Biosensing Strategies Based on Particle Behavior
Abstract
:1. Introduction
2. Principles of Biosensing Based on Various Particle Behaviors
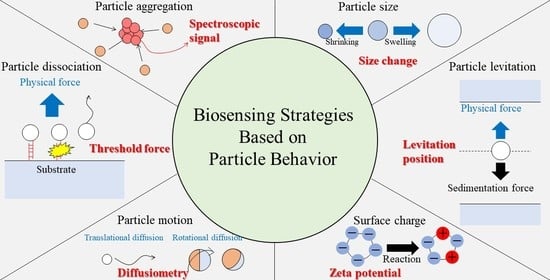
2.1. Particle Aggregation
2.2. Particle Dissociation from Substrates
2.3. Levitation of Particles in Physical Fields
2.4. Change in the Particle Size
2.5. Particle Motion
2.6. Change in the Surface Charge
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Tong, R.-J.; Xia, F.; Peng, Y. Current status of optical fiber biosensor based on surface plasmon resonance. Biosens. Bioelectron. 2019, 142, 111505. [Google Scholar] [CrossRef] [PubMed]
- Low, S.S.; Chen, Z.; Li, Y.; Lu, Y.; Liu, Q. Design principle in biosensing: Critical analysis based on graphitic carbon nitride (G-C3N4) photoelectrochemical biosensor. TrAC Trends Anal. Chem. 2021, 145, 116454. [Google Scholar] [CrossRef]
- Wang, X.; Lu, X.; Chen, J. Development of biosensor technologies for analysis of environmental contaminants. Trends Environ. Anal. Chem. 2014, 2, 25–32. [Google Scholar] [CrossRef]
- Guo, X. Surface plasmon resonance based biosensor technique: A review. J. Biophoton. 2012, 5, 483–501. [Google Scholar] [CrossRef]
- Kumai, M.; Kozuka, S.; Hashimoto, T.; Suzuki, I.; Hayashita, T. Glucose Recognition by a Supramolecular Complex of Boronic. Anal. Sci. 2012, 28, 121–126. [Google Scholar] [CrossRef] [Green Version]
- Fukuhara, G.; Inoue, Y. Peptide chirality sensing by a cyclodextrin-polythiophene conjugate. Chem. Eur. J. 2012, 18, 11459–11464. [Google Scholar] [CrossRef]
- Ma, D.; Zhang, L.-M. Novel biosensing platform based on self-assembled supramolecular hydrogel. Mater. Sci. Eng. 2013, 33, 2632–2638. [Google Scholar] [CrossRef]
- Fukuhara, G. Smart Polymer chemosensors: Signal-amplification systems with allosterism. Polym. J. 2021, 53, 1325–1334. [Google Scholar] [CrossRef]
- Barrow, S.J.; Kasera, S.; Rowlannd, M.J.; del Barrio, J.; Scherman, O.A. Cucurbituril-Based Molecular Recognition. Chem. Rev. 2015, 115, 12320–12406. [Google Scholar] [CrossRef] [Green Version]
- Berchmans, S.; Venkatesan, M.; Vusa, C.S.R.; Arumugam, P. PAMAM Dendrimer Modified Reduced Graphene Oxide Postfunctionalized by Horseradish Peroxidase for Biosensing H2O2. Methods Enzymol. 2018, 609, 143–170. [Google Scholar] [CrossRef]
- Liu, X.; Luo, L.; Ding, Y.; Xu, Y.; Li, F. Hydrogen peroxide biosensor based on the immobilization of horseradish peroxidase on γ-Al2O3 nanoparticles/chitosan film-modified electrode. J. Solid State Electrochem. 2011, 15, 447–453. [Google Scholar] [CrossRef]
- Rasheed, T.; Hassan, A.A.; Kausar, F.; Sher, F.; Bilal, M.; Iqbal, H.M.N. Carbon nanotubes assisted analytical detection—Sensing/delivery cues for environmental and biomedical monitoring. TrAC Trends Anal. Chem. 2020, 132, 116066. [Google Scholar] [CrossRef]
- Fu, L.-H.; Qi, C.; Lin, J.; Huang, P. Catalytic chemistry of glucose oxidase in cancer diagnosis and treatment. Chem. Soc. Rev. 2018, 47, 6454–6472. [Google Scholar] [CrossRef] [PubMed]
- Dongare, P.R.; Gore, A.H. Recent Advances in Colorimetric and Fluorescent Chemosensors for Ionic Species: Design, Principle and Optical Signalling Mechanism. ChemistrySelect 2021, 6, 5657–5669. [Google Scholar] [CrossRef]
