Electrochemical Sensors for Antibiotic Susceptibility Testing: Strategies and Applications
Abstract
:1. Introduction
2. Electrochemical Sensors for Determining the Antibiotic Resistance Level of Bacteria
Electrochemical Sensing Strategy for Determining Antibiotic Resistance/Susceptibility
3. Current Electrochemical Sensors for the Detection and Monitoring of Antibiotic Resistance in Pathogenic Bacteria
3.1. Detection of Antibiotic-Resistant Genes
3.2. Measurement of the Change in Impedance Caused by Cell Lysis
3.3. Measurement of Current Response Caused by Changes in Membrane Properties
3.4. Assessment of the Redox Change of a Redox-Active Molecule Resulting from the Bacterial Metabolic Activity
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States 2019; Centers for Disease Control and Prevention (CDC), U.S. Department of Health and Human Services: Atlanta, GA, USA, 2019.
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Burden of AMR Collaborative Group. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Brogan, D.M.; Mossialos, E. A critical analysis of the review on antimicrobial resistance report and the infectious disease financing facility. Glob. Health 2016, 12, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, H.; Colodner, R.; Halachmi, S.; Segal, E. Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sens. 2018, 3, 2202–2217. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Zhang, Y.; Cao, Z.H.; Wang, X.Y.; Ma, B.; Wu, W.B.; Hu, N.; Huo, Z.Y.; Yuan, Q.B. Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: Prevalence and antibiotic resistance profiles. Sci. Total Environ. 2019, 651 Pt 2, 1946–1957. [Google Scholar] [CrossRef]
- Gilligan, P.H. Identification of pathogens by classical clinical tests. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Narayana Iyengar, S.; Dietvorst, J.; Ferrer-Vilanova, A.; Guirado, G.; Muñoz-Berbel, X.; Russom, A. Toward rapid detection of viable bacteria in whole blood for early sepsis diagnostics and susceptibility testing. ACS Sens. 2021, 6, 3357–3366. [Google Scholar] [CrossRef]
- Schuetz, A.N. Antimicrobial resistance and susceptibility testing of anaerobic bacteria. Clin. Infect. Dis. 2014, 59, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef]
- Chen, C.; Hong, W. Recent development of rapid antimicrobial susceptibility testing methods through metabolic profiling of bacteria. Antibiotics 2021, 10, 311. [Google Scholar] [CrossRef]
- Khan, Z.A.; Siddiqui, M.F.; Park, S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics 2019, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Majdinasab, M.; Mitsubayashi, K.; Marty, J.L. Optical and electrochemical sensors and biosensors for the detection of quinolones. Trends Biotechnol. 2019, 37, 898–915. [Google Scholar] [CrossRef]
