Biodetection Techniques for Quantification of Chemokines
Abstract
:1. Introduction
2. Classification of Chemokines
3. Chemokine Biomarkers and Related Diseases
3.1. Cancer
3.2. Autoimmune Diseases
4. Biodetection Methods for Determination of Chemokines
4.1. Electrochemical Techniques
4.2. Optical Techniques
5. Advantages and Disadvantages of Electrochemical and Optical Biosensors
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zlotnik, A.; Yoshie, O. The chemokine superfamily revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Bacon, K.B.; Harrison, J.K. Chemokines and their receptors in neurobiology: Perspectives in physiology and homeostasis. J. Neuroimmun. 2000, 104, 92–97. [Google Scholar] [CrossRef]
- Charo, I.F.; Ransohoff, R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Eng. J. Med. 2006, 354, 610–621. [Google Scholar] [CrossRef]
- Walz, D.A.; Wu, V.Y.; de Lamo, R.; Dene, H.; McCoy, L.E. Primary structure of human platelet factor 4. Thrombosis Res. 1977, 11, 893–898. [Google Scholar] [CrossRef]
- Yoshimura, T.; Matsushima, K.; Tanaka, S.; Robinson, E.A.; Appella, E.; Oppenheim, J.J.; Leonard, E.J. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc. Natl. Acad. Sci. USA 1987, 84, 9233–9237. [Google Scholar] [CrossRef] [Green Version]
- Cocchi, F.; DeVico, A.L.; Garzino-Demo, A.; Arya, S.K.; Gallo, R.C.; Lusso, P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+T cells. Science 1995, 270, 1811–1815. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef]
- Callewaare, C.; Banisadr, G.; Rostène, W.; Parsadaniantz, S.M. Chemokines and chemokine receptors in the brain: Implication in neuroendocrine regulation. J. Mol. Edocrinol. 2007, 38, 355–363. [Google Scholar] [CrossRef]
- Moser, B.; Wolf, M.; Walz, A.; Loetscher, P. Chemokines: Multiple levels of leukocyte control. Trends Immunol. 2004, 25, 75–84. [Google Scholar] [CrossRef]
- Kimura, Y.N.; Watari, K.; Fotovati, A.; Hosoi, F.; Yasumoto, K.; Izumi, H.; Kohno, K.; Umezawa, K.; Iguchi, H.; Shirouzu, K.; et al. Inflammatory stimuli from macrophages and cancer cells synergistically promote tumor growth and angiogenesis. Cancer Sci. 2007, 98, 2009–2018. [Google Scholar] [CrossRef]
- Rotondi, M.; Chiovato, L.; Romagnani, S.; Serio, M.; Romagnani, P. Role of chemokines in endocrine autoimmune diseases. Endocrine Rev. 2007, 28, 492–520. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.J.; Kelvin, D.J. Cytokines, chemokines and their receptors. In Madame Curie Bioscience Dataabase [Internet]; Landes Bioscience: Austin, TX, USA, 2000–2013. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6294/ (accessed on 27 June 2022).
- Unver, N.; Esendagli, G.; Yilmaz, G.; Guc, D. CXCL7-induced macrophage infiltration in lung tumor is independent of CXCR2 expression: CXCL7-induced macrophage chemotaxis in LLC tumors. Cytokine 2015, 75, 330–337. [Google Scholar] [CrossRef]
- Jin, T.; Xu, X.; Hereld, D. Chemotaxis, chemokine receptors and human disease. Cytokine 2008, 44, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.-D.; Song, X.-Y.; Yang, P.-F.; Ai, Q.-D.; Wang, Y.-Y.; Feng, X.-Y.; He, X.; Chen, N.-H. Progress in pharmacological research of chemokine like factor 1 (CKLF1). Cytokine 2018, 102, 41–50. [Google Scholar] [CrossRef]
- Proudfoot, A.E.I.; Johnson, Z.; Bonvin, P.; Handel, T.M. Glycosaminoglycan interactions with chemokines add complexity to a complex system. Pharmaceuticals 2017, 10, 70. [Google Scholar] [CrossRef] [Green Version]
- Stone, M.J. Regulation of chemokine-receptor interactions and functions. Int. J. Mol. Sci. 2017, 18, 2415. [Google Scholar] [CrossRef] [Green Version]
- Lindley, I.J.D. 8. Interleukin-8. In Cytokines Handbook of Immunopharmacology; Academic Press: Cambridge, MA, USA, 1998; pp. 125–140. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O.; Nomiyama, H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006, 7, 243. [Google Scholar] [CrossRef]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta-Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [Green Version]
- Panda, S.; Padhiary, S.K.; Routray, S. Chemokines accentuating protumoral activities in oral cancer microenvironment possess an imperious stratagem for therapeutic resolutions. Oral Oncol. 2016, 60, 8–17. [Google Scholar] [CrossRef]
- Gerszten, R.E.; Mach, F.; Sauty, A.; Rosenzweig, A.; Luster, A.D. Chemokines, leukocytes, and atherosclerosis. J. Lab. Clin. Med. 2000, 136, 87–92. [Google Scholar] [CrossRef]
- Rot, A.; von Andrian, U.H. Chemokines in innate and adaptive host defense: Basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 2004, 22, 891–928. [Google Scholar] [CrossRef]
- Butera, D.; Marukian, S.; Iwamaye, A.E.; Hembrador, E.; Chambers, T.J.; di Bisceglie, A.M.; Charles, E.D.; Talal, A.H.; Jacobson, I.M.; Rice, C.M.; et al. Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood 2005, 106, 1175–1182. [Google Scholar] [CrossRef] [Green Version]
