Development of ANN Models for Prediction of Physical and Chemical Characteristics of Oil-in-Aqueous Plant Extract Emulsions Using Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Plants, Sunflower Oil, and Pea Protein Powder
2.1.2. Chemicals
2.2. Methods
2.2.1. Solid-Liquid Extraction Procedure
2.2.2. Pea Protein Dispersion in Aqueous Plant Extracts
2.2.3. Preparation of Oil-in-Water Emulsions Containing Oregano/Rosemary Extracts
2.2.4. Dry Matter Content Measurement
2.2.5. Zeta Potential Measurement
2.2.6. The Average Feret Droplet Diameter Measurement
2.2.7. Measurement of Total Polyphenolic Content and Antioxidant Activity of the Prepared Emulsions
2.2.8. Near-Infrared Spectroscopy (NIR)
2.2.9. NIR Data Preprocessing
2.2.10. Artificial Neural Network (ANN) Modeling
3. Results and Discussion
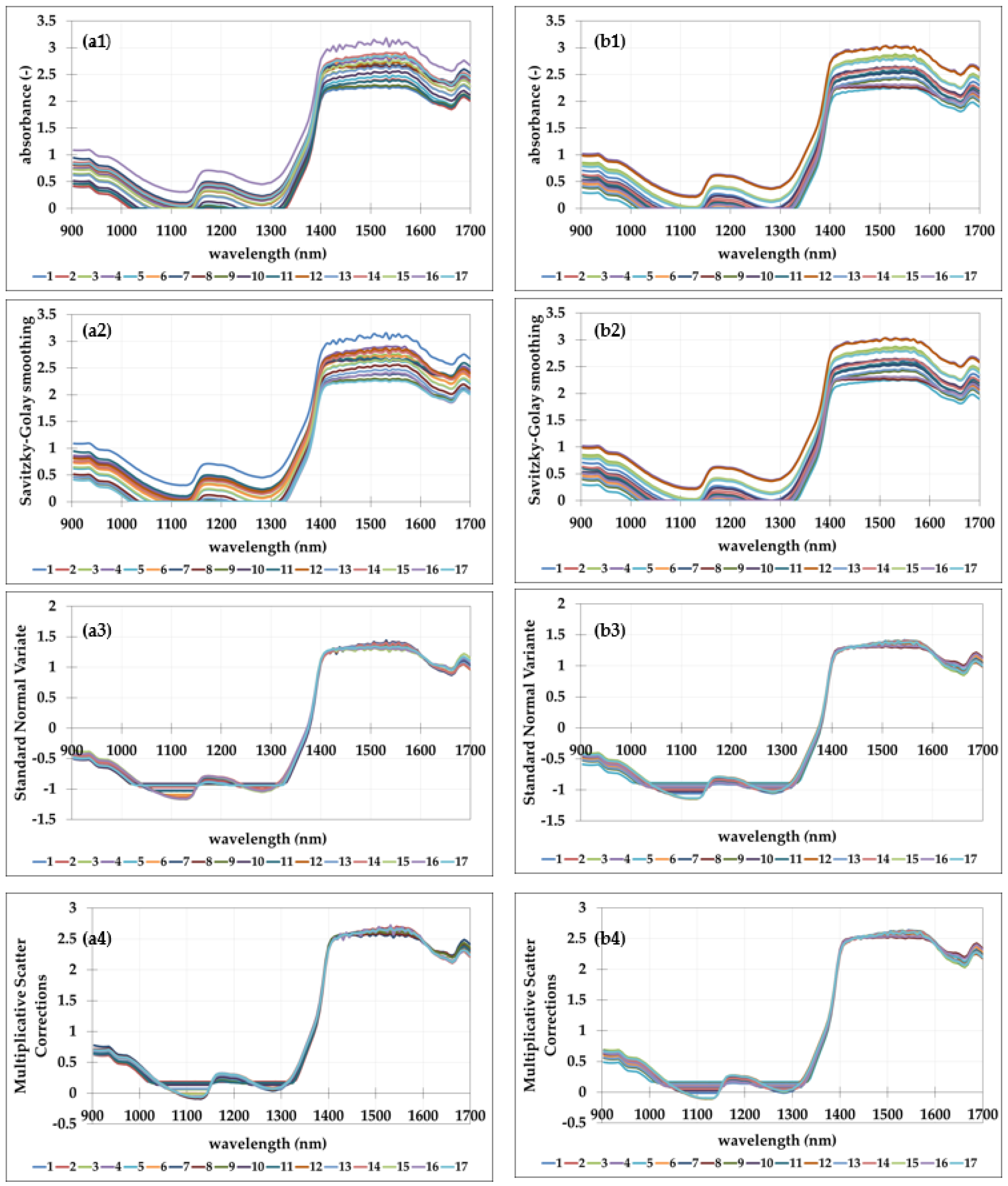
3.1. NIR Spectra of the Prepared Emulsions
3.2. ANN Modeling of Oil-in-Aqueous Oregano/Rosemary Extract Emulsions Selected Physical and Chemical Properties Based on NIR Spectra
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chakrabartty, I.; Mohanta, Y.K.; Nongbet, A.; Mohanta, T.K.; Mahanta, S.; Das, N.; Saravanan, M.; Sharma, N. Exploration of Lamiaceae in cardio vascular diseases and functional foods: Medicine as food and food as medicine. Front. Pharmacol. 2022, 13, 2041. [Google Scholar] [CrossRef] [PubMed]
- Michel, J.; Abd Rani, N.Z.; Husain, K. A Review on the potential use of medicinal plants from Asteraceae and Lamiaceae plant family in cardiovascular diseases. Front. Pharmacol. 2020, 11, 852. [Google Scholar] [CrossRef]
- Hossain, M.B.; Barry-Ryan, C.; Martin-Diana, A.B.; Brunton, N.P. Effect of drying method on the antioxidant capacity of six Lamiaceae herbs. Food Chem. 2010, 123, 85–91. [Google Scholar] [CrossRef]
- Munin, A.; Edwards-Lévy, F. Encapsulation of natural polyphenolic compounds: A review. Pharmaceutics 2011, 3, 793–829. [Google Scholar] [CrossRef]
- Dawurung, C.J.; Nguyen, M.T.H.; Pengon, J.; Dokladda, K.; Bunyong, R.; Rattanajak, R.; Kamchonwongpaisan, S.; Nguyen, P.T.M.; Pyne, S.G. Isolation of bioactive compounds from medicinal plants used in traditional medicine: Rautandiol B, a potential lead compound against Plasmodium falciparum. BMC Complement Med. Ther. 2021, 21, 231. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Bhandari, B. Encapsulation of polyphenols–A review. Trends Food Sci. Technol. 2010, 21, 510–523. [Google Scholar] [CrossRef]
- Benita, S. Microencapsulation: Methods and Industrial Applications, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Shanthi, C.N.; Gupta, D.; Kumar Mahato, A. Traditional and emerging applications of microspheres: A Review. Int. J. Pharm Tech. Res. 2010, 2, 675–681. [Google Scholar]
- Lu, W.; Kelly, A.L.; Miao, S. Emulsion-based encapsulation and delivery systems for polyphenols. Trends Food Sci. Technol. 2016, 47, 1–9. [Google Scholar] [CrossRef]
