Automatic Analysis of Isothermal Amplification via Impedance Time-Constant-Domain Spectroscopy: A SARS-CoV-2 Case Study
Abstract
1. Introduction
2. Theoretical Background
2.1. Electrochemical Impedance
2.2. Distribution of Relaxation Times Model
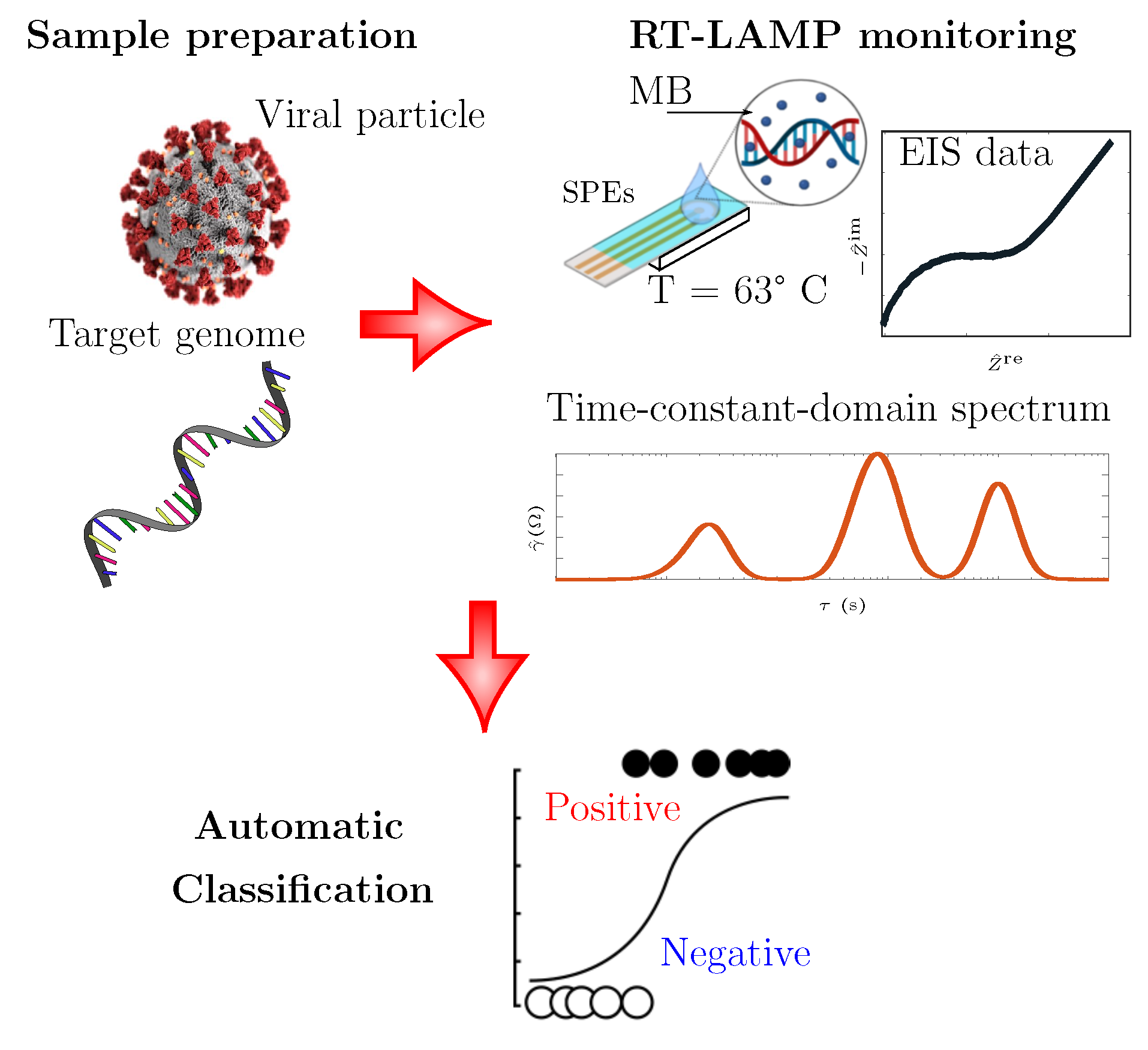
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Sample Collection and Preparation
3.3. RT-LAMP Assay
3.4. Electrochemical Impedance Measurements
3.5. Time-Constant-Domain Spectroscopy
3.6. Classification Algorithm
4. Results
4.1. Performance to Detect the SARS-CoV-2 Genome
4.1.1. Impedance Measurements of RT-LAMP Reactions
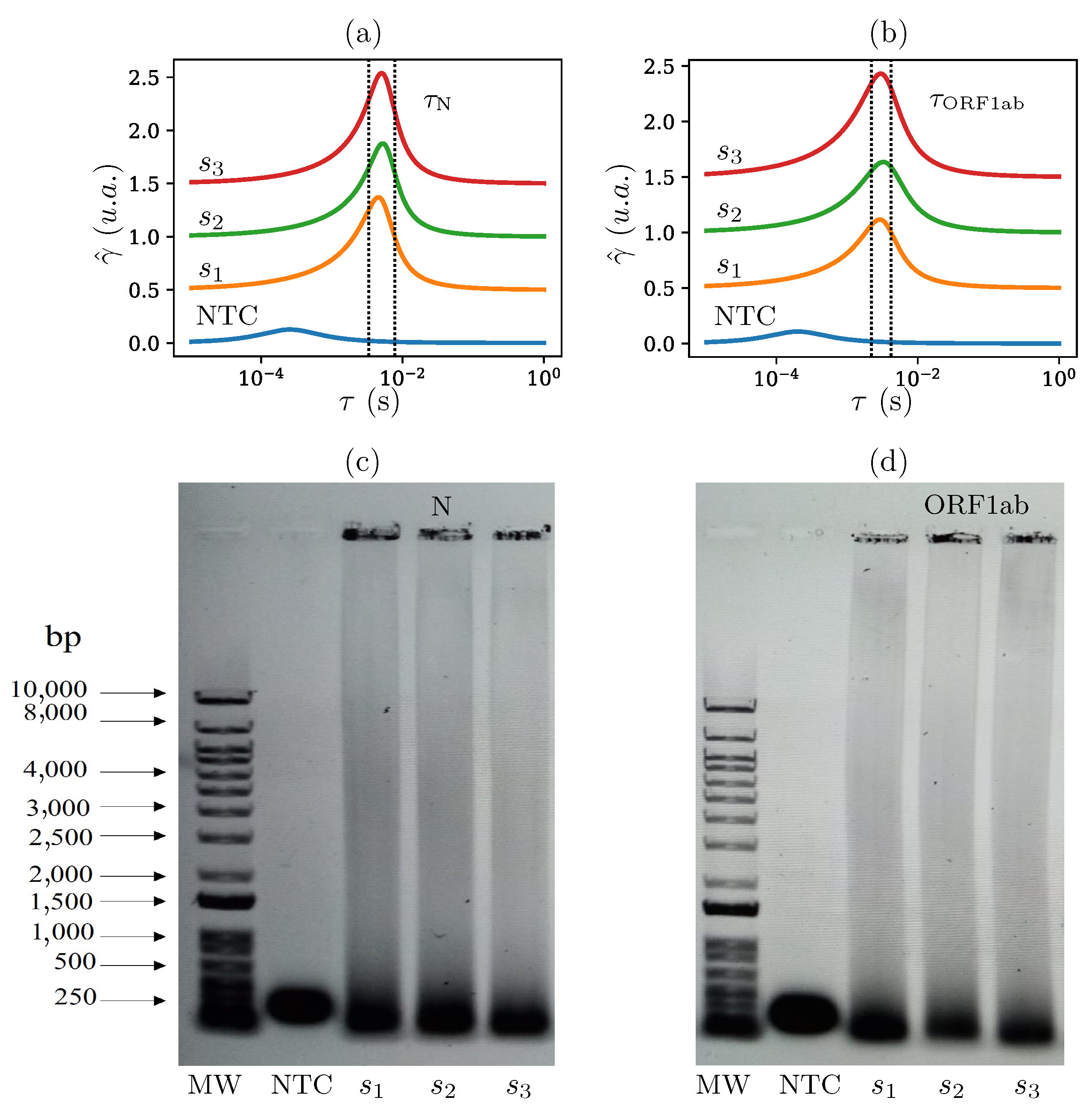
4.1.2. Time-Constant-Domain Spectroscopy of RT-LAMP Reactions
4.2. Detecting SARS-CoV-2 Genome in Wastewater Samples
4.3. Automatic Classification of Samples
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 Variants—Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med. 2021, 384, 1866–1868. [Google Scholar] [CrossRef] [PubMed]
- Alcoba-Florez, J.; Gil-Campesino, H.; de Artola, D.G.M.; González-Montelongo, R.; Valenzuela-Fernández, A.; Ciuffreda, L.; Flores, C. Sensitivity of different RT-qPCR solutions for SARS-CoV-2 detection. Int. J. Infect. Dis. 2020, 99, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Sil, B.K.; Jahan, N.; Haq, M.A.; Oishee, M.J.; Ali, T.; Khandker, S.S.; Kobatake, E.; Mie, M.; Khondoker, M.U.; Jamiruddin, M.R.; et al. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS ONE 2021, 16, 1–16. [Google Scholar] [CrossRef]
- Hao, Z.; Chen, H.; Shi, X.; Tan, W.; Zhu, G. Fabrication for paper-based microfluidic analytical devices and saliva analysis application. Microfluid. Nanofluidics 2021, 25, 80. [Google Scholar] [CrossRef]
- Kumblathan, T.; Liu, Y.; Uppal, G.K.; Hrudey, S.E.; Li, X.F. Wastewater-Based Epidemiology for Community Monitoring of SARS-CoV-2: Progress and Challenges. ACS Environ. Au 2021, 1, 18–31. [Google Scholar] [CrossRef]
- Shrestha, S.; Yoshinaga, E.; Chapagain, S.K.; Mohan, G.; Gasparatos, A.; Fukushi, K. Wastewater-Based Epidemiology for Cost-Effective Mass Surveillance of COVID-19 in Low- and Middle-Income Countries: Challenges and Opportunities. Water 2021, 13, 2897. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Ongerth, J.E.; Danielson, R.E. RT qLAMP—Direct Detection of SARS-CoV-2 in Raw Sewage. J. Biomol. Tech. 2021, 32, 206–213. [Google Scholar] [CrossRef]
- Rezaei, M.; Razavi Bazaz, S.; Morshedi Rad, D.; Shimoni, O.; Jin, D.; Rawlinson, W.; Ebrahimi Warkiani, M. A Portable RT-LAMP/CRISPR Machine for Rapid COVID-19 Screening. Biosensors 2021, 11, 369. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- Naikoo, G.A.; Awan, T.; Hassan, I.U.; Salim, H.; Arshad, F.; Ahmed, W.; Asiri, A.M.; Qurashi, A. Nanomaterials-Based Sensors for Respiratory Viral Detection: A Review. IEEE Sens. J. 2021, 21, 17643–17656. [Google Scholar] [CrossRef]
- González-González, E.; Lara-Mayorga, I.M.; Rodríguez-Sánchez, I.P.; Zhang, Y.S.; Martínez-Chapa, S.O.; Santiago, G.T.d.; Alvarez, M.M. Colorimetric loop-mediated isothermal amplification (LAMP) for cost-effective and quantitative detection of SARS-CoV-2: The change in color in LAMP-based assays quantitatively correlates with viral copy number. Anal. Methods 2021, 13, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Serna, B.E.; Ramírez-Chavarría, R.G.; Castillo-Villanueva, E.; Carrillo-Reyes, J.; Ramírez-Zamora, R.M.; Buitrón, G.; Alvarez-Icaza, L. Label-free and portable field-effect sensor for monitoring RT-LAMP products to detect SARS-CoV-2 in wastewater. Talanta 2023, 253, 124060. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Ramanujam, A.; Neyhouse, B.; Keogh, R.A.; Muthuvel, M.; Carroll, R.K.; Botte, G.G. Rapid electrochemical detection of Escherichia coli using nickel oxidation reaction on a rotating disk electrode. Chem. Eng. J. 2021, 411, 128453. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Chaudhary, N.; Kim, H.; Jang, J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta 2019, 1064, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Electrochemical impedance-based DNA sensor using pyrrolidinyl peptide nucleic acids for tuberculosis detection. Anal. Chim. Acta 2018, 1044, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.W.; Yang, Z.W.; Liu, X.L.; Zhang, L.W.; Guo, W.B.; Zhang, J.; Ding, L.H. 2D Co metal-organic framework nanosheet as an oxidase-like nanozyme for sensitive biomolecule monitoring. Rare Met. 2023, 42, 797–805. [Google Scholar] [CrossRef]
- Ding, L.; Yan, F.; Zhang, Y.; Liu, L.; Yu, X.; Liu, H. Microflowers Comprised of Cu/CuxO/NC Nanosheets as Electrocatalysts and Horseradish Peroxidase Mimics. ACS Appl. Nano Mater. 2020, 3, 617–623. [Google Scholar] [CrossRef]
