M.; Cao, H.; Yin, F.; Gu, H.; Xu, F. Fluorescence Resonance Energy Transfer Aptasensor of Ochratoxin A Constructed Based on Gold Nanorods and DNA Tetrahedrons. J. Agric. Food Chem. 2022, 70, 10662–10668. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Ma, C.; Yan, Y.; Chen, J. Detection of DNA Methyltransferase Activity via Fluorescence Resonance Energy Transfer and Exonuclease-Mediated Target Recycling. Biosensors 2022, 12, 395. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Zheng, H.; Almashriqi, H.S.; Lv, W.; Zhai, H. DNA-Templated Gold Nanoclusters for Fluorescence Resonance Energy Transfer-Based Human Serum Albumin Detection. J. Anal. Chem. 2022, 77, 216–223. [Google Scholar]
- Liu, J.; Wang, C.; Jiang, Y.; Hu, Y.; Li, J.; Yang, S.; Li, Y.; Yang, R.; Tan, W.; Huang, C.Z. Graphene Signal Amplification for Sensitive and Real-Time Fluorescence Anisotropy Detection of Small Molecules. Anal. Chem. 2013, 85, 1424–1430. [Google Scholar] [CrossRef]
- Fan, Y.-L.; Liu, Z.-Y.; Zeng, Y.-M.; Huang, L.-Y.; Li, Z.; Zhang, Z.-L.; Pang, D.-W.; Tian, Z.-Q. A near-infrared-II fluorescence anisotropy strategy for separation-free detection of adenosine triphosphate in complex media. Talanta 2021, 223, 121721. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ravelet, C.; Perrier, S.; Guieu, V.; Fiore, E.; Peyrin, E. Single-Stranded DNA Binding Protein-Assisted Fluorescence Polarization Aptamer Assay for Detection of Small Molecules. Anal. Chem. 2012, 84, 7203–7211. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zheng, C.; Cansiz, S.; Wu, C.; Xu, J.; Cui, C.; Liu, Y.; Hou, W.; Wang, Y.; Zhang, L.; et al. Self-assembly of DNA Nanohydrogels with Controllable Size and Stimuli-Responsive Property for Targeted Gene Regulation Therapy. J. Am. Chem. Soc. 2015, 137, 1412–1415. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, D.; Sim, S.; Niwa, T.; Taguchi, H.; Aida, T. Protein Nanotube Selectively Cleavable with DNA: Supramolecular Polymerization of “DNA-Appended Molecular Chaperones”. J. Am. Chem. Soc. 2018, 140, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tian, C.; Guo, F.; Liu, Z.; Jiang, W.; Mao, C. DNA-Directed Three-Dimensional Protein Organization. Angew. Chem. Int. Ed. 2012, 51, 3382–3385. [Google Scholar] [CrossRef] [PubMed]
- Sanghamitra, N.J.M.; Ueno, T. Expanding coordination chemistry from protein to protein assembly. Chem. Commun. 2013, 49, 4114–4126. [Google Scholar] [CrossRef]
- Pieters, B.J.G.E.; van Eldijk, M.B.; Nolte, R.J.M.; Mecinovic, J. Natural supramolecular protein assemblies. Chem. Soc. Rev. 2016, 45, 24–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgilis, E.; Abdelghani, M.; Pille, J.; Aydinlioglu, E.; van Hest, J.C.M.; Lecommandoux, S.; Garanger, E. Nanoparticles based on natural, engineered or synthetic proteins and polypeptides for drug delivery applications. Int. J. Pharm. 2020, 586, 119537. [Google Scholar] [CrossRef] [PubMed]
- Mo, F.; Jiang, K.; Zhao, D.; Wang, Y.; Song, J.; Tan, W. DNA hydrogel-based gene editing and drug delivery systems. Adv. Drug Deliv. Rev. 2021, 168, 79–98. [Google Scholar] [CrossRef]