- Fukuhara, G. Allosteric signal-amplification sensing with polymer-based supramolecular hosts. J. Incl. Phenom. Macrocycl. Chem. 2019, 93, 127–143. [Google Scholar] [CrossRef]
- Fukuhara, G. Analytical supramolecular chemistry: Colorimetric and fluorimetric chemosensors. J. Photochem. Photobiol. C 2020, 42, 100340. [Google Scholar] [CrossRef]
- Cao, D.; Zhu, L.; Liu, Z.; Lin, W. Through bond energy transfer (TBET)-based fluorescent chemosensors. J. Photochem. Photobiol. C 2020, 44, 100371. [Google Scholar] [CrossRef]
- Móczár, I.; Huszthy, P. Optically active crown ether-based fluorescent sensor molecules: A mini-review. Chirality 2019, 31, 97–109. [Google Scholar] [CrossRef]
- Ji, W.; Li, L.; Zhang, Y.; Wang, X.; Ozaki, Y. Recent advances in surface-enhanced Raman scattering-based sensors for the detection of inorganic ions: Sensing mechanism and beyond. J. Raman Spectrosc. 2021, 52, 468–481. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, Z.; Li, Q.; Wang, R.; Yu, F. Toward Sensitive and Reliable Surface-Enhanced Raman Scattering Imaging: From Rational Design to Biomedical Applications. ACS Sens. 2021, 6, 3912–3932. [Google Scholar] [CrossRef]
- Sun, X.-T.; Liu, M.; Xu, Z.-R. Microfluidic fabrication of multifunctional particles and their analytical applications. Talanta 2014, 121, 163–177. [Google Scholar] [CrossRef]
- Jones, O.G.; McClements, D.J. Functional Biopolymer Particles: Design, Fabrication, and Applications. Compr. Rev. Food Sci. Food Saf. 2010, 9, 374–397. [Google Scholar] [CrossRef]
- Tardy, B.L.; Mattos, B.D.; Otoni, C.G.; Beaumont, M.; Majoinen, J.; Kämäräinen, T.; Rojas, O.J. Deconstruction and Reassembly of Renewable Polymers and Biocolloids into Next Generation Structured Materials. Chem. Rev. 2021, 121, 14088–14188. [Google Scholar] [CrossRef]
- Shan, S.; Lai, W.; Xiong, Y.; Wei, H.; Xu, H. Novel strategies to enhance lateral flow immunoassay sensitivity for detecting foodborne pathogens. J. Agric. Food Chem. 2015, 63, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Xing, K.-Y.; Shan, S.; Liu, D.-F.; Lai, W.-H. Recent advances of lateral flow immunoassay for mycotoxins detection. TrAC Trends Anal. Chem. 2020, 133, 116087. [Google Scholar] [CrossRef]
- Fong, L.-K.; Wang, Z.; Schatz, G.C.; Luijten, E.; Mirkin, C.A. The Role of Structural Enthalpy in Spherical Nucleic Acid Hybridization. J. Am. Chem. Soc. 2018, 140, 6226–6230. [Google Scholar] [CrossRef]
- Semenova, M. Protein–polysaccharide associative interactions in the design of tailor-made colloidal particles. Curr. Opin. Colloid Interface Sci. 2017, 28, 15–21. [Google Scholar] [CrossRef]
- Porter, C.L.; Crocker, J.C. Directed assembly of particles using directional DNA interactions. Curr. Opin. Colloid Interface Sci. 2017, 30, 34–44. [Google Scholar] [CrossRef]
- Buyong, M.R.; Kayani, A.A.; Hamzah, A.A.; Yeop, M.B. Dielectrophoresis Manipulation: Versatile Lateral and Vertical Mechanisms. Biosensors 2019, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Miyagawa, A.; Okada, T. Particle Manipulation with External Field; From Recent Advancement to Perspectives. Anal. Sci. 2021, 37, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Brook, M.A.; Li, Y. Design of Gold Nanoparticle-Based Colorimetric Biosensing Assays. ChemBioChem 2008, 9, 2363–2371. [Google Scholar] [CrossRef]