- Ding, L.; Zhang, L.; Yang, H.; Liu, H.; Ge, S.; Yu, J. Electrochemical biosensor for p53 gene based on HRP-mimicking DNAzyme-catalyzed deposition of polyaniline coupled with hybridization chain reaction. Sens. Actuators B Chem. 2018, 268, 210–216. [Google Scholar] [CrossRef]
- Strong, M.E.; Richards, J.R.; Torres, M.; Beck, C.M.; La Belle, J.T. Faradaic electrochemical impedance spectroscopy for enhanced analyte detection in diagnostics. Biosens. Bioelectron. 2021, 177, 112949. [Google Scholar] [CrossRef] [PubMed]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical Impedance Spectroscopy (EIS): Principles, Construction, and Biosensing Applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef] [PubMed]
- Stupin, D.D.; Kuzina, E.A.; Abelit, A.A.; Emelyanov, A.K.; Nikolaev, D.M.; Ryazantsev, M.N.; Koniakhin, S.V.; Dubina, M.V. Bioimpedance Spectroscopy: Basics and Applications. ACS Biomater. Sci. Eng. 2021, 7, 1962–1986. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, C.; Mei, W.; Guo, M.; Yang, Y. Equivalent circuit models for a biomembrane impedance sensor and analysis of electrochemical impedance spectra based on support vector regression. Med. Biol. Eng. Comput. 2019, 57, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Van Haeverbeke, M.; Stock, M.; De Baets, B. Practical Equivalent Electrical Circuit Identification for Electrochemical Impedance Spectroscopy Analysis With Gene Expression Programming. IEEE Trans. Instrum. Meas. 2021, 70, 1–12. [Google Scholar] [CrossRef]
- Kežionis, A.; Kazakevičius, E. Some features of the analysis of broadband impedance data using distribution of relaxation times. Electrochim. Acta 2020, 349, 136379. [Google Scholar] [CrossRef]
- Li, X.; Ahmadi, M.; Collins, L.; Kalinin, S.V. Deconvolving distribution of relaxation times, resistances and inductance from electrochemical impedance spectroscopy via statistical model selection: Exploiting structural-sparsity regularization and data-driven parameter tuning. Electrochim. Acta 2019, 313, 570–583. [Google Scholar] [CrossRef]
- Saccoccio, M.; Wan, T.H.; Chen, C.; Ciucci, F. Optimal regularization in distribution of relaxation times applied to electrochemical impedance spectroscopy: Ridge and Lasso regression methods—A theoretical and experimental Study. Electrochim. Acta 2014, 147, 470–482. [Google Scholar] [CrossRef]