- Qin, N.; Zhao, P.; Ho, E.A.; Xin, G.; Ren, C.L. Microfluidic technology for antibacterial resistance study and antibiotic susceptibility testing: Review and perspective. ACS Sens. 2021, 6, 3–21. [Google Scholar] [CrossRef]
- Yoo, S.M.; Lee, S.Y. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef]
- Yin, X.; Hou, T.; Huang, B.; Yang, L.; Li, F. Aptamer recognition-trigged label-free homogeneous electrochemical strategy for an ultrasensitive cancer-derived exosome assay. Chem. Commun. 2019, 55, 13705–13708. [Google Scholar] [CrossRef]
- Gai, P.; Gu, C.; Hou, T.; Li, F. Ultrasensitive self-powered aptasensor based on enzyme biofuel cell and DNA bioconjugate, A facile and powerful tool for antibiotic residue detection. Anal. Chem. 2017, 89, 2163–2169. [Google Scholar] [CrossRef]
- Chang, J.; Lv, W.; Li, Q.; Li, H.; Li, F. One-step synthesis of methylene blue-encapsulated zeolitic imidazolate framework for dual-signal fluorescent and homogeneous electrochemical biosensing. Anal. Chem. 2020, 92, 8959–8964. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef]
- Huang, L.; Tian, S.; Zhao, W.; Liu, K.; Guo, J. Electrochemical vitamin sensors: A critical review. Talanta 2021, 222, 121645. [Google Scholar] [CrossRef]
- Zhang, Y.; Lei, Y.; Lu, H.; Shi, L.; Wang, P.; Ali, Z.; Li, J. Electrochemical detection of bisphenols in food: A review. Food Chem. 2021, 346, 128895. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Beni, V.; Zourob, M. Nanomaterial application in bio/sensors for the detection of infectious diseases. Talanta 2021, 230, 122026. [Google Scholar] [CrossRef]
- Garkani Nejad, F.; Tajik, S.; Beitollahi, H.; Sheikhshoaie, I. Magnetic nanomaterials based electrochemical (bio)sensors for food analysis. Talanta 2021, 228, 122075. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Thong, K.L.; Thiha, A.; Dinshaw, I.J.; Lai, C.W.; Ibrahim, F.; Leo, B.F. Carbon Nanomaterial-based electrochemical biosensors for foodborne bacterial detection. Crit. Rev. Anal. Chem. 2019, 49, 510–533. [Google Scholar] [CrossRef]
- Trotter, M.; Borst, N.; Thewes, R.; von Stetten, F. Review, Electrochemical DNA sensing-Principles, commercial systems, and applications. Biosens. Bioelectron. 2020, 154, 112069. [Google Scholar] [CrossRef]
- Lee, V.B.C.; Mohd-Naim, N.F.; Tamiya, E.; Ahmed, M.U. Trends in paper-based electrochemical biosensors, from design to application. Anal. Sci. 2018, 34, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Karbelkar, A.A.; Furst, A.L. Electrochemical diagnostics for bacterial infectious diseases. ACS Infect. Dis. 2020, 6, 1567–1571. [Google Scholar] [CrossRef]