- Shah, B.M.; Palakurthi, S.S.; Khare, T.; Khare, S.; Palakurthi, S. Natural proteins and polysaccharides in the development of micro/nano delivery systems for the treatment of inflammatory bowel disease. Int. J. Biol. Macromol. 2020, 165, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.V.; Zareie, H.M.; Sarikaya, M. Chimeric Peptide-Based Biomolecular Constructs for Versatile Nucleic Acid Biosensing. ACS Appl. Mater. Interfaces 2022, 14, 23164–23181. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Williams, J.Z.; Chang, R.; Li, Z.; Burnett, C.E.; Hernandez-Lopez, R.; Setiady, I.; Gai, E.; Patterson, D.M.; Yu, W.; et al. DNA scaffolds enable efficient and tunable functionalization of biomaterials for immune cell modulation. Nat. Nanotechnol. 2021, 16, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Fang, T.; Pang, T.; Chen, Z.; Cheng, L.; Ma, D.; Xi, Z. The Potential Application of Branch-PCR Assembled PTEN Gene Nanovector in Lung Cancer Gene Therapy. Chembiochem 2022, 23, e202200387. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lv, J.; Huang, H.; Xu, H.; Zhang, J.; Xue, C.; Zhang, S.; Wu, Z.-S. Structure-switchable aptamer-arranged reconfigurable DNA nanonetworks for targeted cancer therapy. Nanomedicine 2022, 43, 102553. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Jiang, Y.; Lu, C. Self-Assembled ATP-Responsive DNA Nanohydrogel for Specifically Activated Fluorescence Imaging and Chemotherapy in Cancer Cells. Anal. Chem. 2022, 94, 10221–10226. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, T.; Mo, F.; Yu, W.; Shen, Y.; Jiang, Q.; Wang, F.; Liu, X. Programmable Assembly of Multivalent DNA-Protein Superstructures for Tumor Imaging and Targeted Therapy. Angew. Chem. Int. Ed. 2022, 61, e202211505. [Google Scholar] [CrossRef]
- Huang, W.; Zhan, D.; Xie, Y.; Li, X.; Lai, G. Dual CHA-mediated high-efficient formation of a tripedal DNA walker for constructing a novel proteinase-free dual-mode biosensing strategy. Biosens. Bioelectron. 2022, 197, 113708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Waseem, M.; Zeng, Z.; Xu, J.; Chen, C.; Liu, Y.; Zhai, J.; Xia, R. MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 2022, 233, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, S.; Oh, S.J.; Han, S.M.; Moon, S.Y.; Kang, B.; Seo, S.B.; Jang, S.; Son, S.U.; Jung, J.; et al. miRNA sensing hydrogels capable of self-signal amplification for early diagnosis of Alzheimer’s disease. Biosens. Bioelectron. 2022, 209, 114279. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, H.; Liu, T.; Liang, C.; Luo, J. A knowledge-driven network for fine-grained relationship detection between miRNA and disease. Brief. Bioinform. 2022, 23, bbac058. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, S.; Wang, H.; Yu, K.; Guan, Y.; Liu, X.; Li, N.; Liu, F. DNA Dendrimer-Streptavidin Nanocomplex: An Efficient Signal Amplifier for Construction of Biosensing Platforms. Anal. Chem. 2017, 89, 6907–6914. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, D.; Luo, Z.; Duan, Y. A dual-functional fluorescent biosensor based on enzyme-involved catalytic hairpin assembly for the detection of APE1 and miRNA-21. Analyst 2022, 147, 2834–2842. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, M.; Jin, Y.; Li, B. Rapid and enzyme-free signal amplification for fluorescent detection of microRNA via localized catalytic hairpin assembly on gold nanoparticles. Talanta 2022, 242, 123142. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Hu, T.; Fang, Y.; Xiang, X.; Ma, C. A fluorescence strategy for the rapid detection of miRNA-21 based on G-quadruplex and cyclic amplification signal. Anal. Biochem. 2022, 652, 114775. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pan, M.; Zou, Z.-Q.; Fan, L.; Liu, X.-Q. Hybridization Chain Reaction-Mediated Luminescent Silver Nanocluster System for Amplified Detection of miRNA-21. Chin. J. Anal. Chem. 2020, 48, 1193–1201. [Google Scholar]