- Mannelli, I.; Marco, M.-P. Recent advances in analytical and bioanalysis applications of noble metal nanorods. Anal. Bioanal. Chem. 2010, 398, 2451–2469. [Google Scholar] [CrossRef]
- Tauran, Y.; Brioude, A.; Coleman, A.W.; Rhimi, M.; Kim, B. Molecular recognition by gold, silver and copper nanoparticles. World J. Biol. Chem. 2013, 4, 35–63. [Google Scholar] [CrossRef] [Green Version]
- Jazayeri, M.H.; Aghaie, T.; Avan, A.; Vatankhah, A.; Ghaffari, M.R.S. Colorimetric detection based on gold nano particles (GNPs): An easy, fast, inexpensive, low-cost and short time method in detection of analytes (protein, DNA, and ion). Sens. Bio-Sens. Res. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Tang, Z.; Takarada, T.; Maeda, M. Non-Cross-Linking Aggregation of DNA-Carrying Polymer Micelles Triggered by Duplex Formation. Langmuir 2018, 34, 14899–14910. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Yang, X. Aptamer-based colorimetric biosensing of dopamine using unmodified gold nanoparticles. Sens. Actuators B 2011, 156, 95–99. [Google Scholar] [CrossRef]
- Das, C.M.; Kong, K.V.; Yong, K.-T. Diagnostic plasmonic sensors: Opportunities and challenges. Chem. Commun. 2022, 58, 9573–9585. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Bhaskar, S.; Moronshing, M.; Srinivasan, V.; Badiya, P.K.; Subramaniam, C.; Ramamurthy, S.S. Silver Soret Nanoparticles for Femtomolar Sensing of Glutathione in a Surface Plasmon-Coupled Emission Platform. ACS Appl. Nano Mater. 2020, 3, 4329–4341. [Google Scholar] [CrossRef]
- Rai, A.; Bhaskar, S.; Genesh, K.M.; Ramamurthy, S.S. Engineering of coherent plasmon resonances from silver soret colloids, graphen oxide and Nd2O3 nanohybrid architectures studied in mobile phone-based surface plasmon-coupled emission platform. Mater. Lett. 2021, 304, 130632. [Google Scholar] [CrossRef]
- Rai, A.; Bhaskar, S.; Genesh, K.M.; Ramamurthy, S.S. Hottest Hotspot from the coldest cold: Welcome to Nano 4.0. ACS Appl. Nano Mater. 2022, 5, 12245–12264. [Google Scholar] [CrossRef]
- Jha, M.K.; Babu, B.; Parker, B.J.; Surendran, J.; Cameron, N.R.; Shaijumon, M.M.; Subramaniam, C. hierarchically Engineered Nanocarbon Florets as Bifunctional Electrode Materials for Adsorptive and Intercalative Energy Storage. ACS Appl. Mater. Interfaces 2020, 12, 42669–42677. [Google Scholar] [CrossRef]
- Shi, L.; Liu, M.; Zhang, L.; Tian, Y. A Liquid Interfacial SERS Platform on a Nanoparticle Array Stabilized by Rigid Probes for the Quantification of Norepinephrine in Rat Brain Microdialysates. Angew. Chem. Int. Ed. 2022, 61, e202117125. [Google Scholar] [CrossRef]
- Xu, L.-J.; Zong, C.; Zheng, X.-S.; Hu, P.; Feng, J.-M.; Ren, B. Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal. Chem. 2014, 86, 2238–2245. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, Y.; Okada, T. Quantification using statistical parameters derived from signal intensity distributions in surface enhanced Raman scattering (SERS). Anal. Chim. Acta 2021, 1181, 338931. [Google Scholar] [CrossRef]
- Fukunaga, Y.; Harada, M.; Okada, T. Surface-enhanced Raman scattering of DNA bases using frozen silver nanoparticle dispersion as a platform. Microchim. Acta 2021, 188, 406. [Google Scholar] [CrossRef]