- Kang, M.; Jo, Y.; Mun, C.W.; Yeom, J.; Park, J.S.; Jung, H.S.; Kim, D.-H.; Park, S.-G.; Yoo, S.M. Nanoconfined 3D redox capacitor-based electrochemical sensor for ultrasensitive monitoring of metabolites in bacterial communication. Sens. Actuators B—Chem. 2021, 345, 130427. [Google Scholar] [CrossRef]
- Zhang, R.; Belwal, T.; Li, L.; Lin, X.; Xu, Y.; Luo, Z. Nanomaterial-based biosensors for sensing key foodborne pathogens, Advances from recent decades. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1465–1487. [Google Scholar] [CrossRef] [PubMed]
- Wongkaew, N.; Simsek, M.; Griesche, C.; Baeumner, A.J. Functional nanomaterials and nanostructures enhancing electrochemical biosensors and lab-on-a-chip performances, recent progress, applications, and future perspective. Chem. Rev. 2019, 119, 120–194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, G.; Li, H.; Du, D.; Lin, Y. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal. Chem. 2015, 87, 230–249. [Google Scholar] [CrossRef] [PubMed]
- Amali, R.K.A.; Lim, H.N.; Ibrahim, I.; Huang, N.M.; Zainal, Z.; Ahmad, S.A.A. Significance of nanomaterials in electrochemical sensors for nitrate detection, A review. Trends Environ. Anal. Chem. 2021, 31, e00135. [Google Scholar] [CrossRef]
- Kang, M.; Mun, C.; Jung, H.S.; Ansah, I.B.; Kim, E.; Yang, H.; Payne, G.F.; Kim, D.H.; Park, S.G. Tethered molecular redox capacitors for nanoconfinement-assisted electrochemical signal amplification. Nanoscale 2020, 12, 3668–3676. [Google Scholar] [CrossRef]
- Lemay, S.G.; Moazzenzade, T. Single-entity electrochemistry for digital biosensing at ultralow concentrations. Anal. Chem. 2021, 93, 9023–9031. [Google Scholar] [CrossRef]
- Nguyen, T.H.T.; Lee, J.; Kim, H.Y.; Nam, K.M.; Kim, B.K. Current research on single-entity electrochemistry for soft nanoparticle detection, Introduction to detection methods and applications. Biosens. Bioelectron. 2020, 151, 111999. [Google Scholar] [CrossRef]
- Zhang, J.-H.; Zhou, Y.-G. Nano-impact electrochemistry, Analysis of single bioentities. TrAC—Trends Anal. Chem. 2020, 123, 115768. [Google Scholar] [CrossRef]
- Couto, R.A.S.; Chen, L.; Kuss, S.; Compton, R.G. Detection of Escherichia coli bacteria by impact electrochemistry. Analyst 2018, 143, 4840–4843. [Google Scholar] [CrossRef]