- Boukamp, B.A. Fourier transform distribution function of relaxation times; application and limitations. Electrochim. Acta 2015, 154, 35–46. [Google Scholar] [CrossRef]
- Ciucci, F.; Chen, C. Analysis of electrochemical impedance spectroscopy data using the distribution of relaxation times: A Bayesian and hierarchical Bayesian approach. Electrochim. Acta 2015, 167, 439–454. [Google Scholar] [CrossRef]
- Liu, J.; Ciucci, F. The Deep-Prior Distribution of Relaxation Times. J. Electrochem. Soc. 2020, 167, 026506. [Google Scholar] [CrossRef]
- Maradesa, A.; Py, B.; Quattrocchi, E.; Ciucci, F. The probabilistic deconvolution of the distribution of relaxation times with finite Gaussian processes. Electrochim. Acta 2022, 413, 140119. [Google Scholar] [CrossRef]
- Ramírez-Chavarría, R.; Sánchez-Pérez, C.; Matatagui, D.; Qureshi, N.; Pérez-García, A.; Hernández-Ruíz, J. Ex-vivo biological tissue differentiation by the distribution of relaxation times method applied to electrical impedance spectroscopy. Electrochim. Acta 2018, 276, 214–222. [Google Scholar] [CrossRef]
- Shi, F.; Kolb, J.F. Enhanced resolution impedimetric analysis of cell responses from the distribution of relaxation times. Biosens. Bioelectron. 2020, 157, 112149. [Google Scholar] [CrossRef]
- Ramírez-Chavarría, R.G.; Sánchez-Pérez, C.; Romero-Ornelas, L.; Ramón-Gallegos, E. Time-Constant-Domain Spectroscopy: An Impedance-Based Method for Sensing Biological Cells in Suspension. IEEE Sens. J. 2021, 21, 185–192. [Google Scholar] [CrossRef]
- Saxena, R.; Srivastava, S. An insight into impedimetric immunosensor and its electrical equivalent circuit. Sens. Actuators B Chem. 2019, 297, 126780. [Google Scholar] [CrossRef]
- Vivier, V.; Orazem, M.E. Impedance Analysis of Electrochemical Systems. Chem. Rev. 2022, 122, 11131–11168. [Google Scholar] [CrossRef]
- Carrillo-Reyes, J.; Barragán-Trinidad, M.; Buitrón, G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process. Eng. 2021, 40, 101815. [Google Scholar] [CrossRef]