- McClements, D.J.; Li, Y. Structured emulsion-based delivery systems: Controlling the digestion and release of lipophilic food components. Adv. Colloid Interface Sci. 2010, 159, 213–228. [Google Scholar] [CrossRef]
- Tan, C.; McClements, D.J. Application of advanced emulsion technology in the food industry: A review and critical evaluation. Foods 2021, 10, 812. [Google Scholar] [CrossRef]
- Sirovec, S.; Jurinjak Tušek, A.; Benković, M.; Valinger, D.; Sokač Cvetinić, T.; Gajdoš Kljusurić, J.; Jurina, T. Emulsification of rosemary and oregano aqueous extracts and their in vitro bioavailability. Plants 2022, 11, 3372. [Google Scholar] [CrossRef] [PubMed]
- Kutzli, I.; Griener, D.; Gibis, M.; Grossmann, L.; Baier, S.K.; Weiss, J. Improvement of emulsifying behavior of pea proteins as plant-based emulsifiers via Maillard-induced glycation in electrospun pea protein–maltodextrin fibers. Food Funct. 2020, 11, 4049–4056. [Google Scholar] [CrossRef]
- Östbring, K.; Nilsson, K.; Ahlström, C.; Fridolfsson, A.; Rayner, M. Emulsifying and anti-oxidative properties of proteins extracted from industrially cold-pressed rapeseed press-cake. Foods 2020, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Araujo, A.M.; Santos, L.M.; Fortuny, M.; Melo, R.L.F.V.; Coutinho, R.C.C.; Santos, A.F. Evaluation of water content and average droplet size in water-in-crude oil emulsions by means of near-infrared spectroscopy. Energy Fuels 2008, 22, 3450–3458. [Google Scholar] [CrossRef]
- Pedro, A.M.K.; Ferreira, M.M.C. The use of near-infrared spectroscopy and chemometrics for determining the shelf-life of products. Appl. Spectrosc. 2009, 63, 1308–1314. [Google Scholar] [CrossRef]
- Borges, G.R.; Farias, G.B.; Braz, T.M.; Santos, L.M.; Amaral, M.J.; Fortuny, M.; Franceschi, E.; Dariva, C.; Santos, A.F. Use of near infrared for evaluation of droplet size distribution and water content in water-in-crude oil emulsions in pressurized pipeline. Fuel 2015, 147, 43–52. [Google Scholar] [CrossRef]
- Dinache, A.; Tozar, T.; Smarandache, A.; Andrei, I.R.; Nistorescu, S.; Nastasa, V.; Staicu, A.; Pascu, M.L.; Romanitan, M.O. Spectroscopic characterization of emulsions generated with a new laser-assisted device. Molecules 2020, 25, 1729. [Google Scholar] [CrossRef]
- Grgić, F.; Jurina, T.; Valinger, D.; Gajdoš Kljusurić, J.; Jurinjak Tušek, A.; Benković, M. Near-infrared spectroscopy coupled with chemometrics and artificial neural network modeling for prediction of emulsion droplet diameters. Micromachines 2022, 13, 1876. [Google Scholar] [CrossRef]
- Jurinjak Tušek, A.; Jurina, T.; Čulo, I.; Valinger, D.; Gajdoš Kljusurić, J.; Benković, M. Application of NIRs coupled with PLS and ANN modelling to predict average droplet size in oil-in-water emulsions prepared with different microfluidic devices. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 270, 120860. [Google Scholar] [CrossRef]