- Gao, G.; Wang, D.; Brocenschi, R.; Zhi, J.; Mirkin, M.V. Toward the detection and identification of single bacteria by electrochemical collision technique. Anal. Chem. 2018, 90, 12123–12130. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, D.; Liu, Y.; Gao, G.; Zhi, J. Redox activity of single bacteria revealed by electrochemical collision technique. Biosens. Bioelectron. 2021, 176, 112914. [Google Scholar] [CrossRef]
- Cardoso, R.M.; Kalinke, C.; Rocha, R.G.; Dos Santos, P.L.; Rocha, D.P.; Oliveira, P.R.; Janegitz, B.C.; Bonacin, J.A.; Richter, E.M.; Munoz, R. Additive-manufactured (3D-printed) electrochemical sensors: A critical review. Anal. Chim. Acta. 2020, 1118, 73–91. [Google Scholar] [CrossRef]
- Ambaye, A.D.; Kefeni, K.K.; Mishra, S.B.; Nxumalo, E.N.; Ntsendwana, B. Recent developments in nanotechnology-based printing electrode systems for electrochemical sensors. Talanta 2021, 225, 121951. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Ferapontova, E.E. Hybridization biosensors relying on electrical properties of nucleic acids. Electroanalysis 2017, 29, 6–13. [Google Scholar] [CrossRef]
- Ferapontova, E.E. DNA Electrochemistry and Electrochemical sensors for nucleic acids. Annu. Rev. Anal. Chem. 2018, 11, 197–218. [Google Scholar] [CrossRef]
- Paleček, E.; Bartošík, M. Electrochemistry of nucleic acids. Chem. Rev. 2012, 112, 3427–3481. [Google Scholar] [CrossRef]
- Butterworth, A.; Pratibha, P.; Marx, A.; Corrigan, D.K. Electrochemical detection of oxacillin resistance using direct-labeling solid-phase isothermal amplification. ACS Sens. 2021, 6, 3773–3780. [Google Scholar] [CrossRef]
- Ortiz, M.; Jauset-Rubio, M.; Skouridou, V.; Machado, D.; Viveiros, M.; Clark, T.G.; Simonova, A.; Kodr, D.; Hocek, M.; O’Sullivan, C.K. Electrochemical detection of single-nucleotide polymorphism associated with rifampicin resistance in Mycobacterium tuberculosis using solid-phase primer elongation with ferrocene-linked redox-labeled nucleotides. ACS Sens. 2021, 6, 4398–4407. [Google Scholar] [CrossRef]
- Li, X.; Lu, J.; Feng, L.; Zhang, L.; Gong, J. Smart pH-regulated switchable nanoprobes for photoelectrochemical multiplex detection of antibiotic resistance genes. Anal. Chem. 2020, 92, 11476–11483. [Google Scholar] [CrossRef]
- Wan, C.; Qu, A.; Li, M.; Tang, R.; Fu, L.; Liu, X.; Wang, P.; Wu, C. Electrochemical sensor for directional recognition and measurement of antibiotic resistance genes in water. Anal. Chem. 2022, 94, 732–739. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Mergel, O.; Uria, N.; Abramova, N.; van Rijn, P.; Bratov, A. 3D impedimetric sensors as a tool for monitoring bacterial response to antibiotics. Lab Chip 2019, 19, 1436–1447. [Google Scholar] [CrossRef] [Green Version]