- Song, J.; El-Tholoth, M.; Li, Y.; Graham-Wooten, J.; Liang, Y.; Li, J.; Li, W.; Weiss, S.R.; Collman, R.G.; Bau, H.H. Single- and Two-Stage, Closed-Tube, Point-of-Care, Molecular Detection of SARS-CoV-2. Anal. Chem. 2021, 93, 13063–13071. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Chavarría, R.; Quintana-Carapia, G.; Müller, M.I.; Mattila, R.; Matatagui, D.; Sánchez-Pérez, C. Bioimpedance Parameter Estimation using Fast Spectral Measurements and Regularization. IFAC-PapersOnLine 2018, 51, 521–526. [Google Scholar] [CrossRef]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar] [CrossRef]
- Tang, Z.; Cui, J.; Kshirsagar, A.; Liu, T.; Yon, M.; Kuchipudi, S.V.; Guan, W. SLIDE: Saliva-Based SARS-CoV-2 Self-Testing with RT-LAMP in a Mobile Device. ACS Sens. 2022, 7, 2370–2378. [Google Scholar] [CrossRef] [PubMed]
- Nasir, N.; Kansal, A.; Alshaltone, O.; Barneih, F.; Sameer, M.; Shanableh, A.; Al-Shamma’a, A. Water quality classification using machine learning algorithms. J. Water Process. Eng. 2022, 48, 102920. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Chavarría, R.G.; Castillo-Villanueva, E.; Alvarez-Serna, B.E.; Carrillo-Reyes, J.; Torres, L.; Ramírez-Zamora, R.M.; Buitrón, G.; Alvarez-Icaza, L. Automatic Analysis of Isothermal Amplification via Impedance Time-Constant-Domain Spectroscopy: A SARS-CoV-2 Case Study. Chemosensors 2023, 11, 230. https://doi.org/10.3390/chemosensors11040230
Ramírez-Chavarría RG, Castillo-Villanueva E, Alvarez-Serna BE, Carrillo-Reyes J, Torres L, Ramírez-Zamora RM, Buitrón G, Alvarez-Icaza L. Automatic Analysis of Isothermal Amplification via Impedance Time-Constant-Domain Spectroscopy: A SARS-CoV-2 Case Study. Chemosensors. 2023; 11(4):230. https://doi.org/10.3390/chemosensors11040230
Chicago/Turabian StyleRamírez-Chavarría, Roberto G., Elizabeth Castillo-Villanueva, Bryan E. Alvarez-Serna, Julián Carrillo-Reyes, Lizeth Torres, Rosa María Ramírez-Zamora, Germán Buitrón, and Luis Alvarez-Icaza. 2023. "Automatic Analysis of Isothermal Amplification via Impedance Time-Constant-Domain Spectroscopy: A SARS-CoV-2 Case Study" Chemosensors 11, no. 4: 230. https://doi.org/10.3390/chemosensors11040230
APA StyleRamírez-Chavarría, R. G., Castillo-Villanueva, E., Alvarez-Serna, B. E., Carrillo-Reyes, J., Torres, L., Ramírez-Zamora, R. M., Buitrón, G., & Alvarez-Icaza, L. (2023). Automatic Analysis of Isothermal Amplification via Impedance Time-Constant-Domain Spectroscopy: A SARS-CoV-2 Case Study. Chemosensors, 11(4), 230. https://doi.org/10.3390/chemosensors11040230