- Zhao, H.; Qu, Y.; Yuan, F.; Quan, X. A visible and label-free colorimetric sensor for miRNA-21 detection based on peroxidase-like activity of graphene/gold-nanoparticle hybrids. Anal. Methods 2016, 8, 2005–2012. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Z.; Liu, J.-H.; Yu, B.-Y.; Tian, J. Dual signal amplification for microRNA-21 detection based on duplex-specific nuclease and invertase. Rsc Adv. 2020, 10, 11257–11262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Lv, X.; Li, X.; Yao, M.; Li, K.; Deng, Y. Fluorescent microspheres lateral flow assay integrated with Smartphone-based reader for multiple microRNAs detection. Microchem. J. 2022, 179, 107551. [Google Scholar] [CrossRef]
- Seo, S.B.; Hwang, J.-S.; Kim, E.; Kim, K.; Roh, S.; Lee, G.; Lim, J.; Kang, B.; Jang, S.; Son, S.U.; et al. Isothermal amplification-mediated lateral flow biosensors for in vitro diagnosis of gastric cancer-related microRNAs. Talanta 2022, 246, 123502. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Cai, R.; Gui, S.; Zhang, Y.; Wang, X.; Zhou, N. A portable and quantitative detection of microRNA-21 based on cascade enzymatic reactions with dual signal outputs. Talanta 2021, 235, 112802. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, S.; Zou, R.; Chen, C.; Cai, C. An enzyme-free probe based on G-triplex assisted by silver nanocluster pairs for sensitive detection of microRNA-21. Microchim. Acta 2021, 188, 55. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Xiang, L.; Wang, L.; Gong, H.; Chen, F.; Cai, C. pH-responsive DNA hydrogels with ratiometric fluorescence for accurate detection of miRNA-21. Anal. Chim. Acta 2022, 1207, 339795. [Google Scholar] [CrossRef] [PubMed]


| Sample | Add (nM) | Found (nM) | Recovery (%) | RSD(%) (n = 3) |
|---|---|---|---|---|
| 1% Human serum | 2 | 2.2 ± 0.3 | 94.3–119.5 | 0.9 |
| 10 | 9.6 ± 0.7 | 90.2–102.3 | 1.8 | |
| 18 | 18.1 ± 1.8 | 90.7–109.9 | 5.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, T.; Liu, Y.; Xie, J.; Luo, Y.; Mao, K.; Huang, C.; Li, Y.; Zhen, S. Catalyzed Hairpin Assembly-Assisted DNA Dendrimer Enhanced Fluorescence Anisotropy for MicroRNA Detection. Chemosensors 2022, 10, 501. https://doi.org/10.3390/chemosensors10120501
Xie T, Liu Y, Xie J, Luo Y, Mao K, Huang C, Li Y, Zhen S. Catalyzed Hairpin Assembly-Assisted DNA Dendrimer Enhanced Fluorescence Anisotropy for MicroRNA Detection. Chemosensors. 2022; 10(12):501. https://doi.org/10.3390/chemosensors10120501
Chicago/Turabian StyleXie, Tianjin, Yuxin Liu, Jiali Xie, Yujie Luo, Kai Mao, Chengzhi Huang, Yuanfang Li, and Shujun Zhen. 2022. "Catalyzed Hairpin Assembly-Assisted DNA Dendrimer Enhanced Fluorescence Anisotropy for MicroRNA Detection" Chemosensors 10, no. 12: 501. https://doi.org/10.3390/chemosensors10120501
APA StyleXie, T., Liu, Y., Xie, J., Luo, Y., Mao, K., Huang, C., Li, Y., & Zhen, S. (2022). Catalyzed Hairpin Assembly-Assisted DNA Dendrimer Enhanced Fluorescence Anisotropy for MicroRNA Detection. Chemosensors, 10(12), 501. https://doi.org/10.3390/chemosensors10120501