- Li, T.; Wu, X.; Liu, F.; Li, N. Analytical methods based on the light-scattering of plasmonic nanoparticles at the single particle level with dark-field microscopy imaging. Analyst 2017, 142, 248–256. [Google Scholar] [CrossRef]
- Gao, P.F.; Lei, G.; Huang, C.Z. Dark-Field Microscopy: Recent Advances in Accurate Analysis and Emerging Applications. Anal. Chem. 2021, 93, 4707–4726. [Google Scholar] [CrossRef]
- Li, M.-X.; Zhao, W.; Wang, H.; Li, X.-L.; Xu, C.-H.; Chen, H.-Y.; Xu, J.-J. Dynamic Single Molecular Rulers: Toward Quantitative Detection of MicroRNA-21 in Living Cells. Anal. Chem. 2018, 90, 14255–14259. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, K.; Wei, W.; Liu, S. Single-Particle Assay of Poly(ADP-ribose) Polymerase-1 Activity with Dark-Field Optical Microscopy. ACS Sens. 2020, 5, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Dillen, A.; Mohrbacher, A.; Lammertyn, J. A Versatile One-Step Competitive Fiber Optic Surface Plasmon Resonance Bioassay Enabled by DNA Nanotechnology. ACS Sens. 2021, 6, 3677–3684. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lu, P.; Yu, C.; Feng, F.; Li, Q.; Zhan, J.; Xu, M.; Liu, Y.; Yao, L. Force-Coded Strategy for the Simultaneous Detection of Multiple Tumor-Related Proteins. Anal. Chem. 2022, 94, 8992–8998. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, A.; Oshiyama, K.; Nagatomo, S.; Nakatani, K. Semi-quantification of the binding constant based on bond breaking in a combined acoustic-gravitational field. Analyst 2022, 147, 4735–4738. [Google Scholar] [CrossRef]
- Miyagawa, A.; Oshiyama, K.; Nagatomo, S.; Nakatani, K. Zeptomole detection of DNA based on microparticle dissociation from a glass plate in a combined acoustic-gravitational field. Talanta 2022, 238, 123042. [Google Scholar] [CrossRef]
- Ashkin, A. Acceleration and Trapping of Particles by Radiation Pressure. Phys. Rev. Lett. 1970, 24, 156–159. [Google Scholar] [CrossRef] [Green Version]
- Laurell, T.; Petersson, F.; Nilsson, A. Chip integrated strategies for acoustic separation and manipulation of cells and particles. Chem. Soc. Rev. 2007, 36, 492–506. [Google Scholar] [CrossRef]
- Petersson, F.; Åberg, L.; Swärd-Nilsson, A.-M.; Laurell, T. Free Flow Acoustophoresis Microfluidic-Based Mode of Particle and Cell Separation. Anal. Chem. 2007, 79, 5117–5123. [Google Scholar] [CrossRef]
- Yavuz, C.T.; Prakash, A.; Mayo, J.T.; Colvin, V.L. Magnetic separations: From steel plants to biotechnology. Chem. Eng. Sci. 2009, 64, 2510–2521. [Google Scholar] [CrossRef]
- Iranmanesh, M.; Hulliger, J. Magnetic separation: Its application in mining, waste purification, medicine, biochemistry and chemistry. Chem. Soc. Rev. 2017, 46, 5925–5934. [Google Scholar] [CrossRef]
- Ge, S.; Whitesides, G.M. “Axial” Magnetic Levitation Using Ring Magnets Enables Simple Density-Based Analysis, Separation, and Manipulation. Anal. Chem. 2018, 90, 12239–12245. [Google Scholar] [CrossRef]
- Nemiroski, A.; Kumar, A.A.; Soh, S.; Harburg, D.V.; Yu, H.-D.; Whitesides, G.M. High-Sensitivity Measurement of Density by Magnetic Levitation. Anal. Chem. 2016, 88, 2666–2674. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Semenov, S.N.; Nagarkar, A.A.; Milette, J.; Christodouleas, D.C.; Yuan, L.; Whitesides, G.M. Magnetic Levitation To Characterize the Kinetics of Free-Radical Polymerization. J. Am. Chem. Soc. 2017, 139, 18688–18697. [Google Scholar] [CrossRef] [PubMed]