- Ruffini, P.A.; Morandi, P.; Cabioglu, N.; Altundag, K.; Cristofanilli, M. Manipulating the chemokine-chemokine receptor network to treat cancer. Cancer 2007, 109, 2392–2404. [Google Scholar] [CrossRef]
- Saruhan-Direskeneli, G.; Yentür, S.P.; Akman-Demir, G.; Isik, N.; Serdaroğlu, P. Cytokines and chemokines in neuro-Behçet’s disease compared to multiple sclerosis and other neurological diseases. J. Neuroimmunol. 2003, 145, 127–134. [Google Scholar] [CrossRef]
- Godessart, N.; Kunkel, S.L. Chemokines in autoimmune disease. Curr. Opin. Immunol. 2001, 13, 670–675. [Google Scholar] [CrossRef]
- Collins, P.J.; McCully, M.L.; Martínez-Muñoz, L.; Santiago, C.; Wheeldon, J.; Caucheteux, S.; Thelen, S.; Cecchinato, V.; Laufer, J.M.; Purvanov, V.; et al. Epithelial chemokine CXCL14 synergizes with CXCL12 via allosteric modulation of CXCR4. Faseb J. 2017, 31, 3084–3097. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.T.; Wong, Y.K.; Hsiao, H.Y.; Wang, Y.W.; Chan, M.Y.; Chang, K.W. Evaluation of saliva and plasma cytokine biomarkers in patients with oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2018, 47, 699–707. [Google Scholar] [CrossRef]
- Mrugacz, M.; Zelazowska, B.; Bakunowicz-Lazarczyk, A.; Kaczmarski, M.; Wysocka, J. Elevated tear fluid levels of MIP-1alpha in patients with cystic fibrosis. J. Interferon Cytokine Res. 2007, 27, 491–495. [Google Scholar] [CrossRef]
- Gornowicz, A.; Bielawska, A.; Bielawski, K.; Grabowska, S.Z.; Wójcicka, A.; Zalewska, M.; Maciorkowska, E. Pro-inflammatory cytokines in saliva of adolescents with dental caries disease. Ann. Agric. Environ. Med. 2012, 19, 711–716. [Google Scholar]
- Ujhelyi, B.; Gogolak, P.; Erdei, A.; Nagy, V.; Balazs, E.; Rajnavolgyi, E.; Berta, A.; Nagy, E.V. Graves’ Orbitopathy results in profound changes in tear composition: A study of plasminogen activator inhibitor-1 and seven cytokines. Thyroid 2012, 22, 407–414. [Google Scholar] [CrossRef]
- Thorman, R.; Lundahl, J.; Yucel-Lindberg, T.; Hylander, B. Inflammatory cytokines in saliva: Early signs of metabolic disorders in chronic kidney disease. A controlled cross sectional study. Oral Surg. Oral Med. Oral Pathol. Oral Radio 2010, 110, 597–604. [Google Scholar] [CrossRef]
- Kishazi, E.; Dor, M.; Eperon, S.; Oberic, A.; Turck, N.; Hamedani, M. Differential profiling of lacrimal cytokines in patients suffering from thyroid-associated orbitopathy. Sci. Rep. 2018, 8, 10792. [Google Scholar] [CrossRef] [Green Version]
- Robison, H.M.; Chapman, C.A.; Zhou, H.; Erskine, C.L.; Theel, E.; Peikert, T.; Arlehamn, C.S.L.; Sette, A.; Bushell, C.; Welge, M.; et al. Risk assessment of latent tuberculosis infection through a multiplexed cytokine biosensor assay and machine learning feature selection. Sci. Rep. 2021, 11, 20544. [Google Scholar] [CrossRef]
- He, W.Z.Z.; Yi, L.; Mao, S.; Li, H.; Lin, J.-M. A dual-functional microfluidic chip for on-line detection of interleukin-8 based on rolling circle amplification. Biosens. Bioelectron. 2018, 102, 652–660. [Google Scholar] [CrossRef]
- Zhou, W.; Mahshid, S.S.; Wang, W.; Vallée-Bélisle, A.; Zandstra, P.W.; Sargent, E.H.; Kelley, S.O. Steric hindrance assay for secreted factors in stem cell culture. ACS Sens. 2017, 2, 495–500. [Google Scholar] [CrossRef]
- Kramer, J.M.; Klimatcheva, E.; Rothstein, T.L. CXCL13 is elevated in Sjögren’s syndrome in mice and humans and is implicated in disease pathogenesis. J. Leukoc. Biol. 2013, 94, 1079–1089. [Google Scholar] [CrossRef] [Green Version]
- Alosaimi, B.; Hamed, M.E.; Naeem, A.; Alsharef, A.A.; al Qahtani, S.Y.; al Dosari, K.M.; Alamri, A.A.; Al-Eisa, K.; Khojah, T.; Assiri, A.M.; et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 2020, 126, 154895. [Google Scholar] [CrossRef]
- Jagannath, B.; Pali, M.; Lin, K.-C.; Sankhala, D.; Naraghi, P.; Muthukumar, S.; Prasad, S. Novel approach to track the lifecycle of inflammation from chemokine expression to inflammatory proteins in sweat using electrochemical biosensor. Adv. Mater. Technol. 2022, 2101356. [Google Scholar] [CrossRef]
- Hernández-Molina, G.; Michel-Peregrina, M.; Hernández-Ramírez, D.F.; Sánchez-Guerrero, J.; Llorente, L. Chemokine saliva levels in patients with primary Sjögren’s syndrome, associated Sjögren’s syndrome, pre-clinical Sjögren’s syndrome and systemic autoimmune diseases. Rheumatology 2011, 50, 1288–1292. [Google Scholar] [CrossRef] [Green Version]
- Jekarl, D.W.; Kim, J.Y.; Ha, J.H.; Lee, S.; Yoo, J.; Kim, M.; Kim, Y. Diagnosis and prognosis of sepsis based on use of cytokines, chemokines, and growth factors. Dis. Markers 2019, 1089107. [Google Scholar] [CrossRef]