- Safavieh, M.; Pandya, H.J.; Venkataraman, M.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Singh, A.; Prabhakar, D.; Chug, M.K.; Shafiee, H. Rapid real-time antimicrobial susceptibility testing with electrical sensing on plastic microchips with printed electrodes. ACS Appl. Mater. Interfaces 2017, 9, 12832–12840. [Google Scholar] [CrossRef] [Green Version]
- Dizaji, A.N.; Ali, Z.; Ghorbanpoor, H.; Ozturk, Y.; Akcakoca, I.; Avci, H.; Dogan Guzel, F. Electrochemical-based “antibiotsensor” for the whole-cell detection of the vancomycin-susceptible bacteria. Talanta 2021, 234, 122695. [Google Scholar] [CrossRef] [PubMed]
- Hannah, S.; Addington, E.; Alcorn, D.; Shu, W.; Hoskisson, P.A.; Corrigan, D.K. Rapid antibiotic susceptibility testing using low-cost, commercially available screen-printed electrodes. Biosens. Bioelectron. 2019, 145, 111696. [Google Scholar] [CrossRef] [PubMed]
- Guimerà, A.; Gabriel, G.; Prats-Alfonso, E.; Abramova, N.; Bratov, A.; Villa, R. Effect of surface conductivity on the sensitivity of interdigitated impedimetric sensors and their design considerations. Sens. Actuators B—Chem. 2015, 207, 1010–1018. [Google Scholar] [CrossRef]
- Bratov, A.; Abramova, N. Response of a microcapillary impedimetric transducer to changes in surface conductance at liquid/solid interface. J. Colloid Interface Sci. 2013, 403, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.P.; Sharma, S.; Mehrotra, T.; Das, R.; Kumar, R.; Singh, R.; Roy, I.; Basu, T. Rapid electrochemical monitoring of bacterial respiration for Gram-positive and Gram-negative microbes: Potential application in antimicrobial susceptibility testing. Anal. Chem. 2020, 92, 4266–4274. [Google Scholar] [CrossRef]
- Jo, N.; Kim, B.; Lee, S.M.; Oh, J.; Park, I.H.; Lim, K.J.; Shin, J.S.; Yoo, K.H. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens. Bioelectron. 2018, 102, 164–170. [Google Scholar] [CrossRef]
- Li, C.; Sun, F. Graphene-assisted sensor for rapid detection of antibiotic resistance in Escherichia coli. Front. Chem. 2021, 9, 696906. [Google Scholar] [CrossRef]
- Besant, J.D.; Sargent, E.H.; Kelley, S.O. Rapid electrochemical phenotypic profiling of antibiotic-resistant bacteria. Lab Chip 2015, 15, 2799–2807. [Google Scholar] [CrossRef]
- Crane, B.; Hughes, J.; Neale, S.J.R.; Rashid, M.; Linton, P.E.; Banks, C.E.; Shaw, K. Rapid antibiotic susceptibility testing using resazurin bulk modified screen-printed electrochemical sensing platforms. Analyst 2021, 146, 5574–5583. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, D.; Mishra, K.P.; Kaur, G.; Dhull, N.; Tomar, M.; Gupta, V.; Kumar, B.; Ganju, L. Rapid antibiotic susceptibility testing by resazurin using thin film platinum as a bio-electrode. J. Microbiol. Methods 2019, 162, 69–76. [Google Scholar] [CrossRef]
- Bolotsky, A.; Muralidharan, R.; Butler, D.; Root, K.; Murray, W.; Liu, Z.; Ebrahimi, A. Organic redox-active crystalline layers for reagent-free electrochemical antibiotic susceptibility testing (ORACLE-AST). Biosens. Bioelectron. 2021, 172, 112615. [Google Scholar] [CrossRef]
- Ren, Y.; Ji, J.; Sun, J.; Pi, F.; Zhang, Y.; Sun, X. Rapid detection of antibiotic resistance in Salmonella with screen printed carbon electrodes. J. Solid State Electrochem. 2020, 24, 1539–1549. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.; Lee, J.; Shin, S.; Kim, Y.T. Development of a telemetric, miniaturized electrochemical amperometric analyzer. Sensors 2017, 17, 2416. [Google Scholar] [CrossRef] [Green Version]
- Chae, Y.K.; Kim, S.H.; Ellinger, J.E.; Markley, J.L. Dosage effects of salt and pH stresses on Saccharomyces cerevisiae as monitored via metabolites by using two dimensional NMR Spectroscopy. Bull. Korean Chem. Soc. 2013, 34, 3602–3608. [Google Scholar] [CrossRef] [Green Version]
- Ranganathan, S.; Kuo, T.C.; McCreery, R.L. Facile preparation of active glassy carbon electrodes with activated carbon and organic solvents. Anal. Chem. 1999, 71, 3574–3580. [Google Scholar] [CrossRef]
- Woods, L.A.; Powell, P.R.; Paxon, T.L.; Ewing, A.G. Analysis of mammalian cell cytoplasm with electrophoresis in nanometer inner diameter capillaries. Electroanalysis 2005, 17, 1192–1197. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.L.; Steele, T.W.J.; Stuckey, D.C. Metabolic reduction of resazurin; location within the cell for cytotoxicity assays. Biotechnol. Bioeng. 2018, 115, 351–358. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple applications of Alamar blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef]
- Chotinantakul, K.; Suginta, W.; Schulte, A. Advanced amperometric respiration assay for antimicrobial susceptibility testing. Anal. Chem. 2014, 86, 10315–10322. [Google Scholar] [CrossRef]
- Katseli, V.; Economou, A.; Kokkinos, C. Single-step fabrication of an integrated 3D-printed device for electrochemical sensing applications. Electrochem. Commun. 2019, 103, 100–103. [Google Scholar] [CrossRef]

| (Working) Electrode | Antibiotics Tested | Target Species | Sensing Approach | Assay Time | Sample | Feature | Reference |
|---|---|---|---|---|---|---|---|
| Screen printed gold electrode (SPGE) | Oxacillin | Escherichia coli | Chronoamperometry | 60 min | None | Use of amperometric change via HPR-mediated TMB oxidation. Use of recombinase and polymerase-aided isothermal amplification. LoD: 319 CFU/mL. | [47]; Figure 1A |