- Mirica, K.A.; Shevkoplyas, S.S.; Phillips, S.T.; Gupta, M.; Whitesides, G.M. Measuring Densities of Solids and Liquids Using Magnetic Levitation: Fundamentals. J. Am. Chem. Soc. 2009, 131, 10049–10058. [Google Scholar] [CrossRef] [PubMed]
- Bwambok, D.K.; Thuo, M.M.; Atkinson, M.B.; Mirica, K.A.; Shapiro, N.D.; Whitesides, G.M. Paramagnetic ionic liquids for measurements of density using magnetic levitation. Anal. Chem. 2013, 85, 8442–8447. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Nemiroski, A.; Mirica, K.A.; Mace, C.R.; Hennek, J.W.; Kumar, A.A.; Whitesides, G.M. Magnetic Levitation in Chemistry, Materials Science, and Biochemistry. Angew. Chem. Int. Ed. 2020, 59, 17810–17855. [Google Scholar] [CrossRef]
- Ozefe, F.; Yildiz, A.A. Smartphone-assisted Hepatitis C detection assay based on magnetic levitation. Analyst 2020, 145, 5816–5825. [Google Scholar] [CrossRef] [PubMed]
- Yaman, S.; Tekin, H.C. Magnetic Susceptibility-Based Protein Detection Using Magnetic Levitation. Anal. Chem. 2020, 92, 12556–12563. [Google Scholar] [CrossRef]
- Andersen, M.S.; Howard, E.; Lu, S.; Richard, M.; Gregory, M.; Ogembo, G.; Mazor, O.; Gorelik, P.; Shapiro, N.I.; Sharda, A.V.; et al. Detection of membrane-bound and soluble antigens by magnetic levitation. Lab Chip 2017, 17, 3462–3473. [Google Scholar] [CrossRef]
- Miyagawa, A. Acoustic Levitation-Based Trace-Level Biosensing: Design of Detection Systems and Applications to Real Samples; Springer Singapore Pre Ltd.: Singapore, 2021. [Google Scholar] [CrossRef]
- Miyagawa, A.; Okada, Y.; Okada, T. Aptamer-Based Sensing of Small Organic Molecules by Measuring Levitation Coordinate of Single Microsphere in Combined Acoustic-Gravitational Field. ACS Omega 2020, 5, 3542–3549. [Google Scholar] [CrossRef]
- Miyagawa, A.; Harada, M.; Okada, T. Multiple MicroRNA Quantification Based on Acoustic Levitation of Single Microspheres after One-Pot Sandwich Interparticle Hybridizations. Anal. Chem. 2018, 90, 13729–13735. [Google Scholar] [CrossRef]
- Miyagawa, A.; Harada, M.; Okada, T. Zeptomole Biosensing of DNA with Flexible Selectivity Based on Acoustic Levitation of a Single Microsphere Binding Gold Nanoparticles by Hybridization. ACS Sens. 2018, 3, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, A.; Harada, M.; Okada, T. Zeptomole Detection Scheme Based on Levitation Coordinate Measurements of a Single Microparticle in a Coupled Acoustic-Gravitational Field. Anal. Chem. 2018, 90, 2310–2316. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, A.; Inoue, Y.; Harada, M.; Okada, T. Acoustic Sensing Based on Density Shift of Microspheres by Surface Binding of Gold Nanoparticles. Anal. Sci. 2017, 33, 939–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Lu, J. A Review on Particle Size Effect in Metal-Catalyzed Heterogeneous Reactions. Chin. J. Chem. 2020, 38, 1422–1444. [Google Scholar] [CrossRef]
- Le Goff, G.C.; Srinivas, R.L.; Hill, W.A.; Doyle, P.S. Hydrogel microparticles for biosensing. Eur. Polym. J. 2015, 72, 386–412. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, A.; Haag, R.; Schedler, U. Hydrogels and Their Role in Biosensing Applications. Adv. Healthc. Mater. 2021, 10, 2100062. [Google Scholar] [CrossRef]