- Poeta, V.M.; Massara, M.; Capucetti, A.; Bonecchi, R. Chemokines and chemokine receptors: New targets for cancer immunotherapy. Front. Immunol. 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karin, N. Chemokines and cancer: New immune checkpoints for cancer therapy. Curr. Op. Immunol. 2018, 51, 140–145. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Simińska, D.; Bohatyrewicz, R.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CC chemokines in a tumor: A review of pro-cancer and anti-cancer properties of the ligands of receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci. 2020, 21, 8412. [Google Scholar] [CrossRef]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC chemokines in a tumor: A review of pro-cancer and anti-cancer properties of receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef]
- Waugh, D.J.J.; Wilson, C.; Seaton, A.; Maxwell, P.J. Multi-faceted roles for CXC-chemokines in prostate cancer progression. Front. Biosci. 2008, 13, 4595–4604. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Riese, D.J., II; Shen, J. The role of the CXCL12/CXCR4/CXCR7 chemokine axis in cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef]
- Singh, J.K.; Simoes, B.M.; Howell, S.J.; Farnie, G.; Clarke, R.B. Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells. Breast Cancer Res. 2013, 15, 210. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Gomes-Giacoia, E.; Dai, Y.; Lawton, A.; Miyake, M.; Furuya, H.; Goodison, S.; Rosser, C.J. Validation and clinicopathologic associations of a urine-based bladder cancer biomarker signature. Diagn. Pathol. 2014, 9, 200. [Google Scholar] [CrossRef] [Green Version]
- Margel, D.; Pesvner-Fischer, M.; Baniel, J.; Yossepowitch, O.; Cohen, I.R. Stress proteins and cytokines are urinary biomarkers for diagnosis and staging of bladder cancer. Eur. Urol. 2011, 59, 113–119. [Google Scholar] [CrossRef]
- Samarendra, H.; Jones, K.; Petrinic, T.; Silva, M.A.; Reddy, S.; Soonawalla, Z.; Gordon-Weeks, A. A meta-analysis of CXCL12 expression for cancer prognosis. Br. J. Cancer 2017, 117, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Domanska, U.M.; Kruizinga, R.C.; Nagengast, W.B.; Timmer-Bosscha, H.; Huls, G.; de Vries, E.G.E.; Walenkamp, A.M.E. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur. J. Cancer 2013, 49, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Bissonnette, M.; Khare, S. CXCL12-CXCR4/CXCR7 axis in colorectal cancer: Therapeutic target in preclinical and clinical studies. Int. J. Mol. Sci. 2021, 22, 7371. [Google Scholar] [CrossRef]
- Ieranò, C.; D’Alterio, C.; Giarra, S.; Napolitano, M.; Rea, G.; Portella, L.; Santagata, A.; Trotta, A.M.; Barbieri, A.; Campani, V.; et al. CXCL12 loaded-dermal filler captures CXCR4 expressing melanoma circulating tumor cells. Cell Death Dis. 2019, 10, 562. [Google Scholar] [CrossRef] [PubMed]
- Mortezaee, K. CXCL12/CXCR4 axis in the microenvironment of solid tumors: A critical mediator of metastasis. Life Sci. 2020, 249, 117534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xi, L.; Gooding, W.; Godfrey, T.E.; Ferrisa, R.L. Chemokine receptors 6 and 7 identify a metastatic expression pattern in squamous cell carcinoma of the head and neck. Adv. Otorhinolaryngol. 2005, 62, 121–133. [Google Scholar] [PubMed]
- Chen, K.; Bao, Z.; Tang, P.; Gong, W.; Yoshimura, T.; Wang, J.M. Chemokines in homeostasis and diseases. Cell. Mol. Immunol. 2018, 15, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, T.; Shetty, V.; Yamaguchi, M. Salivary cytokine panel indicative of non-small cell lung cancer. J. Int. Med. Res. 2018, 46, 3570–3582. [Google Scholar] [CrossRef] [PubMed]
- Katsura, M.; Shoji, F.; Okamoto, T.; Shimamatsu, S.; Hirai, F.; Toyokawa, G.; Morodomi, Y.; Tagawa, T.; Oda, Y.; Maehara, Y. Correlation between CXCR4/ CXCR7/ CXCL12 chemokine axis expression and prognosis in lymph-node-positive lung cancer patients. Cancer Sci. 2018, 109, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Rinaldi, C.; Corrigan, D.K.; Dennany, L.; Jarrett, R.F.; Lake, A.; Baker, M.J. Development of an electrochemical CCL17/TARC biosensor toward rapid triage and monitoring of classic Hodgkin lymphoma. ACS Sens. 2021, 6, 3262–3272. [Google Scholar] [CrossRef] [PubMed]
- Elemam, N.M.; Hannawi, S.; Maghazachi, A.A. Role of chemokines and chemokine receptors in rheumatoid arthritis. Immunotargets Ther. 2020, 9, 43–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, L.; Adlard, N.; Biehl, M.; Juarez, M.; Smallie, T.; Snow, M.; Buckley, C.D.; Raza, K.; Filer, A.; Scheel-Toellner, D. Expression of chemokines CXCL4 and CXCL7 by synovial macrophages defines an early stage of rheumatoid arthritis. Ann. Rheum. Dis. 2016, 75, 763–771. [Google Scholar] [CrossRef] [Green Version]
- Moadab, F.; Khorramdelazad, H.; Abbasifard, M. Role of CCL2/CCR2 axis in the immunopathogenesis of rheumatoid arthritis: Latest evidence and therapeutic approaches. Life Sci. 2021, 269, 119034. [Google Scholar] [CrossRef] [PubMed]