| Gold electrode | Rifampicin | Mycobacterium tuberculosis | Square wave voltammetry (SWV) | 20 min hybridization with the target, 5 min solid-phase primer elongation | None | Use of Klenow (exo-) DNA polymerase-mediated primer elongation reaction. Immobilization of thiolated primer on the surface of gold electrode. Use of ferrocene-labeled dNTPs. LoD: 3 pM (2.7 × 106 DNA copies) of target DNAs | [48]; Figure 1B |
| pH-controlled metal ion-embedded nanosphere, which was immobilized on the ZnS modified fluorine tin oxide (FTO) substrate | Penicillin | E. coli | Photoelectrochemistry | Plasmid | Use of change in pH. Linear range: 0.001 μM to 10 μM. | [49]; Figure 1C | |
| Gold electrode with surface graphene ink | Ampicillin | Impedance | None | Linear range: 6.3− 900.0 ng/mL. Detection of DNAs in water. | [50] | ||
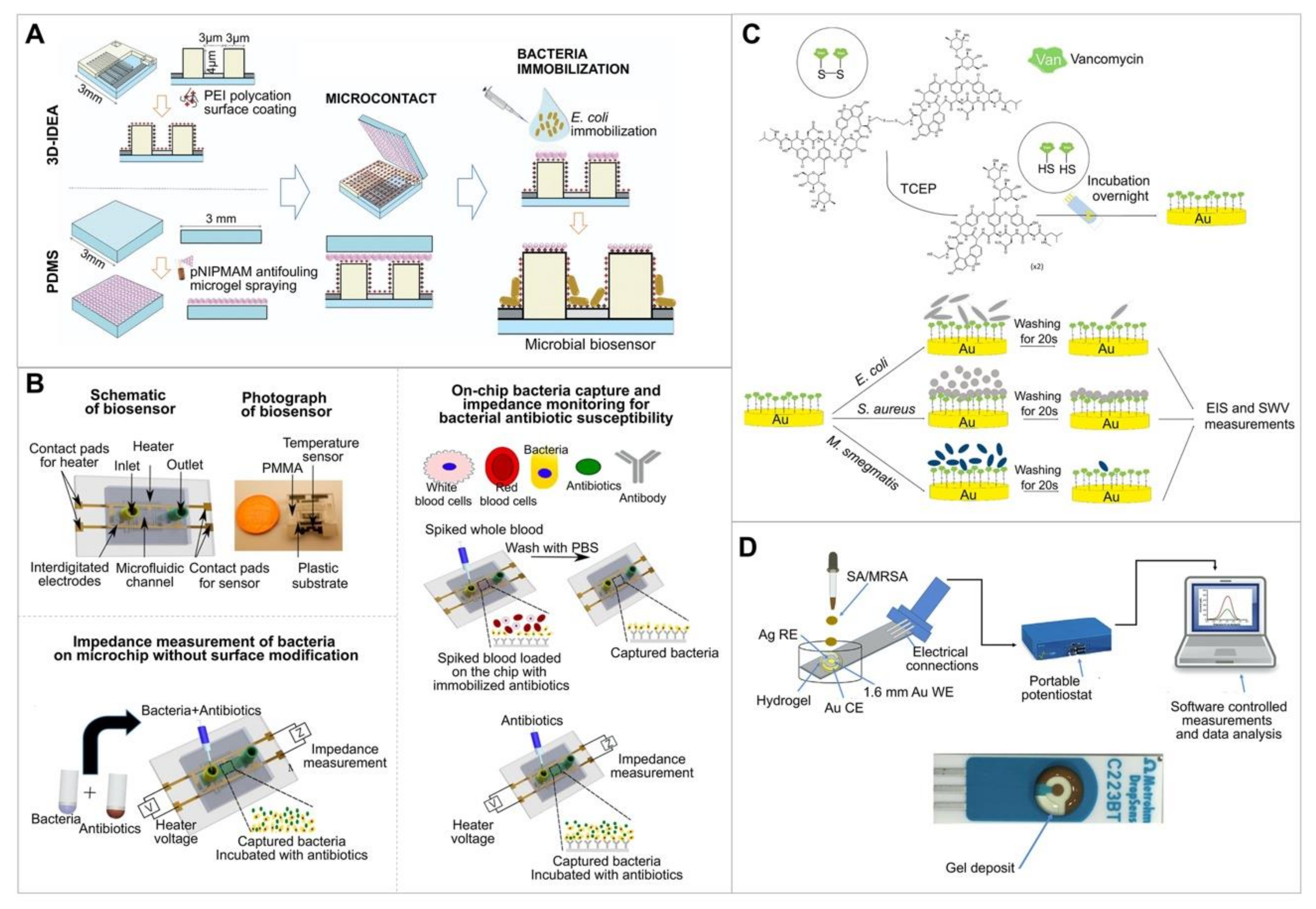
| Tantalum silicide electrode | Ampicillin | E. coli | Impedance | 60–120 min | None | Immobilization of bacteria on the three-dimensional interdigitated electrode array impedimetric transducer. Enables multiple detection or monitoring using multielectrode by separating via the insulating barriers. | [51]; Figure 2A |
| Silver interdigitated carbon working electrode (WE), counter electrode (CE), and reference electrode (RE). Plastic microchips with printed electrodes | Ampicillin, erythromycin, ciprofloxacin, methicillin, daptomycin, gentamicin | E. coli, Methicillin-resistant Staphylococcus aureus (MRSA) | Impedance | <90 min | Whole blood, human urine | Immobilization of bacteria on the electrode by using an antibody. No dye to interfere with cellular processes. | [52]; Figure 2B |
| SPGE immobilized with thiolated vancomycin, which was used as the binder with Gram-positive bacteria | Vancomycin | S. aureus | Impedance | None | Linear range: 101–108 CFU/mL. LoD: < 39 CFU/mL. | [53]; Figure 2C | |
| Gold WE, gold CE, silver RE | Amoxicillin, oxacillin | S. aureus, MRSA | Impedance and differential pulse voltammetry (DPV) | <45 min | None | Drop-coating of three electrodes with a hydrogel containing agarose, LB growth media, ferri-ferro cyanide redox mediator, and antibiotics. Enables a low-cost and mass production of electrode. | [54]; Figure 2D |
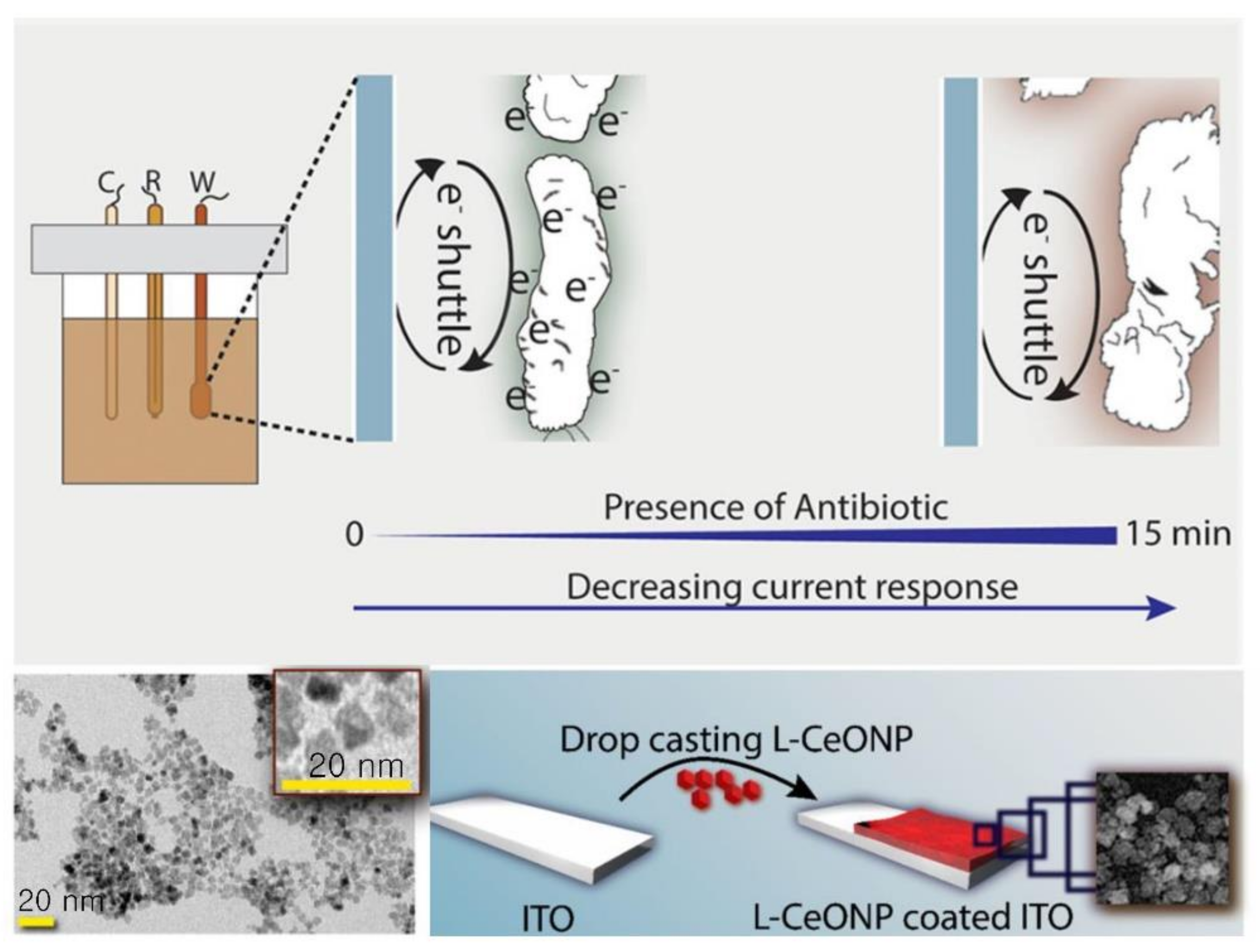
| L-lysine coated cerium oxide nanoparticle coated indium tin oxide (ITO) | Ciprofloxacin, cefixime, amoxycillin | E. coli | Cyclic voltammetry (CV) | Measurement time: 15 min | None | Use of electron-transfer rate due to different membrane properties of Gram (+) and (−) bacteria. | [57]; Figure 3 |
| Miniaturized incubation chamber containing working, counter, and reference electrodes | Ampicillin, ciprofloxacin | E. coli, Klebsiella pneumoniae | DPV | 30 min | Human urine | Incubation of living cells with antibiotics and redox reporter molecules in a miniaturized chamber. | [60] |
| Gold electrode pattened on a glass substrate | Gentamicin | E. coli, S. aureus | Capacitance | None | Use of bacterial-specific aptamer-functionalized electrode. Measurement of bacterial growth curves for 10 CFU/mL. | [58] | |
| Glassy carbon electrode | Ofloxacin, penicillin, cefepime | E. coli | DPV | None | Immobilization of cells with graphene dispersion on the electrode surface. | [59] | |
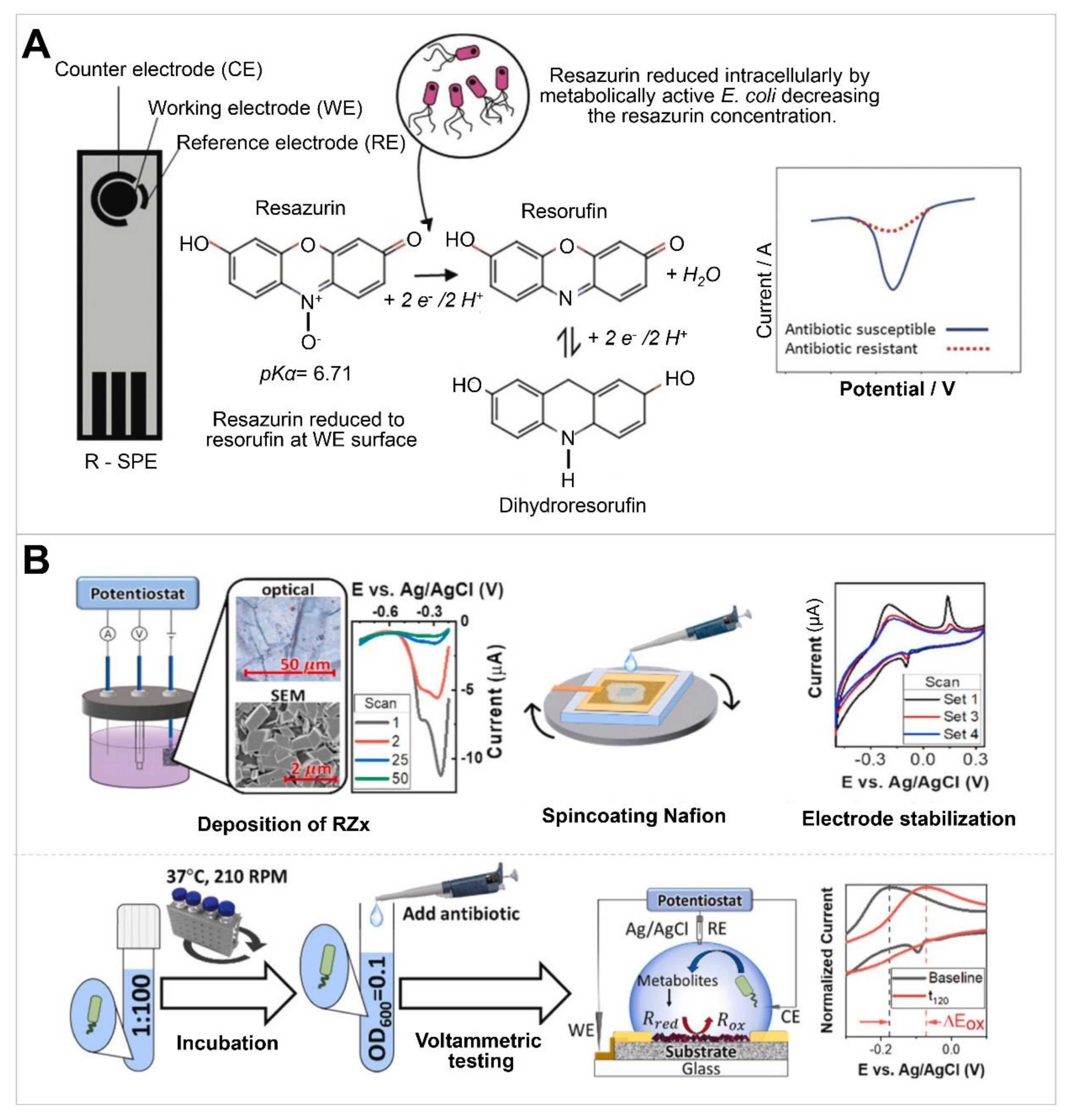
| Screen-printed carbon-graphite electrode | Gentamycin | E. coli | DPV | 90 min | Artificial urine | Use of resazurin-containing electrode. LoD: 15.6 μM | [61]; Figure 4A |
| Platinum WE and CE, Ag/AgCl RE | Ampicillin, kanamycin, tetracycline | E. coli, K. pneumoniae | DPV, detection of resazurin | 4 h | None | Reusable biosensor. Electrode type incompatible with miniaturization. | [62] |
| Nafion-coated organic redox-active crystal layers on planar pyrolytic graphite sheets as the sensing platform | Ampicillin, kanamycin | E. coli | DPV | 60 min | Whole blood, milk | Use of change in pH via cell proliferation. Assay time: 60 min. Stable (<12% degradation in ~60 d). Detection range: 0.001–10 μM, 0 μg/mL and 16 μg/mL. | [63]; Figure 4B |
| Screen-printed carbon electrode modified with multi-welled carbon tube and gold nanoparticle | Ofloxacin, penicillin | Salmonella gallinarum | DPV | 60 min | Egg | Single-step modification. Modification process currently incompatible with bulk modification. Multi-channel. LoD: 100 CFU/mL. | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Yoo, S. Electrochemical Sensors for Antibiotic Susceptibility Testing: Strategies and Applications. Chemosensors 2022, 10, 53. https://doi.org/10.3390/chemosensors10020053
Kim D, Yoo S. Electrochemical Sensors for Antibiotic Susceptibility Testing: Strategies and Applications. Chemosensors. 2022; 10(2):53. https://doi.org/10.3390/chemosensors10020053
Chicago/Turabian StyleKim, Dongmin, and Seungmin Yoo. 2022. "Electrochemical Sensors for Antibiotic Susceptibility Testing: Strategies and Applications" Chemosensors 10, no. 2: 53. https://doi.org/10.3390/chemosensors10020053
APA StyleKim, D., & Yoo, S. (2022). Electrochemical Sensors for Antibiotic Susceptibility Testing: Strategies and Applications. Chemosensors, 10(2), 53. https://doi.org/10.3390/chemosensors10020053