- Ong, C.-B.; Annuar, M.S.M.A. Hydrogels Responsive Towards Important Biological-Based Stimuli. Polym. Sci. B 2022, 64, 271–286. [Google Scholar] [CrossRef]
- Culver, H.R.; Clegg, J.R.; Peppas, N.A. Analyte-Responsive Hydrogels: Intelligent Materials for Biosensing and Drug Delivery. Acc. Chem. Res. 2017, 50, 170–178. [Google Scholar] [CrossRef]
- Park, H.-I.; Park, S.-Y. Smart Fluorescent Hydrogel Glucose Biosensing Microdroplets with Dual-Mode Fluorescence Quenching and Size Reduction. ACS Appl. Mater. Interfaces 2018, 10, 30172–30179. [Google Scholar] [CrossRef]
- Krisch, E.; Gyarmati, B.; Barczikai, D.; Lapeyre, V.; Szilágyi, B.Á.; Ravaine, V.; Szilágyi, A. Poly(aspartic acid) hydrogels showing reversible volume change upon redox stimulus. Eur. Polym. J. 2018, 105, 459–468. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Yu, C.-N.; Zheng, M.; Kim, H.; Eggleston, M.S. Microparticle-Based Biochemical Sensing Using Optical Coherence Tomography and Deep Learning. ACS Nano 2021, 15, 9764–9774. [Google Scholar] [CrossRef] [PubMed]
- Mortelmans, T.; Kazazis, D.; Padeste, C.; Berger, P.; Li, X.; Ekinci, Y. Poly(methyl methacrylate)-Based Nanofluidic Device for Rapid and Multiplexed Serological Antibody Detection of SARS-CoV-2. ACS Appl. Nano Mater. 2022, 5, 517–526. [Google Scholar] [CrossRef]
- He, H.; Nie, R.; Lu, P.; Peng, X.; Li, X.; Chen, Y. Low-Cost and Convenient Microchannel Resistance Biosensing Platform by Directly Translating Biorecognition into a Current Signal. Anal. Chem. 2021, 93, 15049–15057. [Google Scholar] [CrossRef]
- Einstein, A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905, 322, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Clayton, K.N.; Berglund, G.D.; Linnes, J.C.; Kinzer-Ursem, T.L.; Wereley, S.T. DNA Microviscosity Characterization with Particle Diffusometry for Downstream DNA Detection Applications. Anal. Chem. 2017, 89, 13334–13341. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wereley, S.T.; Linnes, J.C.; Kinzer-Ursem, T.L. Measurement of Protein-Protein Interaction Dynamics Using Microfluidics and Particle Diffusometry. Anal. Chem. 2022, 94, 15655–15662. [Google Scholar] [CrossRef]
- Das, D.; Chen, W.-L.; Chuang, H.-S. Rapid and Sensitive Pathogen Detection by DNA Amplification Using Janus Particle-Enabled Rotational Diffusometry. Anal. Chem. 2021, 93, 13945–13951. [Google Scholar] [CrossRef]
- Chen, W.-L.; Chuang, H.-S. Trace Biomolecule Detection with Functionalized Janus Particles by Rotational Diffusion. Anal. Chem. 2020, 92, 12996–13003. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Vermaas, R.; Yan, J.; de Jong, A.M.; Prins, M.W.J. Click-Coupling to Electrostatically Grafted Polymers Greatly Improves the Stability of a Continuous Monitoring Sensor with Single-Molecule Resolution. ACS Sens. 2021, 6, 1980–1986. [Google Scholar] [CrossRef]
- Yan, J.; van Smeden, L.; Merkx, M.; Zijlstra, P.; Prins, M.W.J. Continuous Small-Molecule Monitoring with a Digital Single-Particle Switch. ACS Sens. 2020, 5, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.E.; Spoelstra, L.R.; Berendsen, J.T.W.; Loessberg-Zahl, J.T.; Eijkel, J.C.T.; Segerink, L.I. A Multiplexable Plasmonic Hairpin-DNA Sensor Based On Target-specific Tether Dynamics. ACS Sens. 2021, 6, 4297–4303. [Google Scholar] [CrossRef] [PubMed]
- Kremser, L.; Blaas, D.; Kenndler, E. Capillary electrophoresis of biological particles: Viruses, bacteria, and eukaryotic cells. Electrophoresis 2004, 25, 2282–2291. [Google Scholar] [CrossRef]
- Østergaard, J.; Heegaard, N.H. Capillary electrophoresis frontal analysis: Principles and applications for the study of drug-plasma protein binding. Electrophoresis 2003, 24, 2903–2913. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Shan, X.; Wang, S.; Tao, N. Quantifying Ligand-Protein Binding Kinetics with Self-Assembled Nano-oscillators. Anal. Chem. 2019, 91, 14149–14156. [Google Scholar] [CrossRef]
- Ma, G.; Wan, Z.; Zhu, H.; Tao, N. Roles of entropic and solvent damping forces in the dynamics of polymer tethered nanoparticles and implications for single molecule sensing. Chem. Sci. 2020, 11, 1283–1289. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.-L.; Yang, Y.; Wang, S.; Liu, X.-W. Surface Plasmon Resonance Microscopy: From Single-Molecule Sensing to Single-Cell Imaging. Angew. Chem. Int. Ed. 2020, 59, 1776–1785. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, G.; Dong, W.; Wan, Z.; Wang, S.; Tao, N. Plasmonic scattering imaging of single proteins and binding kinetics. Nat. Methods 2020, 17, 1010–1017. [Google Scholar] [CrossRef]
- Li, Z.; Huang, X.; Liu, H.; Luo, F.; Qiu, B.; Lin, Z.; Chen, H. Electrochemiluminescence Biosensor for Hyaluronidase Based on the Adjustable Electrostatic Interaction between the Surface-Charge-Controllable Nanoparticles and Negatively Charged Electrode. ACS Sens. 2022, 7, 2012–2019. [Google Scholar] [CrossRef]
- Brown, M.A.; Bossa, G.V.; May, S. Emergence of a Stern Layer from the Incorporation of Hydration Interactions into the Gouy-Chapman Model of the Electrical Double Layer. Langmuir 2015, 31, 11477–11483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, M.; Nymeyer, H.; Zhou, H.-X. Test of the Gouy-Chapman theory for a charged lipid membrane against explicit-solvent molecular dynamics simulations. Phys. Rev. Lett. 2008, 101, 038103. [Google Scholar] [CrossRef]
- Hagiya, K.; Miyagawa, A.; Nagatomo, S.; Nakatani, K. Direct Quantification of Proteins Modified on a Polystyrene Microparticle Surface Based on zeta Potential Change. Anal. Chem. 2022, 94, 6304–6310. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, S.; Kambhampati, N.A.V.; Ganesh, K.M.; Sharma, M.; Srinivasan, V.; Ramamurthy, S.S. Metal-Free Graphene Pxide-Based Tunable Soliton and Plasmon Engineering for Biosensing Applications. ACS Appl. Mater. Interfaces 2021, 13, 17046–17061. [Google Scholar] [CrossRef] [PubMed]
- Venugopalan, P.; Kumar, S. Highly Sensitive Plasmonic Sensor with Au Bow Tie Nanoantennas on SiO2 Nanopillar Arrays. Chemosensors 2023, 11, 121. [Google Scholar] [CrossRef]
- Bhaskar, S.; Das, P.; Srinivasan, V.; Bhaktha, S.B.N.; Ramamurthy, S.S. Plasmonic-Silver Sorets and Dielectric Nd2O3 nanorods for Ultrasensitive Photonic Crystal-Coupled Emission. Mater. Res. Bull. 2022, 145, 111558. [Google Scholar] [CrossRef]
- Bhaskar, S.; Srinivasan, V.; Ramamurthy, S.S. Nd2O3-Ag Nanostructures for Plasmonic Biosensing, Antimicrobial, and Anticancer Applications. ACS Appl. Nano Mater. 2023, 6, 1129–1145. [Google Scholar] [CrossRef]









| Particle Behavior | Particle Type | Detection Method | Target | Ref |
|---|---|---|---|---|
| Aggregation | AuNP | SERS | Norepinephrine | [44] |
| AuNP | SERS | Lysozyme, avidin, BSA, cytochrome c, and hemoglobin | [45] | |
| AgNP | SERS | DNA bases | [46,47] | |
| AuNP | LSPR wavelength | MicroRNA-21 | [50] | |
| AuNP | LSPR wavelength | Polymerase activity | [51] | |
| Dissociation | AuNP | SPR wavelength | ATP, thrombin, and single strand DNA (ssDNA) | [52] |
| Magnetic microparticle | Optical observation | Mucin-1 glycoprotein and human epidermal growth factor receptor | [53] | |
| Silica and polystyrene (PS) microparticles and AuNPs | Optical observation | Double-strand DNA (dsDNA) | [54,55] | |
| Levitation | PS microparticle | Optical observation | HCV | [67] |
| PS microparticle and magnetic nanoparticle | Optical and fluorescence observation | BSA and mouse immunoglobulin G | [68] | |
| Polymethylmethacrylate (PMMA) microparticle | Optical observation | T-cell antigen CD3, eosinophil antigen Siglec-8, red blood cell antigens CD35 and RhD, red blood cell-bound Ep-stein-Barr viral particle, and soluble IL-6 | [69] | |
| PS and PMMA microparticles and AuNP | Optical observation | Avidin–biotin complex, ssDNA, dsDNA, microRNA-21 and microRNA-122, ATP, dopamine, and ampicillin | [72,73,74,75] | |
| Size | Poly(acrylic acid) hydrogel | Optical observation and fluorescence intensity | Glucose | [81] |
| Poly(aspartic acid) hydrogel | Optical observation | Glucose | [82] | |
| Poly(acrylamide) hydrogel | Optical coherent tomography | Glucose | [83] | |
| Magnetic nanoparticle | Fluorescence observation | SARS-CoV-2 and influenza A | [84] | |
| PS and magnetic microparticle | Electric resistance | Procalcitonin, chlorpyrifos, and L. monocytogenes | [85] | |
| Motion | PS nanoparticle | Fluorescence observation | S. aureus and K. Pneumoniae | [86] |
| Polymer nanoparticle | Fluorescence observation | Avidin–biotin complex | [88] | |
| Janus microparticle and PS nanoparticle | Fluorescence observation | DNA and tumor necrosis factor-α cytokine target | [89,90] | |
| Magnetic microparticle | Optical observation | ssDNA and creatinine | [91,92] | |
| AuNP | Plasmon imaging | ssDNA | [93] | |
| Surface charge | Silica microparticle | Plasmon imaging | BSA, nanodisc encapsulated membrane protein KcsA-Kv1.3 | [96,97] |
| Silica nanoparticle | Electrochemiluminescence | Hyaluronidase | [100] | |
| PS microparticle | Zeta potential | BSA, myoglobin, Lysozyme | [103] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyagawa, A.; Okada, T. Biosensing Strategies Based on Particle Behavior. Chemosensors 2023, 11, 172. https://doi.org/10.3390/chemosensors11030172
Miyagawa A, Okada T. Biosensing Strategies Based on Particle Behavior. Chemosensors. 2023; 11(3):172. https://doi.org/10.3390/chemosensors11030172
Chicago/Turabian StyleMiyagawa, Akihisa, and Tetsuo Okada. 2023. "Biosensing Strategies Based on Particle Behavior" Chemosensors 11, no. 3: 172. https://doi.org/10.3390/chemosensors11030172
APA StyleMiyagawa, A., & Okada, T. (2023). Biosensing Strategies Based on Particle Behavior. Chemosensors, 11(3), 172. https://doi.org/10.3390/chemosensors11030172