- Liou, L.-B.; Fang, Y.-F.; Tan, C.F.; Lai, J.-H.; Jang, S.-s.; Tsai, P.-H.; Yeh, T.-c. A new laboratory surrogate (Monocyte Chemotactic Protein-1) for Disease Activity Score28: A favourable indicator for remission in rheumatoid arthritis. Sci. Rep. 2020, 10, 8238. [Google Scholar] [CrossRef] [PubMed]
- Blaschke, S.; Koziolek, M.; Schwarz, A.; Benohr, P.; Middel, P.; Schwarz, G.; Hummel, K.M.; Muller, G.A. Proinflammatory role of fractalkine (CX3CL1) in rheumatoid arthritis. J. Rheumatol. 2003, 30, 1918–1927. [Google Scholar]
- Demirdöğen, B.C. A literature review of biosensors for multiple sclerosis: Towards personalized medicine and point-of-care testing. Mult. Scler. Relat. Disord. 2021, 48, 102675. [Google Scholar] [CrossRef]
- Comabella, M.; Montalban, X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol. 2014, 13, 113–126. [Google Scholar] [CrossRef]
- Cui, L.-Y.; Chu, S.-F.; Chen, N.-H. The role of chemokines and chemokine receptors in multiple sclerosis. Int. Immunopharmacol. 2020, 83, 106314. [Google Scholar] [CrossRef]
- Mori, F.; Nisticò, R.; Nicoletti, C.G.; Zagaglia, S.; Mandolesi, G.; Piccinin, S.; Martino, G.; Finardi, A.; Rossini, P.M.; Marfia, G.A.; et al. RANTES correlates with inflammatory activity and synaptic excitability in multiple sclerosis. Mult. Sclerosis J. 2016, 22, 1405–1412. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Khademi, M.; Fugger, L.; Lindhe, Ö.; Novakova, L.; Axelsson, M.; Malmeström, C.; Constantinescu, C.; Lyckee, J.; Piehl, F.; et al. Inflammation-related plasma and CSF biomarkers for multiple sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 12952–12960. Available online: www.pnas.org/cgi/doi/10.1073/pnas.1912839117 (accessed on 27 June 2022). [CrossRef]
- Weetman, A.P.; Bennett, G.L.; Wong, W.L. Thyroid follicular cells produce interleukin-8. J. Clin. Endocrinol. Metab. 2002, 75, 328–330. [Google Scholar] [CrossRef]
- Kasai, K.; Banba, N.; Motohashi, S.; Hattor, Y.; Manaka, K.; Shimoda, S.I. Expression of monocyte chemoattractant protein-1 mRNA and protein in cultured human thyrocytes. FEBS Lett. 1996, 394, 137–140. [Google Scholar] [CrossRef] [Green Version]
- Aust, G.; Steinert, M.; Boltze, C.; Kieseling, A.; Simchen, C. GRO-a in normal and pathological thyroid tissues and its regulation in thyroid-derived cells. J. Endocrinol. 2001, 170, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, S.M.; Fallahi, P.; Ruffilli, I.; Elia, G.; Ragusa, F.; Benvenga, S.; Antonelli, A. The association of other autoimmune diseases in patients with Graves’ disease (with or without ophthalmopathy): Review of the literature and report of a large series. Autoimmun. Rev. 2019, 18, 287–292. [Google Scholar] [CrossRef]
- Garcia-Lopez, M.A.; Sancho, D.; Sanchez-Madrid, F.; Marazuela, M. Thyrocytes from autoimmune thyroid disorders produce the chemokines IP-10 and Mig and attract CXCR3 lymphocytes. J. Clin. Endocrinol. Metab. 2001, 86, 5008–5016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romagnani, P.; Rotondi, M.; Lazzeri, E.; Lasagni, L.; Francalanci, M.; Buonamano, A.; Milani, S.; Vitti, P.; Chiovato, L.; Tonacchera, M.; et al. Expression of IP-10/CXCL10 and MIG/CXCL9 in the thyroid and increased levels of IP-10/CXCL10 in the serum of patients with recent-onset Graves’ disease. Am. J. Pathol. 2002, 161, 195–206. [Google Scholar] [CrossRef]
- Ferrer-Francesch, X.; Caro, P.; Alcalde, L.; Armengol, M.P.; Ashhab, Y.; Lucas-Martin, A.; Martinez-Caceres, E.M.; Pujol-Borrell, M.J.R. Onetube-PCR technique for CCL2, CCL3, CCL4 and CCL5 applied to fine needle aspiration biopsies shows different profiles in autoimmune and non-autoimmune thyroid disorders. J. Endocrinol. Investig. 2006, 29, 342–349. [Google Scholar] [CrossRef]
- Armengol, M.P.; Juan, M.; Lucas-Martin, A.; Fernandez-Figueras, M.T.; Jaraquemada, D.; Gallart, T.; Pujol-Borrell, R. Thyroid autoimmune disease: Demonstration of thyroid antigenspecifi c B cells and recombination-activating gene expression in chemokine-containing active intrathyroidal germinal centers. Am. J. Pathol. 2001, 159, 861–873. [Google Scholar] [CrossRef]
- Kemp, E.H.; Metcalfe, R.A.; Smith, K.A.; Woodroofe, M.N.; Watson, P.F.; Weetman, A.P. Detection and localization of chemokine gene expression in autoimmune thyroid disease. Clin. Endocrinol. 2003, 59, 207–213. [Google Scholar] [CrossRef]
- Masih, M.; Agarwal, S.; Kaur, R.; Gautam, P.K. Role of chemokines in breast cancer. Cytokine 2022, 155, 155909. [Google Scholar] [CrossRef]
- Chen, X.; Jin, R.C.R.; Huang, Z. The role of CXCL chemokine family in the development and progression of gastric cancer. Int. J. Clin. Exp. Pathol. 2020, 13, 484–492. [Google Scholar] [PubMed]