- Mishra, P.; van Dijk, M.; Wintermeyer, C.; Sabater, C.; Bot, A.; Verkleij, T.; Broeze, J. At-line and inline prediction of droplet size in mayonnaise with near-infrared spectroscopy. Infrared Phys. Technol. 2022, 123, 104155. [Google Scholar] [CrossRef]
- Holroyd, S.E. The Use of near Infrared Spectroscopy on Milk and Milk Products. J. Near Infrared Spectrosc. 2013, 21, 311–322. [Google Scholar] [CrossRef]
- Grossi, M.; Di Lecce, G.; Arru, M.; Gallina Toschi, T.; Riccò, B. An opto-electronic system for in-situ determination of peroxide value and total phenol content in olive oil. J. Food Eng. 2015, 146, 1–7. [Google Scholar] [CrossRef]
- Zareef, M.; Chen, Q.; Hassan, M.M.; Arslan, M.; Hashim, M.M.; Ahmad, W.; Kutsanedzie, F.Y.H.; Agyekum, A.A. An Overview on the applications of typical non-linear algorithms coupled with NIR spectroscopy in food analysis. Food Eng. Rev. 2020, 12, 173–190. [Google Scholar] [CrossRef]
- Shalev, N.T.; Ghermani, A.; Tchernov, D.; Shemesh, E.; Israel, A.; Brook, A. NIR spectroscopy and artificial neural network for seaweed protein content assessment in-situ. Comput. Electron. Agric. 2022, 201, 107304. [Google Scholar] [CrossRef]
- Mireei, S.A.; Mohtasebi, S.S.; Massudi, R.; Rafiee, S.; Arabanian, A. Feasibility of near infrared spectroscopy for analysis of date fruits. Int. Agrophys. 2010, 24, 351–356. [Google Scholar]
- Costa, L.R.; Tonoli, G.H.D.; Milagres, F.R.; Hein, P.R.G. Artificial neural network and partial least square regressions for rapid estimation of cellulose pulp dryness based on near infrared spectroscopic data. Carbohydr. Polym. 2019, 224, 115186. [Google Scholar] [CrossRef] [PubMed]
- Jurinjak Tušek, A.; Benković, M.; Belščak Cvitanović, A.; Valinger, D.; Jurina, T.; Gajdoš Kljusurić, J. Kinetics and thermodynamics of the solid-liquid extraction process of total polyphenols, antioxidants and extraction yield from Asteraceae plants. Ind. Crops Prod. 2016, 91, 205–214. [Google Scholar] [CrossRef]
- Sridharan, S.; Meinders, M.B.J.; Bitter, J.H.; Nikiforidis, C.V. Pea flour as stabilizer of oil-in-water emulsions: Protein purification unnecessary. Food Hydrocoll. 2020, 101, 105533. [Google Scholar] [CrossRef]
- Horwitz, W. Agricultural Chemicals, Contaminants, Drugs. In Association of Analytical Chemists: Official Methods of Analysis of AOAC International; AOAC International: Gaithersburg, MD, USA, 2010; Volume I. [Google Scholar]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT–Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Matešić, N.; Jurina, T.; Benković, M.; Panić, M.; Valinger, D.; Gajdoš Kljusurić, J.; Jurinjak Tušek, A. Microwave-assisted extraction of phenolic compounds from Cannabis sativa L.: Optimization and kinetics study. Sep. Sci. Technol. 2021, 56, 2047–2060. [Google Scholar] [CrossRef]
- Kim, W.; Wang, Y.; Selomulya, C. Dairy and plant proteins as natural food emulsifiers. Trends Food Sci. Technol. 2020, 105, 261–272. [Google Scholar] [CrossRef]
- Kumar, M.; Tomar, M.; Punia, S.; