Phosphoric Acid Induced Controllable Nanoparticle Aggregation for Ultrasensitive SERS Detection of Malondialdehyde in a Microfluidic Chip
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Preparation of Uniform Silver NPs (Ag NPs)
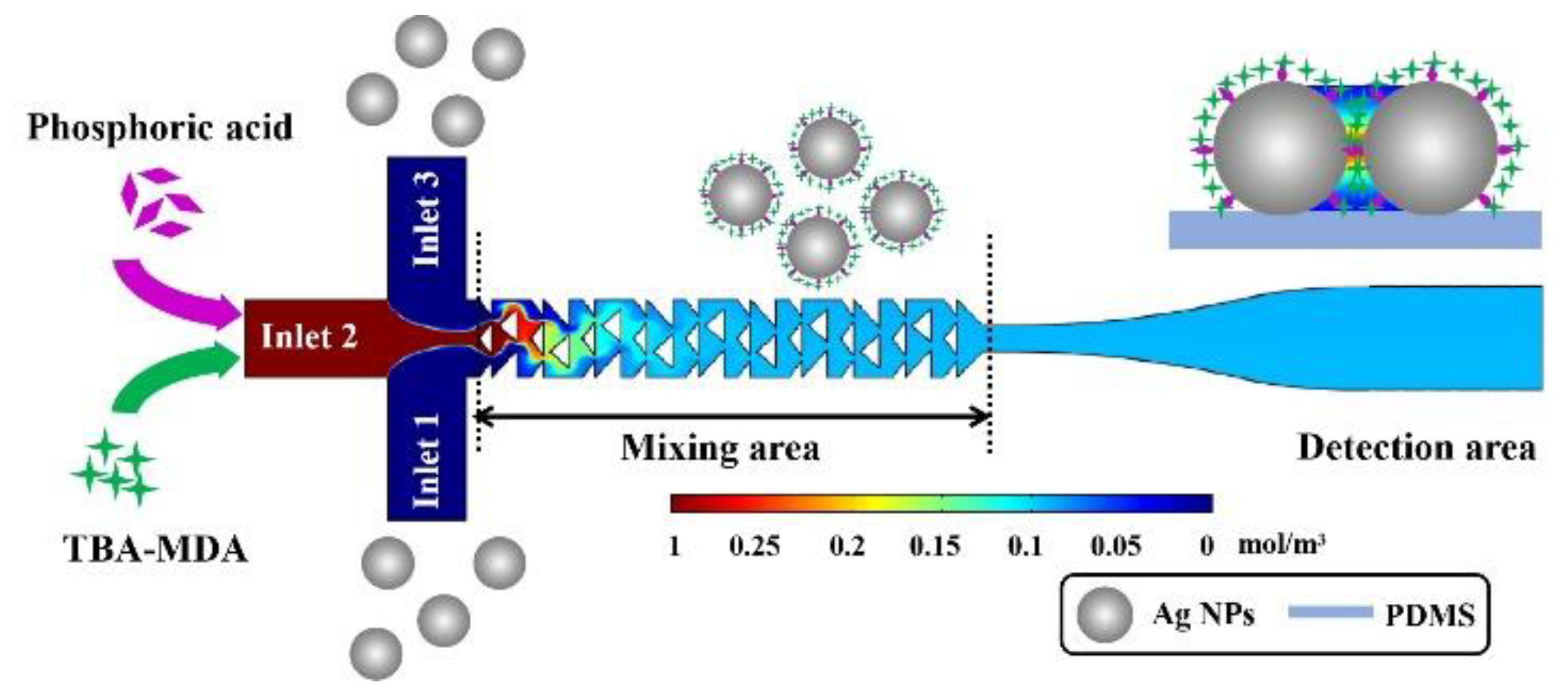
2.3. Fabrication of Cascaded Splitting and Recombination (C-SAR) Microfluidic Mixer
2.4. Preparation of TBA-MDA Adduct and Other Derivatives


2.5. Preparation of Real Samples
2.6. Fabrication of Phosphoric Acid Regulated MAHSO SERS Chip
2.7. SERS Measurement
2.8. Characterization
3. Results and Discussion
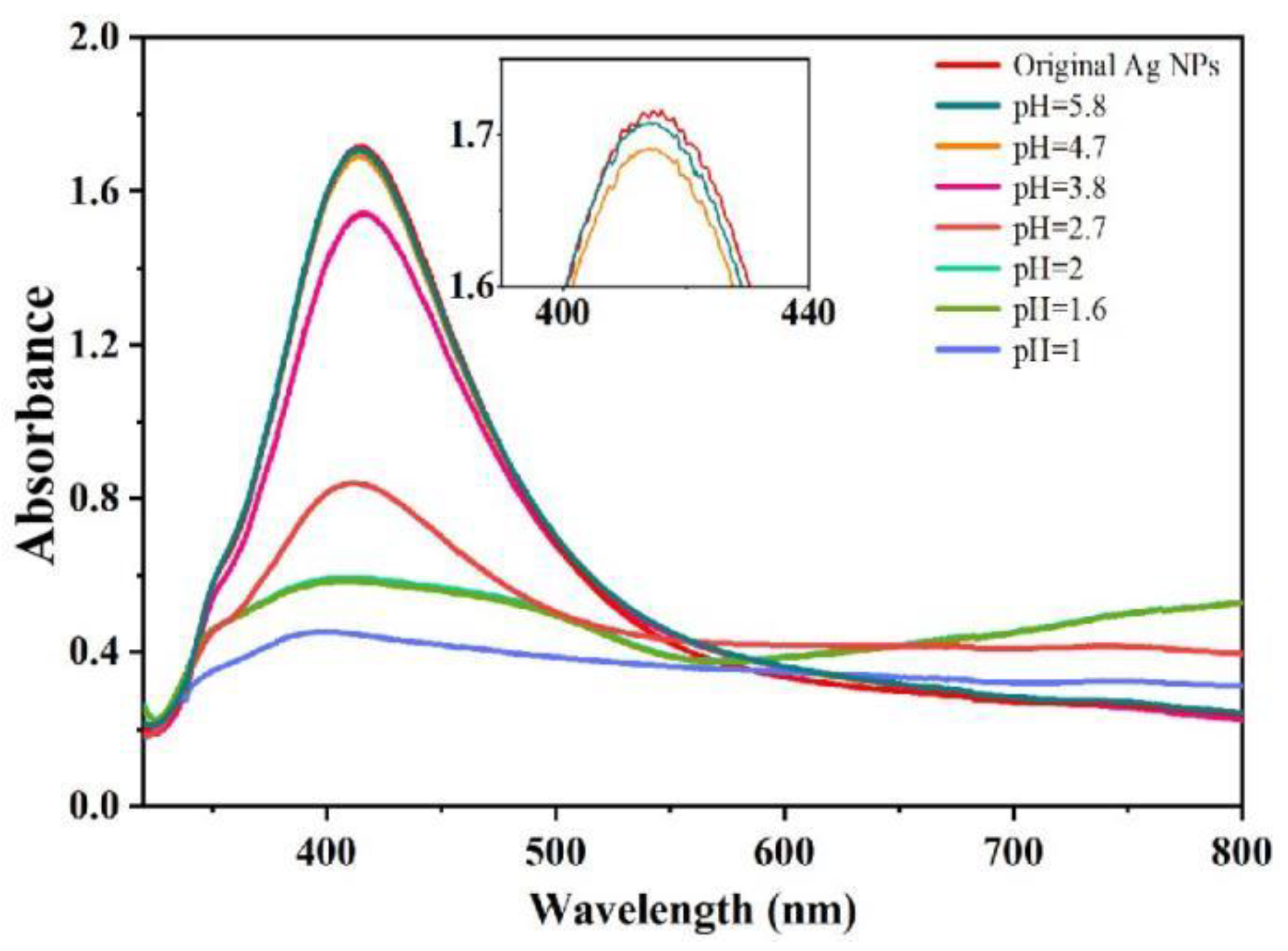
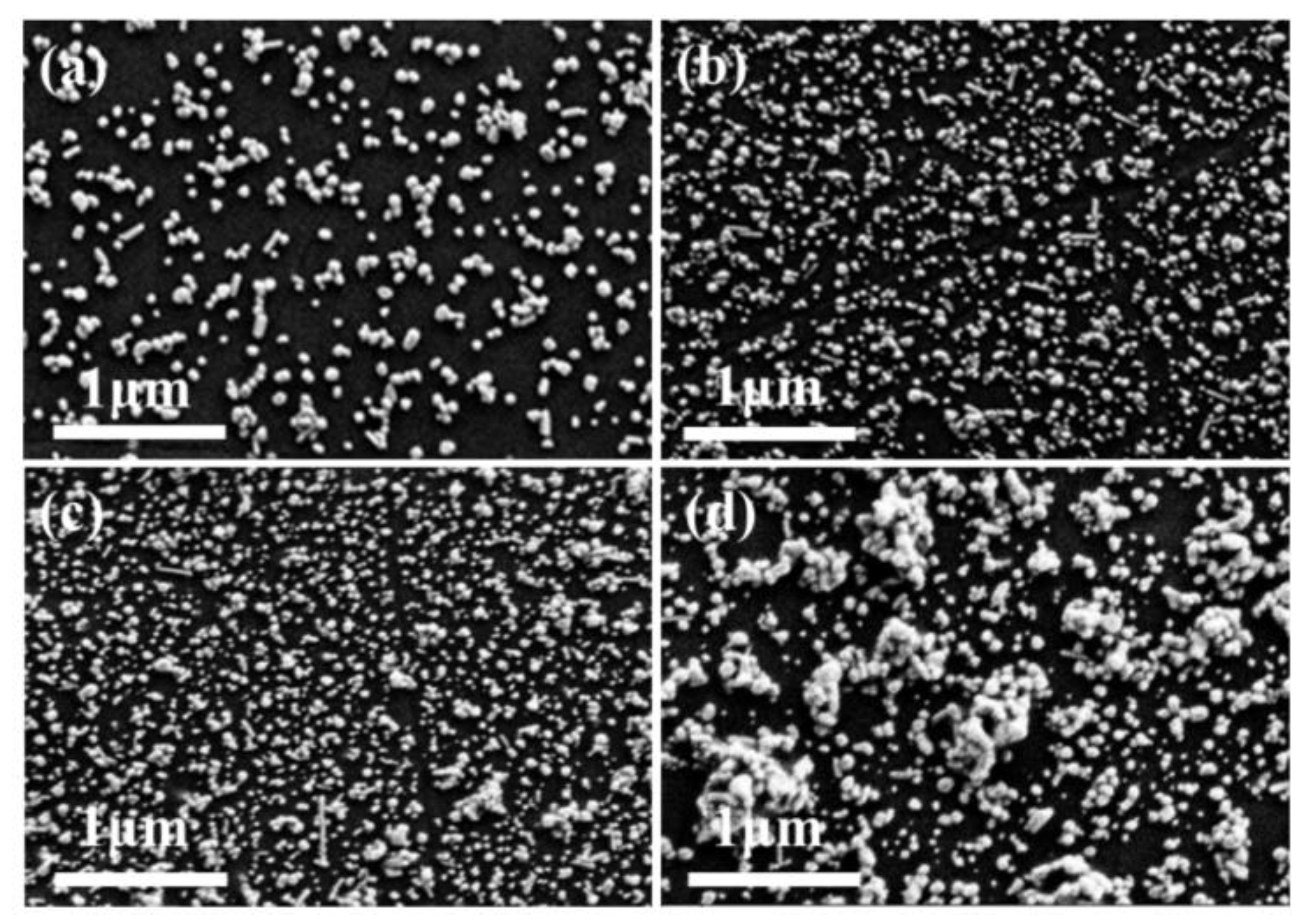
3.1. Phosphoric Acid Induced Ag NPs Aggregation
3.2. Phosphoric Acid Regulated SERS Substrate
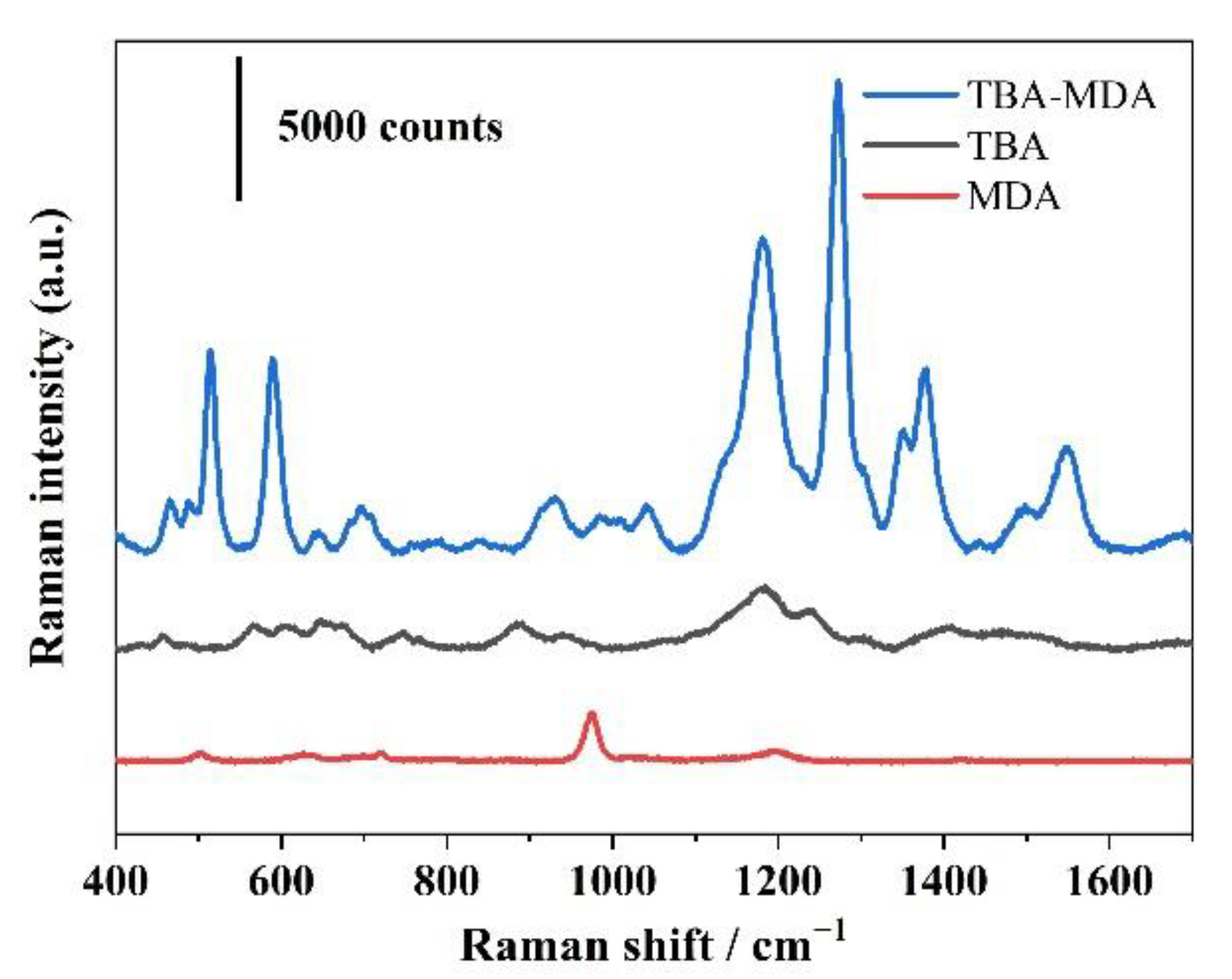
3.3. The Mechanism of Phosphoric Acid Enhanced MAHSO SERS Chip for Ultrasensitive MDA Detection
3.4. Optimization of Phosphoric Acid Concentration
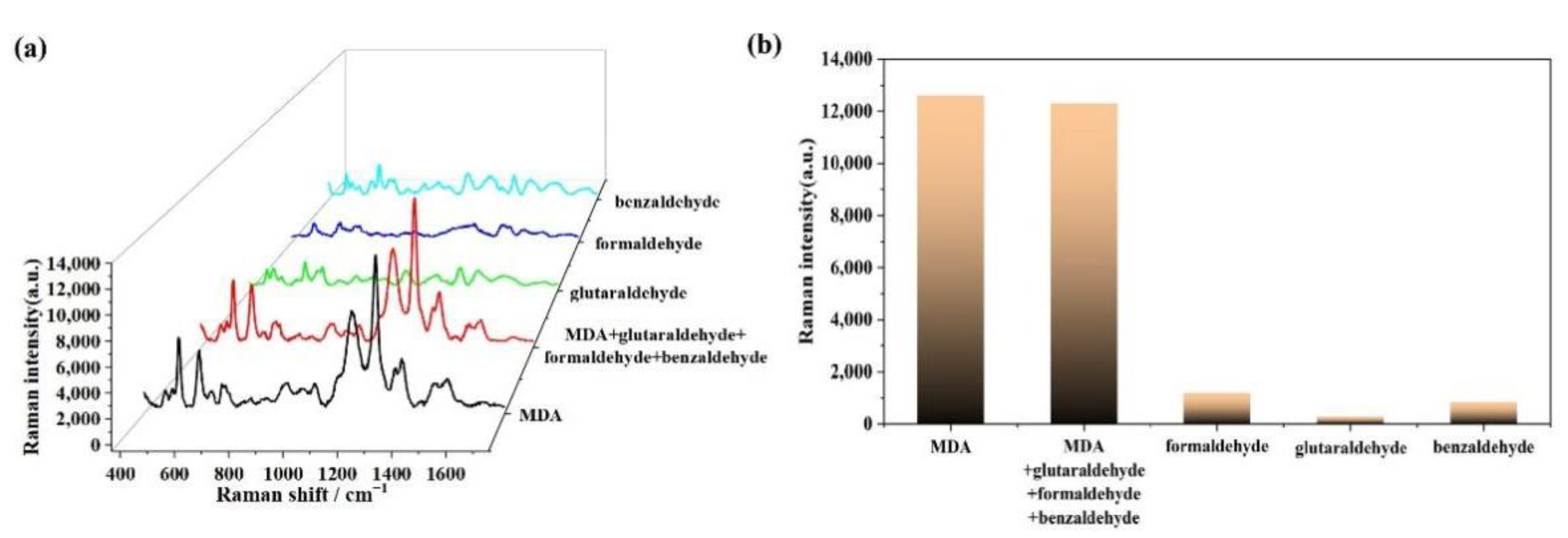
3.5. Specificity and Selectivity of MDA Detection
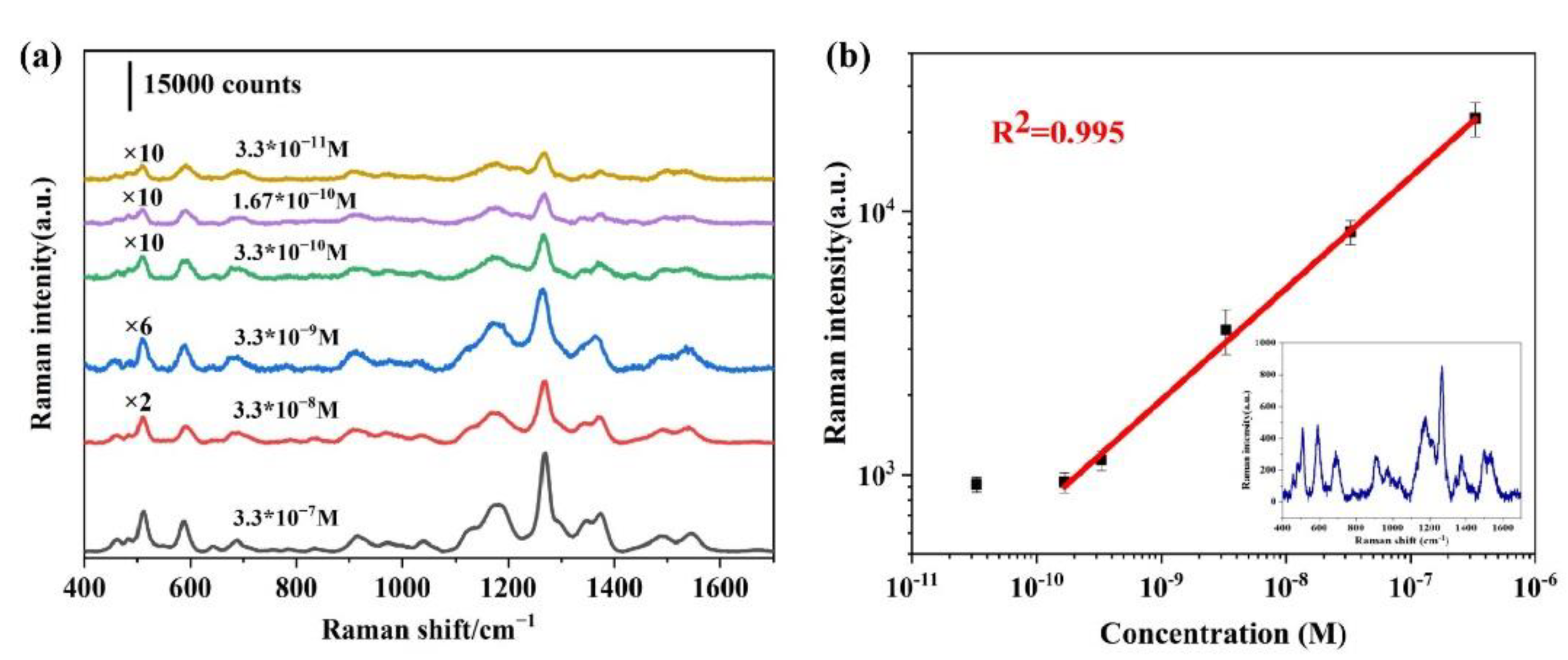
3.6. Sensitivity, Uniformity and Reproducibility
3.7. MDA Detection in Oil Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mateos, R.; Lecumberri, E.; Ramos, S.; Goya, L.; Bravo, L. Determination of malondialdehyde (MDA) by high-performance liquid chromatography in serum and liver as a biomarker for oxidative stress: Application to a rat model for hypercholesterolemia and evaluation of the effect of diets rich in phenolic antioxidants from fruits. J. Chromatgr. B 2005, 827, 76–82. [Google Scholar]
- Guillén-Sans, R.; Guzmán-Chozas, M. Aldehydes in food and its relation with the TBA test for rancidity. Lipid/Fett 1995, 97, 285–286. [Google Scholar] [CrossRef]
- Stefanescu, C.; Ciobica, A. The relevance of oxidative stress status in first episode and recurrent depression. J. Affect. Disord. 2012, 143, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.C.; Zhou, F.L.; He, D.L.; Bai, J.R.; Ding, H.; Wang, X.Y.; Nan, K.J. Oxidative stress in depressive patients with gastric adenocarcinoma. Int. J. Neuropsychopharmacol. 2009, 12, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Michalakeas, C.A.; Parissis, J.T.; Douzenis, A.; Nikolaou, M.; Varounis, C.; Andreadou, I.; Antonellos, N.; Markantonis-Kiroudis, S.; Paraskevaidis, I.; Ikonomidis, I.; et al. Effects of sertraline on circulating markers of oxidative stress in depressed patients with chronic heart failure: A pilot study. J. Card. Fail. 2011, 17, 748–754. [Google Scholar] [CrossRef]
- Singh, Z.; Karthigesu, I.P.; Singh, P.; Kaur, R. Use of malondialdehyde as a biomarker for assessing oxidative stress in different disease pathologies: A review. Iran. J. Public Health 2014, 43, 7–16. [Google Scholar]
- Ghanem, A.A.; Arafa, L.F.; El-Baz, A. Oxidative stress markers in patients with primary open-angle glaucoma. Curr. Eye Res. 2010, 35, 295–301. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Howard, B.; Yatin, S.; Koppal, T.; Drake, J.; Hensley, K.; Aksenov, M.; Aksenova, M.; Subramaniam, R.; Varadarajan, S.; et al. Elevated oxidative stress in models of normal brain aging and Alzheimer’s disease. Life Sci. 1999, 65, 1883–1892. [Google Scholar] [CrossRef]
- Kaykhaii, M.; Yahyavi, H.; Hashemi, M. A simple graphene-based pipette tip solid phase extraction of malondialdehyde from human plasma and its determination by spectrofluorometry. Anal. Bioanal. Chem. 2016, 408, 4907–4915. [Google Scholar] [CrossRef]
- Giera, M.; Kloos, D.P.; Raaphorst, A. Mild and selective labeling of malondialdehyde with 2-aminoacridone: Assessment of urinary malondialdehyde levels. Analyst 2011, 136, 2763–2769. [Google Scholar] [CrossRef]
- Erdelmeier, I.; Gérardmonnier, D.; Yadan, J.C. Reactions of N-methyl-2-phenylindole with malondialdehyde and 4-hydroxyalkenals. Mechanistic aspects of the colorimetric assay of lipid peroxidation. Chem. Res. Toxicol. 1998, 11, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- Korchazhkina, O.; Exley, C.; Spencer, S.A. Measurement by reversed-phase high-performance liquid chromatography of malondialdehyde in normal human urine following derivatisation with 2,4-dinitrophenylhydrazine. J. Chromatogr. B 2003, 794, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, H.; Luo, J. New method for HPLC separation and fluorescence detection of malonaldehyde in normal human plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006, 832, 103–108. [Google Scholar] [CrossRef]
- Korizis, K.N.; Exarchou, A.; Michalopoulos, E.; Georgakopoulos, C.D.; Kolonitsiou, F.; Mantagos, S.; Gartaganis, S.P.; Karamanos, N.K. Determination of malondialdehyde by capillary electrophoresis, application to human plasma and relation of its levels with prematurity. Biomed. Chromatogr 2001, 15, 287–291. [Google Scholar] [CrossRef]
- Zinellu, A.; Sotgia, S.; Deiana, L.; Carru, C. Field-amplified sample injection combined with pressure-assisted capillary electrophoresis UV detection for the simultaneous analysis of allantoin, uric acid, and malondialdehyde in human plasma. Anal. Bioanal. Chem. 2011, 399, 2855–2861. [Google Scholar] [CrossRef]
- Zhang, D.; Haputhanthri, R.; Ansar, S.M.; Vangala, K.; DeSilva, H.I.; Sygula, A.; Saebo, S.; Pittman, C.U. Ultrasensitive detection of malondialdehyde with surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2010, 398, 3193–3201. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.X.; Wang, Z.; Jie, Z.; Gao, R.; Liao, S.; Yang, H.; Wang, F. Gold Nanoparticle Enriched by Q Sepharose Spheres for Chemical Reaction tandem SERS Detection of Malondialdehyde. Sens. Actuators B Chem. 2019, 281, 123–130. [Google Scholar] [CrossRef]
- Lu, H.; Zhu, L.; Zhang, C.; Chen, K.; Cui, Y. Mixing assisted “Hot Spots” Occupying SERS Strategy for Highly Sensitive in-situ Study. Anal. Chem. 2018, 90, 4535–4543. [Google Scholar] [CrossRef]
- Lu, H.; Zhu, L.; Zhang, C.; Wang, Z.; Lv, Y.; Chen, K.; Cui, Y. Highly uniform SERS-active microchannel on hydrophobic PDMS: A balance of high reproducibility and sensitivity for detection of proteins. RSC Adv. 2017, 7, 8771–8778. [Google Scholar] [CrossRef]
- Chen, K.; Lu, H.; Sun, M.; Zhu, L.; Cui, Y. Mixing enhancement of a novel C-SAR microfluidic mixer. Chem. Eng. Res. Des. 2018, 132, 338–345. [Google Scholar] [CrossRef]
- Candon, N.; Tuzmen, N. Very rapid quantification of malondialdehyde (MDA) in rat brain exposed to lead aluminium and phenolic antioxidants by high performance liquid chromatography fluorescence detection. Neurotoxicology 2008, 29, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Sans, R.; Vicario, I.M.; Guzmán-Chozas, M. Further studies and observations on 2-thiobarbituric acid assay (fat autoxidtion) and 2-thiobarbituric acid-aldehyde reactions. Food/Nahrung 1997, 41, 162–166. [Google Scholar] [CrossRef]
- Guillén-Sans, R.; Guzmán-Chozas, M. The Thiobarbituric Acid (TBA) Reaction in Foods: A Review. Crit. Rev. Food Sci. Nutr. 1998, 38, 315–350. [Google Scholar] [CrossRef] [PubMed]
- Sans, R.G.; Chozas, M.G. Historical aspects and applications of barbituric acid derivatives. A review. Die Phamazie 1988, 43, 827–829. [Google Scholar]
- Lane, L.A.; Qian, X.; Nie, S. SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev. 2015, 115, 10489–10529. [Google Scholar] [CrossRef] [PubMed]
- Andreou, C.; Hoonejani, M.R.; Barmi, M.R.; Meysam, R.M.; Martin, M.; Carl, D. Rapid detection of drugs of abuse in saliva using surface enhanced Raman spectroscopy and microfluidics. ACS Nano 2013, 7, 7157–7164. [Google Scholar] [CrossRef]
- Yang, L.; Yu, B.; Pan, L.; Zhou, B.; Tang, X.; Li, S. Sodium Chloride Crystal-Induced SERS Platform for Controlled Highly Sensitive Detection of Illicit Drugs. Chem.–A Eur. J. 2018, 24, 4800–4804. [Google Scholar]
- Yaffe, N.R.; Blanch, E.W. Effects and anomalies that can occur in SERS spectra of biological molecules when using a wide range of aggregating agents for hydroxylamine-reduced and citrate-reduced silver colloids. Vib. Spectrosc. 2008, 48, 196–201. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, Z.W.; Zhu, H.; Hasi, W. Characterization of Chloride-activated Surface Complex and Corresponding Enhancement Mechanism by SERS Saturation Effect. J. Phys. Chem. C 2016, 121, 950–957. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, D.; Ding, M.; Xu, Z.; Zhang, Z.; Li, B.; Shen, D. Controllable Synthesis of Silver Nanoparticle Aggregates for Surface-Enhanced Raman Scattering Studies. J. Phys. Chem. C 2011, 115, 16295–16304. [Google Scholar] [CrossRef]
- Burns, C.; Spendel, W.U.; Puckett, S.; Pacey, G.E. Solution ionic strength effect on gold nanoparticle solution color transition. Talanta 2006, 69, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Nunes, F.; Bonifãcio, L.; Araki, K.; Toma, H. Interaction of 2- and 4-mercaptopyridine with pentacyanoferrates and gold nanoparticles. Inorg. Chem. 2005, 45, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Grotto, D.; Maria, L.; Boeira, S.; Valentini, J.; Charao, M.E.; Moro, A.M.; Nascimento, P.C.; Pomblum, V.J.; Garcia, S.C. Rapid quantification of malondialdehyde in plasma by high performance liquid chromatography-visible detection. J. Pharm. Biomed. Anal. 2007, 43, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; Jung, D.G. Sensitive Analysis of Malondialdehyde in Human Urine by Derivatization with Pentafluorophenylhydrazine then Headspace GC–MS. Chromatographia 2009, 70, 899–903. [Google Scholar] [CrossRef]
- Yuan, L.; Lan, Y.; Han, M.; Bao, J.; Tu, W.; Dai, Z. Label-free and facile electrochemical biosensing using carbon nanotubes for malondialdehyde detection. Analyst 2013, 138, 3131–3134. [Google Scholar] [CrossRef]
- Wang, X.; Liu, X.; Cheng, T.; Li, H.; Yang, X. Development of 4-hydrazinyl-7-nitrobenzofurazan as a fluorogenic probe for detecting malondialdehyde in biological samples. Sens. Actuators B Chem. 2018, 254, 248–254. [Google Scholar] [CrossRef]



| Year | Detection Strategy | Linear Detection Range |
|---|---|---|
| 2007 [33] | UV-Vis | 0.28 μM–6.6 μM |
| 2009 [34] | GC-MS | 0.2 μg/L–20 μg/L |
| 2013 [35] | Electrochemistry | 0.1 μM–90 μM |
| 2018 [36] | Fluorescence | 0.1 μM–20 μM |
| 2019 [17] | SERS | 0.33 μM–3.3 mM |
| this work | SERS | 0.167 nM–0.33 μM |
| Oil Sample | Original Concentration (μM) | Added Concentration (μM) | Measured Concentration (μM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| Sunflower seed oil | 0.1 | 0.1 | 0.195 | 94.9 | 7.7 |
| 0.4 | 0.517 | 104.3 | 8.5 | ||
| 0.7 | 0.868 | 109.7 | 4.2 | ||
| Lard | 0.3 | 0.1 | 0.407 | 106.9 | 4.6 |
| 0.4 | 0.739 | 109.8 | 3.0 | ||
| 0.7 | 0.933 | 90.4 | 3.2 | ||
| Rapeseed oil | 0.2 | 0.1 | 0.306 | 106.2 | 4.3 |
| 0.4 | 0.564 | 90.9 | 5.1 | ||
| 0.7 | 0.880 | 97.5 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Wan, S.; Ruan, X.; Liang, H.; Su, J.; Wang, Z.; Zhu, L. Phosphoric Acid Induced Controllable Nanoparticle Aggregation for Ultrasensitive SERS Detection of Malondialdehyde in a Microfluidic Chip. Chemosensors 2022, 10, 524. https://doi.org/10.3390/chemosensors10120524
Lu Y, Wan S, Ruan X, Liang H, Su J, Wang Z, Zhu L. Phosphoric Acid Induced Controllable Nanoparticle Aggregation for Ultrasensitive SERS Detection of Malondialdehyde in a Microfluidic Chip. Chemosensors. 2022; 10(12):524. https://doi.org/10.3390/chemosensors10120524
Chicago/Turabian StyleLu, Yu, Siying Wan, Xin Ruan, Huijun Liang, Jingting Su, Zhuyuan Wang, and Li Zhu. 2022. "Phosphoric Acid Induced Controllable Nanoparticle Aggregation for Ultrasensitive SERS Detection of Malondialdehyde in a Microfluidic Chip" Chemosensors 10, no. 12: 524. https://doi.org/10.3390/chemosensors10120524
APA StyleLu, Y., Wan, S., Ruan, X., Liang, H., Su, J., Wang, Z., & Zhu, L. (2022). Phosphoric Acid Induced Controllable Nanoparticle Aggregation for Ultrasensitive SERS Detection of Malondialdehyde in a Microfluidic Chip. Chemosensors, 10(12), 524. https://doi.org/10.3390/chemosensors10120524