- McDonough, A.A.; Veiras, L.C.; Minas, J.N.; Ralph, D.L. Considerations when quantitating protein abundance by immunoblot. Am. J. Physiol. Cell. Physiol. 2015, 308, C426–C433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, C.W.; Kilpatrick, L.E.; Pfleger, K.D.G.; Hill, S.J. A nanoluciferase biosensor to investigate endogenous chemokine secretion and receptor binding. iScience 2021, 24, 102011. [Google Scholar] [CrossRef]
- Vega, B.; Calle, A.; Sanchez, A.; Lechuga, L.M.; Ortiz, A.M.; Armelles, G.; Rodríguez-Frade, J.M.; Mellado, M. Real-time detection of the chemokine CXCL12 in urine samples by surface plasmon resonance. Talanta 2013, 109, 209–215. [Google Scholar] [CrossRef]
- Chung, S.; Chandra, P.; Koo, J.P.; Shim, Y.-B. Development of a bifunctional nanobiosensor for screening and detection of chemokine ligand in colorectal cancer cell line. Biosens. Bioelectron. 2018, 100, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, S.; Cadano, D.; Agüí, L.; Barderas, R.; Campuzano, S.; Yañez-Sedeño, P.; Pingarrón, J.M. Click chemistry-assisted antibodies immobilization for immunosensing of CXCL7 chemokine in serum. J. Electroanal. Chem. 2019, 837, 246–253. [Google Scholar] [CrossRef]
- Guerrero, S.; Sánchez-Tirado, E.; Agüí, L.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Simultaneous determination of CXCL7 chemokine and MMP3 metallopro-teinase as biomarkers for rheumatoid arthritis. Talanta 2021, 234, 122705. [Google Scholar] [CrossRef]
- Kamińska, A.; Sprynskyy, M.; Winkler, K.; Szymborski, T. Ultrasensitive SERS immunoassay based on diatom biosilica for detection of interleukins in blood plasma. Anal. Bioanal. Chem. 2017, 409, 6337–6347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydın, E.B.; Sezgintürk, M.K. An impedimetric immunosensor for highly sensitive detection of IL-8 in human serum and saliva samples: A new surface modification method by 6-phosphonohexanoic acid for biosensing applications. Anal. Biochem. 2018, 554, 44–52. [Google Scholar] [CrossRef]
- Aydın, M.; Aydın, E.B.; Sezgintürk, M.K. A highly selective electrochemical immunosensor based on conductive carbon black and star PGMA polymer composite material for IL-8 biomarker detection in human serum and saliva. Biosens. Bioelectron. 2018, 117, 720–728. [Google Scholar] [CrossRef]
- Verbarg, J.; Hadass, O.; Olivo, P.D.; Danielli, A. High sensitivity detection of a protein biomarker interleukin-8 utilizing a magnetic modulation biosensing system. Sens. Actuators B 2017, 241, 614–618. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, R.; Xu, L.; Ning, Y.; Xie, S.; Zhang, G.J. Silicon nanowire biosensor for highly sensitive detection of oral squamous cell carcinoma biomarkers in saliva. Anal. Sci. 2015, 31, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Margulis, M.; Cohen, M.; Burg, S.; Avivi-Mintz, S.; Danielli, A. Optical modulation biosensing system for rapid detection of biological targets at low concentrations. Biomed. Opt. Express 2021, 12, 5338–5350. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jo, E.-J.; Hong, D.; Oh, H.-K.; Lee, K.J.; Shin, Y.-B.; Kim, M.-G. One-pot, solid-phase immunosensing platform consisting of a nanometer-thick Au/TiO2 photocatalytic film and Cy5/capture antibody/gold nanorod conjugates. ACS Appl. Nano Mater. 2021, 4, 5454–5460. [Google Scholar] [CrossRef]
- Aydın, E.B.; Sezgintürk, M.K. Fabrication of electrochemical immunosensor for detection of interleukin 8 biomarker via layer-by-layer self-assembly process on cost-effective fluorine tin oxide electrode. Electroanalysis 2021, 33, 1596–1605. [Google Scholar] [CrossRef]
- Sharma, R.; Deacon, S.E.; Nowak, D.; George, S.E.; Szymonik, M.P.; Tang, A.A.S.; Tomlinson, D.C.; Davies, A.G.; McPherson, M.J.; Wälti, C. Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity. Biosens. Bioelectron. 2016, 80, 607–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan-Nixon, H.A.G.; Singh, N.; Cass, A.E.G. A sensitive impedimetric immuno-sensor for the detection of Interleukin-8 in nasal epithelial lining fluid of asthma patients. Biosens. Bioelectron. X 2022, 10, 100118. [Google Scholar] [CrossRef]
- Torrente-Rodríguez, R.M.; Campuzano, S.; Montiel, V.R.; Gamella, M.; Pingarrón, J.M. Electrochemical bioplatforms for the simultaneous determination of interleukin (IL)-8 mRNA and IL-8 proteinoralcancerbiomarkersinraw saliva. Biosens. Bioelectron. 2016, 77, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Tanak, A.S.; Sardesai, A.; Muthukumar, S.; Krishnan, S.; Striegel, D.A.; Schully, K.L.; Clark, D.V.; Prasad, S. Multiplexed host immune response biosensor for rapid sepsis stratification and endotyping at point-of-care. Biosens. Bioelectron. X 2022, 10, 100144. [Google Scholar] [CrossRef]
- Bonham, A.J.; Paden, N.G.; Ricci, F.; Plaxco, K.W. Detection of IP-10 protein marker in undiluted blood serum via an electrochemical E-DNA scaffold sensor. Analyst 2013, 138, 5580–5583. [Google Scholar] [CrossRef] [Green Version]
- Jagannath, B.; Lin, K.; Pali, M.; Sankhala, D.; Muthukumar, S.; Prasad, S. Temporal profiling of cytokines in passively expressed sweat for detection of infection using wearable device Bioeng. Transl. Med. 2021, 6, e10220. [Google Scholar] [CrossRef]
- Mao, W.; He, J.; Tang, Z.; Zhang, C.; Chen, J.; Li, J.; Yu, C. A sensitive sandwich-type immunosensor for the detection of MCP-1 based on a rGO-TEPA-Thi-Au nanocomposite and novel RuPdPt trimetallic nanoalloy particles. Biosens. Bioelectron. 2019, 131, 67–73. [Google Scholar] [CrossRef]
- Li, Y.; He, J.; Xia, C.; Gao, L.; Yu, C. Ultrasensitive electrochemical immunosensor based on orderly oriented conductive wires for the detection of human monocyte chemotactic protein-1 in serum. Biosens. Bioelectron. 2015, 70, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Valera, E.; Shia, W.W.; Bailey, R.C. Development and validation of an immunosensor for monocyte chemotactic protein 1 using a silicon photonic microring resonator biosensing platform. Clin. Biochem. 2016, 49, 121–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Liu, X.; Zhang, C.; Tang, Z.; Chen, J.; Yu, C. Electrochemical immunosensor for monocyte chemoattractant protein-1 detection based on Pt nanoparticles functionalized single-walled carbon nanohorns. Int. J. Electrochem. Sci. 2018, 13, 3923–3934. [Google Scholar] [CrossRef]
- Aydın, E.B.; Aydın, M.; Sezgintürk, M.K. Electrochemical immunosensor for detection of CCR4 cancer biomarker in human serum: An alternative strategy for modification of disposable ITO electrode. Mol. Biosci. 2021, 21, 2000267. [Google Scholar] [CrossRef]
- Aydın, E.B.; Aydın, M.; Sezgintürk, M.K. Fabrication of electrochemical immunosensor based on acid-substituted poly(pyrrole) polymer modified disposable ITO electrode for sensitive detection of CCR4 cancer biomarker in human serum. Talanta 2021, 222, 121487. [Google Scholar] [CrossRef]
- Hu, B.; Fan, H.; Lv, X.; Chen, S.; Shao, Z. Prognostic significance of CXCL5 expression in cancer patients: A meta-analysis. Cancer Cell Int. 2018, 18, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinchia, J.; Echeverri, D.; Cruz-Pacheco, A.F.; Maldonado, M.E.; Orozco, J. Electrochemical biosensors for determination of colorectal tumor biomarkers. Micromachines 2020, 11, 411. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Jarrett, R.F.; Hjalgrim, H.; Proietti, C.; Chang, E.T.; Smedby, K.E.; Yu, K.J.; Lake, A.; Troy, S.; McAulay, K.A.; et al. Evaluation of the antibody response to the EBV proteome in EBV-associated classical Hodgkin lymphoma. Int. J. Cancer 2020, 147, 608–618. [Google Scholar] [CrossRef]
- Nedeva, C.; Menassa, J.; Puthalakath, H. Sepsis: Inflammation is a necessary evil. Front. Cell Dev. Biol. 2019, 7, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanak, A.S.; Muthukumar, S.; Krishnan, S.; Schully, K.L.; Clark, D.V.; Prasad, S. Multiplexed cytokine detection using electrochemical point-of-care sensing device towards rapid sepsis endotyping. Biosens. Bioelectron. 2021, 171, 112726. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; Koeller, M.; Weisman, M.; Emery, P. New therapies for the treatment of rheumatoid arthritis. Lancet 2007, 370, 1861–1874. [Google Scholar] [CrossRef]
- Ribbens, C.; Porras, M.M.y.; Franchimont, N.; Kaiser, M.-J.; Jaspar, J.-M.; Damas, P.; Houssiau, F.A.; Malaise, M.G. Increased matrix metalloproteinase-3 serum levels in rheumatic diseases: Relationship with synovitis and steroid treatment. Ann. Rheum. Dis. 2002, 61, 161–166. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Li, E.; Liu, Y.; Xie, W.; Huang, C.; Song, J.; Zhang, W.; Zheng, Y.; Wang, H.; Wang, Q. CTAPIII/CXCL7: A novel biomarker for early diagnosis of lung cancer. Cancer Med. 2018, 7, 283–535. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [Green Version]
- Szekanecz, Z.; Vegvari, A.; Szabo, Z.; Koch, A.E. Chemokines and chemokine receptors in arthritis. Front. Biosci. 2010, 2, 153–167. [Google Scholar] [CrossRef] [Green Version]



| Chemokines, Receptors | Disease | Sample | Observations | Ref. |
|---|---|---|---|---|
| CXCL12, CXCL14, CXCR4 | HIV-1 | Peripheral blood mononuclear cells (PBMCs) | CXCL14 synergizes with CXCL12 via allosteric modulation of CXCR4; CXCL14 bound to CXCR4 with high affinity, induced redistribution of cell-surface CXCR4, and enhanced HIV-1 infection by >3-fold | [29] |
| CXCL8, CXCL10 (and pro-inflammatory cytokines) | Oral squamous cell carcinoma | Saliva, plasma | The levels of CXCL8 and CXCL10 were higher in the OSCC patients than in the controls | [30] |
| CCL3, MIP-1α | Cystic fibrosis (CF) | Tears | Patients with severe CF have significantly increased levels of MIP-1 α which correlate negatively with clinical status | [31] |
| CXCL8 (and IL-6, TNF-α) | Dental caries | Saliva | CXCL8 levels significantly higher in dental caries patients | [32] |
| CXCL8, CCL5/ RANTES (IL-1β, IL-6, IL-13, IL-17A, TNF-α) | Graves’ orbitopathy (GO) | Tears | Higher release of CCL5 (RANTES) and cytokines in GO patients | [33] |
| MCP-1, CXCL8 (and IL-6, IL-1β, TNF-α, γ-INF) | CKD (chronic kidney disease) | Saliva | MCP-1 and CXCL8 levels decrease in patients with CKD | [34] |
| CXCL8 (and various cytokines) | Thyroid-associated orbitopathy (TAO) | Tears | CXCL8 levels increase in patients of TAO | [35] |
| CCL2, CCL3, CCL8, CXCL10 (and IL-2, IL-6, IL-15, TNF-α, γ-INF) | Latent tuberculosis infection (LTBI) | Peripheral blood mononuclear cells (PBMCs) | Relevant role of CCL2 relevant for revealing subjects at higher risk of reactivation LTBI | [36] |
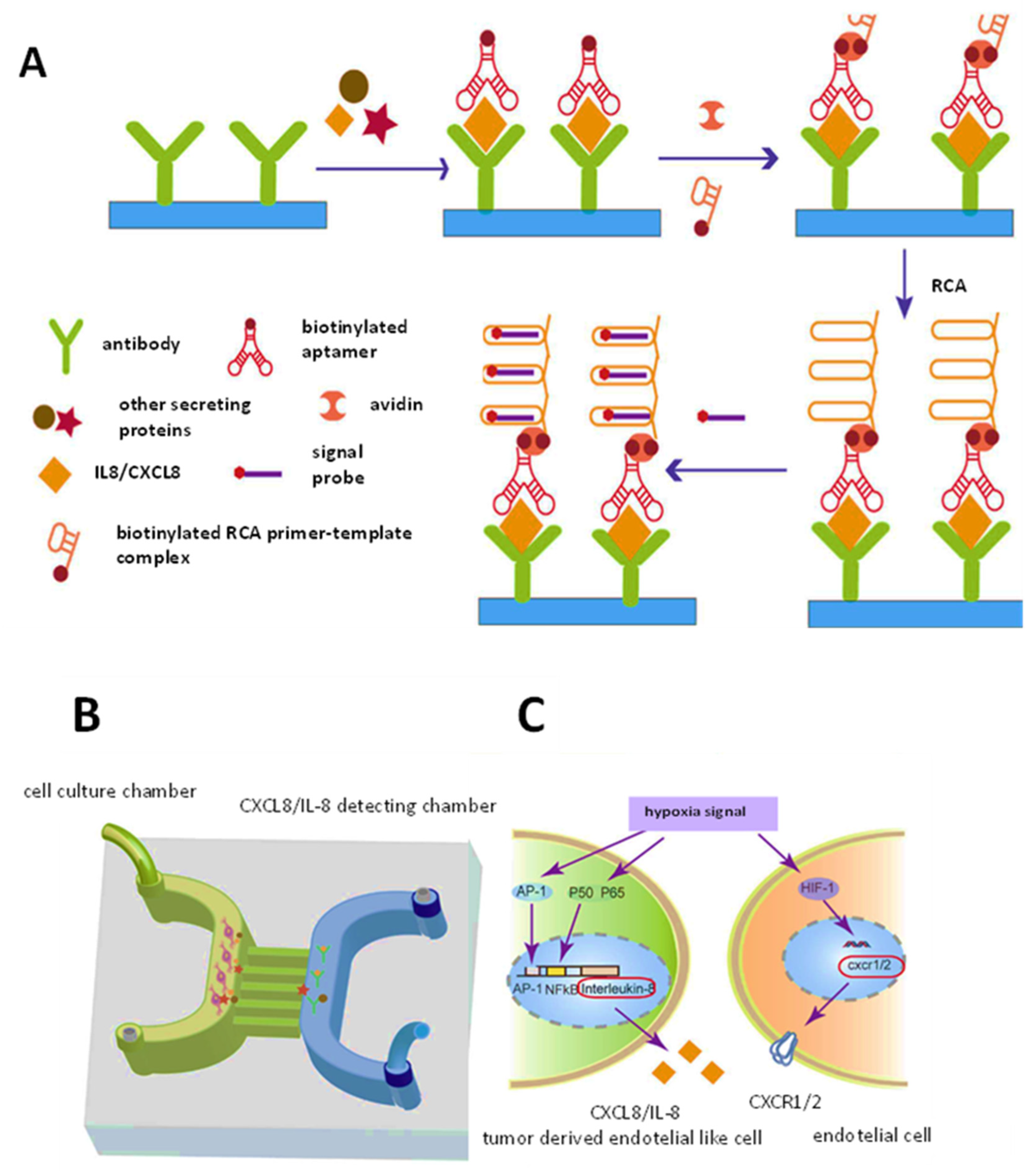
| CXCL8 | Glioblastomas | TDECs and UVECs | CXCL8 plays an important role in the process of glioma stem-like cell vascularization | [37] |
| CCL5/RANTES, MDC (and TGF-β1) | HSC expansion | Stem cell cultures | CCL5, MDC, TGF-β1: secreted factors deleterious to HSC expansion. Significant modulators in stem cell cultures. | [38] |
| CXCL13 | Sjögren’s syndrome (SS) | Serum, saliva | Elevated serum or salivary CXCL13 levels in patients with primary SS or SS | [39] |
| CXCL8 (and IL1α, IL 1β) | MERS-CoV | Sputum, tracheal aspirate | High expression of CXCL8 in the lower respiratory tract of MERS-CoV infected patients | [40] |
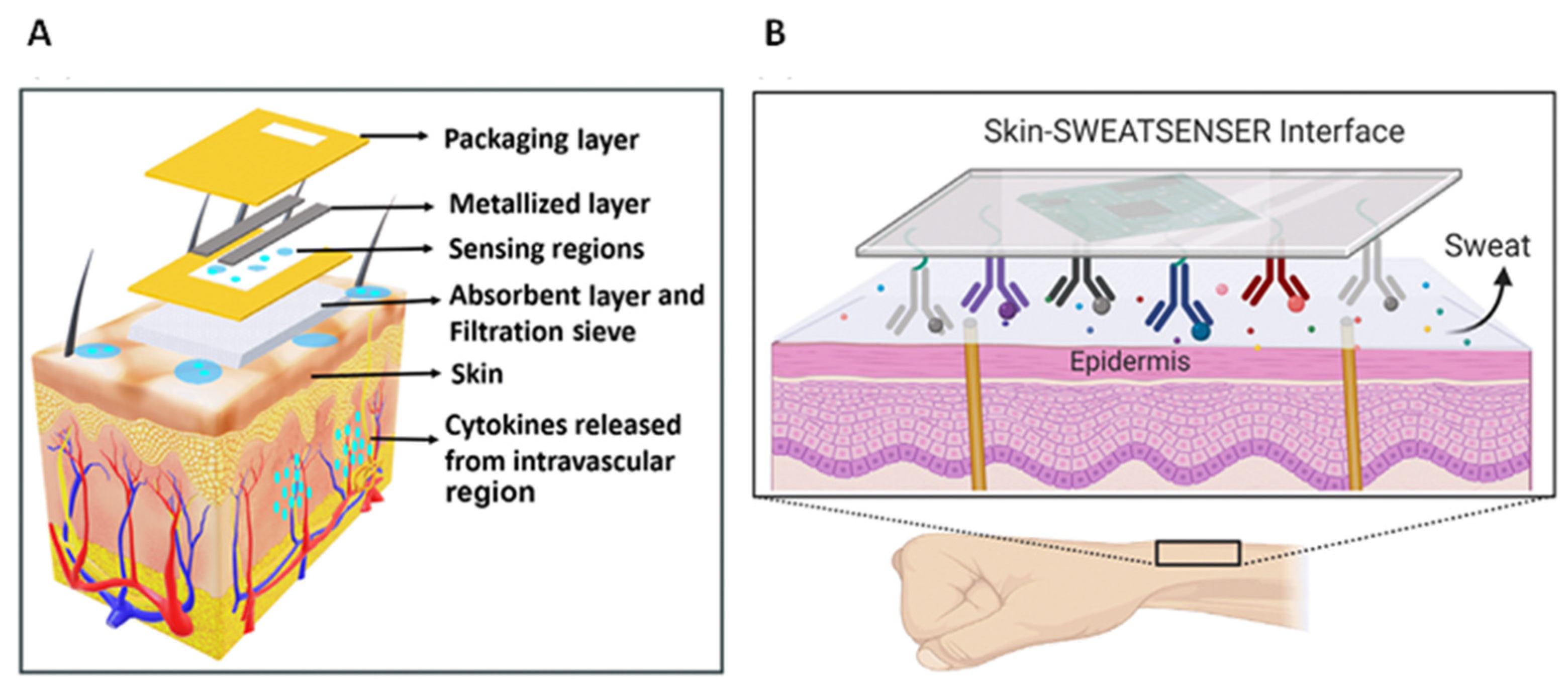
| CXCL8 (and IL-6, IL-10, TNF-α) | infection/ inflammation | Eccrine sweat | CXCL8 levels in sweat can be correlated to that in serum | [41] |
| CXCL13, CXCL10, CCL2, CCL3, CXCL12, CCL5 | Sjögren’s syndrome (SS) | Saliva | Higher levels of CXCL10 and CCL2 in primary SS | [42] |
| 28 cytokines, eight chemokines, and nine growth factors | Sepsis | Serum | Increased levels of CCL2, CCL3, CCL4, CCL5/RANTES, CCL11, CXCL10, CXCL12 in sepsis patients | [43] |
| Type of Biosensor | Technique | Target(s) | Biodetection Principle | Dynamic Range | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|
| Chemokine tagging with nanoluciferase fragment HiBiT and to Alexa-Fluor488-tagged CXCR4 | Luminescence (BRET) | CXCL12 | CRISPR/Cas9 genome editing used to tag the chemokine. CXCL12 secretion monitored in live cells | 1 fM–1 nM | - | live cells | [85] |
| Lentiviral particles with CXCR4 immobilized on MUA SAM onto Au chip | SPR | CXCL12 | Specific CXCL12 binding to LVPX4-coated chip in the biosensor | 5–50 nM | - | urine of RA patients | [86] |
| CXCR2-AuNPs/2,2′:5′,2″-tert-thiophene-3′(p-benzoic acid) | Amperometry | CXCL5 | Selection of the ligand for CXCR2 receptor by EIS | 0.1–10 ng/mL | 0.078 ng/mL | human serum CRC cells | [87] |
| Chip-based gold nanostructured micro-electrode with immobilized thiolated DNA | SWV | CCL5/RANTES CCL22/MDC | Amplified steric hindrance hybridization combining DNA and antibodies signaled by methylene blue | 10 pg/mL–10 ng/mL | 10 pg/mL | stem cell culture | [38] |
| Electrochemical sandwich-type immunosensor with immobilized anti-CXCL7 on IgG-MWCNTs/SPCE | Amperometry | CXCL7 | Electrode modification by click chemistry | 0.5–600 pg/mL | 0.1 pg/mL | human serum form patients with RA | [88] |
| Sandwich-type electrochemical dual immunosensing platform | Amperometry | CXCL7 | Simultaneous determination of CXCL7 and MMP3 with COOH-MBs on SPdCE | 1–75 ng/mL CXCL7 2.0–2000 ng/mL MMP3 | 0.3 ng/mL 0.8 pg/mL | +/− RA human serum | [89] |
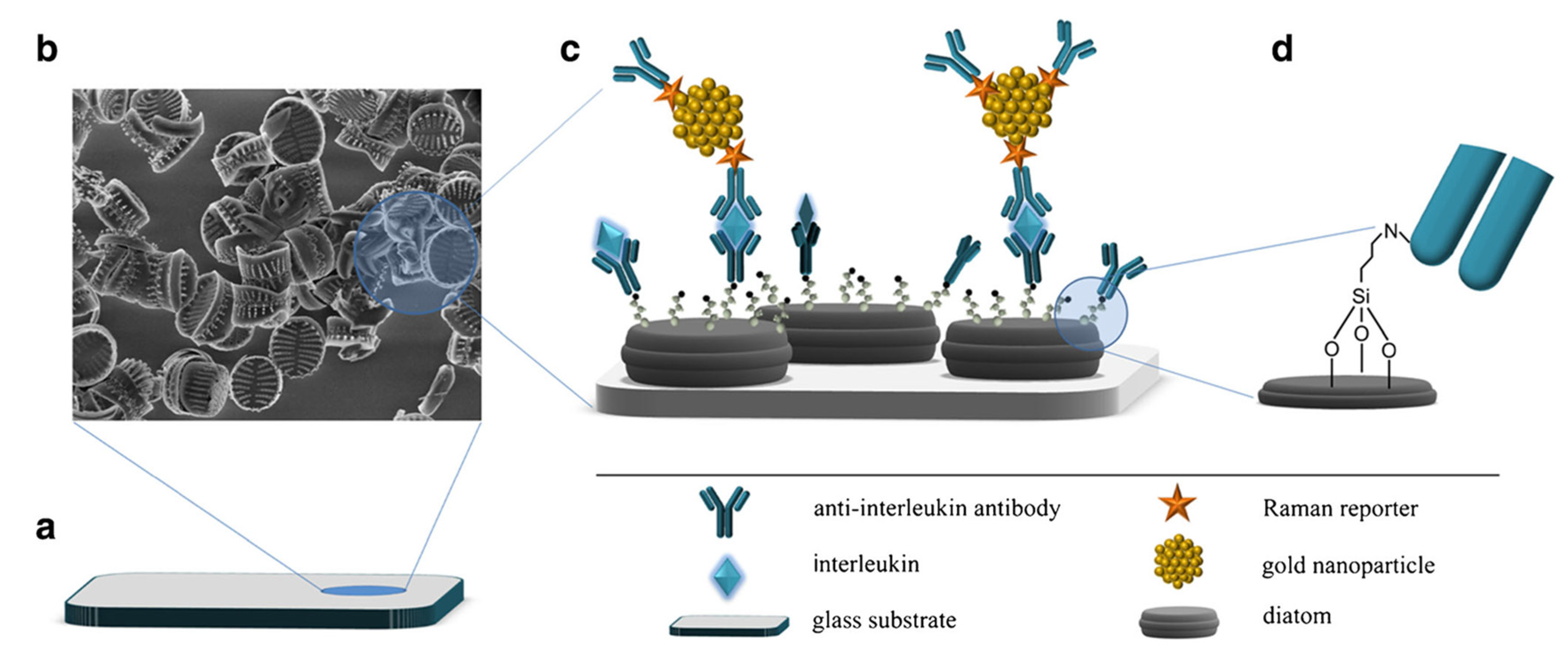
| Sandwich-type with cAb immobilized on diatom biosilica/AuNPs/DTNB | SERS | CXCL8/IL-8 | cAb/AuNPs-DTNB as the Raman reporter | up to 30 ng/mL | 6.2 pg/mL | plasma | [90] |
| Dual-function microfluidic chip. Sandwich-type with Abs and aptamers | Fluorescence | CXCL8/IL-8 | Rolling circle amplification | 7.5–120 pg/mL | 0.84 pg/mL | TDEC and HUVEC | [37] |
| Label-free immunosensor with anti-CXCL8 immobilized on 6-phospho-hexanoic acid/ITO | EIS (charge transfer resistance, ΔRct) | CXCL8/IL-8 | Phosphonic acid covalently bound to hydroxylated ITO. EDC/NHS activation of carboxyl groups for Ab immobilization | 0.02–3 pg/mL | 6 fg/mL | human serum and saliva | [91] |
| Label-free immunosensor with anti-CXCL8 immobilized on SuperP© carbon black/Star polymer/ITO | EIS (charge transfer resistance, ΔRct) | CXCL8/IL-8 | The high conductivity of carbonaceous material enhances electron transfer | 0.01–3 pg/mL | 3.3 fg/mL | human serum and saliva | [92] |
| Sandwich-type fluorometric immunoassay with fluorophore-Avidin-Biotin-CXCL8-cAb-MBs | Fluorescence | CXCL8/IL-8 | Laser excitation of sample fluorophores | up to 5000 ng/L | 0.19 ng/mL | spiked human plasma | [93] |
| SiNMW FET biosensor with Abs immobilized onto APTES and GA modified surface | ΔR in I/V curves | CXCL8/IL-8 | Multiplexed determination of CXCL8 and TNF-α. Current/Resistance increase or decrease depending on pI of proteins | 10 fg/mL–1 ng/mL | 10 fg/mL | saliva | [94] |
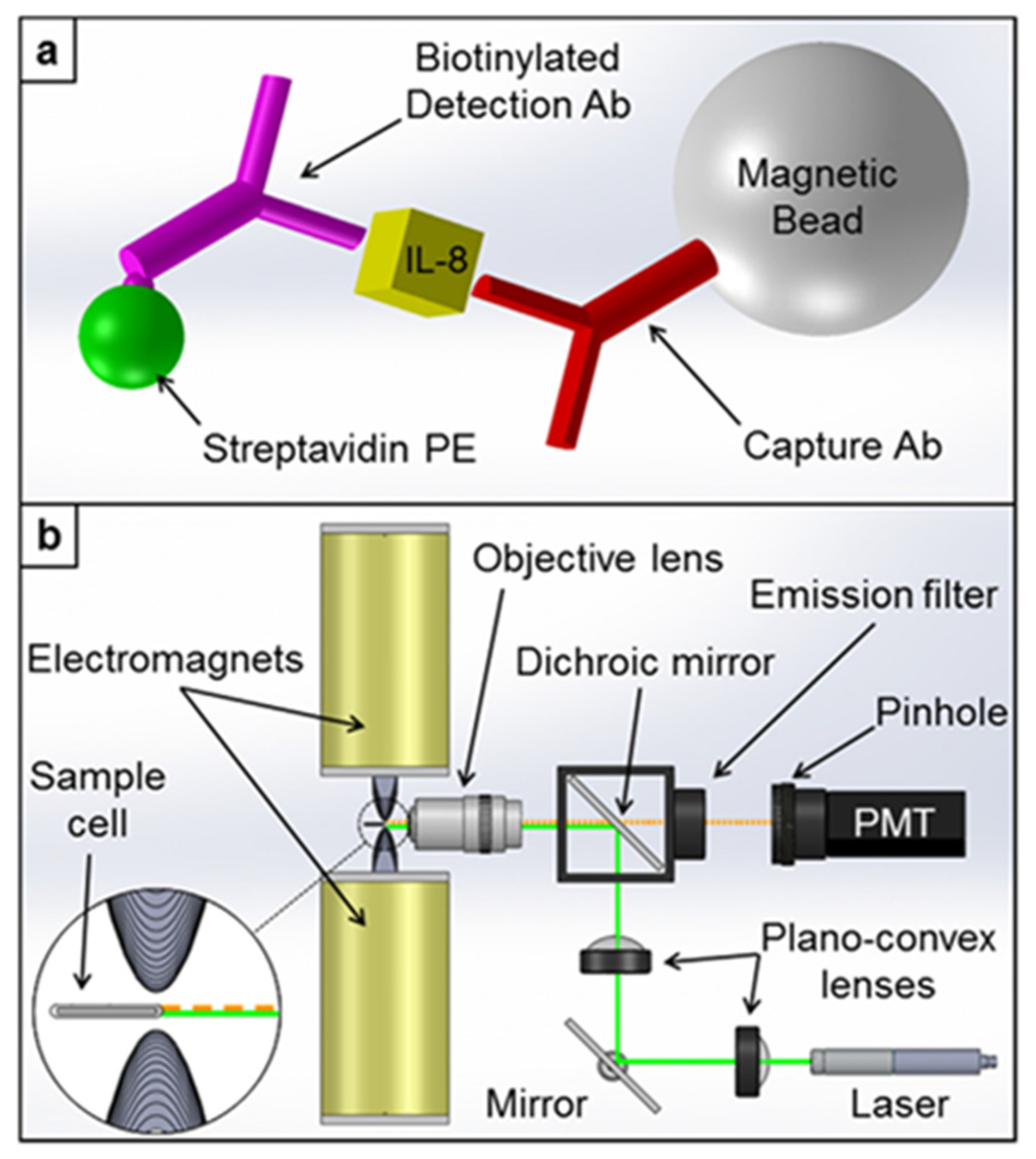
| Optical modulation biosensing (OMB) | Fluorescence | CXCL8/IL-8 | SA-PE-Biotin-Ab2-CXCL8-Ab1-MBs | 0.05–10,000 ng/mL | 0.02 ng/L | - | [95] |
| Fluorometric immunosensor with H2O2 generating Au/TiO2 photocatalytic film and Cy5/cAb/GNR MEF probes | Fluorescence | CXCL8/IL-8 | One-pot, wash-free immunoassay. H2O2 induced chloro-1-naphthol precipitation quench Cy5 fluorescence via FRET | 1–1000 pg/mL | 0.612 pg/mL | serum | [96] |
| Label-free electrochemical immunosensor | EIS (Rct) | CXCL8/IL-8 | Anti-CXCL8 immobilized onto FTO-OH/IPTES platform. | 0.04–2 pg/mL | 11.9 fg/mL | human serum, saliva | [97] |
| Label-free synthetic Ab-mimetic protein-monothiol-alkane-PEG SAM-AuE | EIS (change in phase, Δθ(f)) | CXCL8/IL-8 | Ab-mimetic protein selected via phage display. Capture protein coding region subcloned in top ET11 | 900 fg/mL–900 ng/mL | 90 fg/mL | spiked serum | [98] |
| Label-free immunosensor with cAb immobilized onto pCBMA/AuE | EIS (Rct) | CXCL8/IL-8 | AuE modified by cysteamine adsorption, covalent linking of CB and photo-polymerization. | 55 fM–55 nM | 10 fM | nasal epithelial lining fluid | [99] |
| Electrochemical dual biosensing platform with anti-CXCL8 antibody and specific hairpin DNA sequence | Amperometry | CXCL8/IL-8 CXCL8 mRNA | Simultaneous determination of CXCL8 and CXCL8 mRNA involving COOH-MBs, Strept-MBs and dual SPCEs | 87.9–5000 pg/mL CXCL8; 0.32–7.5 nM CXCL8 mRNA | 72.4 pg/mL 0.21 nM | spiked human saliva | [100] |
| Array of nanofilm ZnO/metal electrodes functionalized with specific Abs | EIS | CXCL8/IL-8 CXCL10/IP-10 | Simultaneous determination of CXCL8 CXCL10, IL-6, IL-10 and TRAIL | 0.1–5000 pg/mL CXCL8 1–2000 pg/mL CXCL10 | ~1 pg/mL | sepsis patients´ serum | [101] |
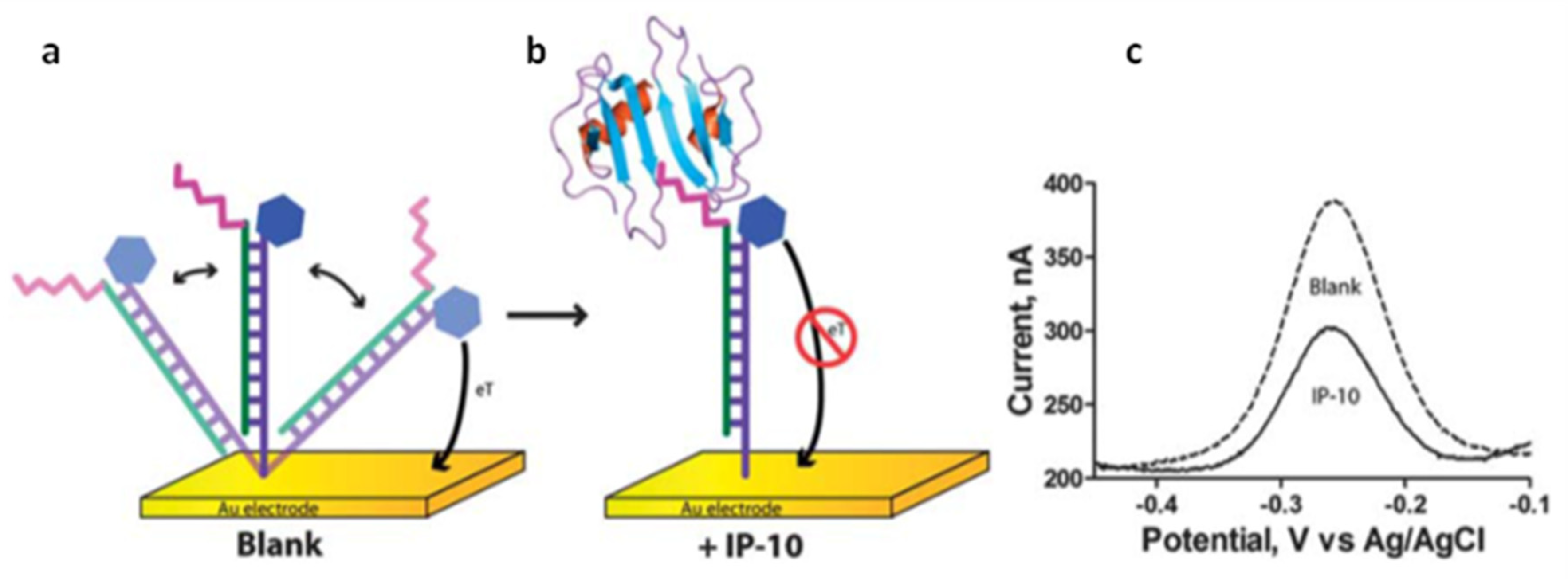
| Electrochemical DNA biosensor. Immobilized DNA strand with a distal methylene blue reporter on SAM-AuE. | Amperometry | CXCL10 /IP-10 | Hybridization to recognition strand with target binding peptide. Current decrease as increase target concentration | 1–2000 nM | ~60 pM | serum | [102] |
| DTSSP/Abs solutions immobilized onto ZnO electrodes | EIS | CXCL10/IP-10 | Simultaneous determination of CXCL10, TRAIL and CRP | up to 500 pg/mL | <2 pg/mL | human sweat | [103] |
| Sulfo-LC-SPDP/Ab solution immobilized onto AuE. Sandwich-type electrochemical immunoassay. | Amperometry | CCL17/TARC | HRP-Strept-Biotin-Ab2 conjugates for TMB/H2O2 detection | 387–50,000 pg/mL | 194 pg/mL | serum from patients with cHL | [62] |
| Sandwich-type electrochemical immunosensor with cAb immobilized on rGO-(rGO-TEPAThi-Au)/GCE | Amperometry | CCL2/MCP1 | Signal amplification with Ab2-RuPdPt | 20 fg/mL–1000 pg/mL | 8.9 fg/mL | spiked serum | [104] |
| Label-free electrochemical immuno-sensor with cAb immobilized on Au @Pt-CA-AuE | DPV | CCL2/MCP1 | Decrease in peak current of Fe(CN)63−/4− as increased CCL2 concentration | 0.09–360 pg/mL | 0.03 pg/mL | spiked serum | [105] |
| Sandwich-type immunosensor with cAb immobilized onto a silicon photonic micro ring resonator. | Res λ shift | CCL2/MCP1 | Shifts in resonance wavelength are related to the target concentration | 84.3–1582.1 pg/mL | 0.5 pg/mL | spiked serum | [106] |
| Label-free electrochemical immunosensor with cAb immobilized on PtNPs/SWCNHs | Amperometry | CCL2/MCP1 | Reduction current of H2O2 decrease as increased CCL2 concentration | 0.06–450 pg/mL | 0.02 pg/mL | serum | [107] |
| Label-free anti-CCR4-PPyr-CSsg-ITO immunosensor | EIS | CCR4 | Amino groups of anti-CCR4 antibodies bound covalently to succinimide groups of PPyr-CSsg polymer | 0.024–12 pg/mL | 7.3 fg/mL | human serum | [108] |
| Label-free anti-CCR4-P(Pyr-Pac)-ITO | EIS | CCR4 | Amino groups of anti-CCR4 antibodies bound covalently to carboxyl groups of P(Pyr-Pac) polymer | 0.02–8 pg/mL | 6.4 fg/mL | human serum | [109] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Tirado, E.; Agüí, L.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Biodetection Techniques for Quantification of Chemokines. Chemosensors 2022, 10, 294. https://doi.org/10.3390/chemosensors10080294
Sánchez-Tirado E, Agüí L, González-Cortés A, Yáñez-Sedeño P, Pingarrón JM. Biodetection Techniques for Quantification of Chemokines. Chemosensors. 2022; 10(8):294. https://doi.org/10.3390/chemosensors10080294
Chicago/Turabian StyleSánchez-Tirado, Esther, Lourdes Agüí, Araceli González-Cortés, Paloma Yáñez-Sedeño, and José M. Pingarrón. 2022. "Biodetection Techniques for Quantification of Chemokines" Chemosensors 10, no. 8: 294. https://doi.org/10.3390/chemosensors10080294
APA StyleSánchez-Tirado, E., Agüí, L., González-Cortés, A., Yáñez-Sedeño, P., & Pingarrón, J. M. (2022). Biodetection Techniques for Quantification of Chemokines. Chemosensors, 10(8), 294. https://doi.org/10.3390/chemosensors10080294









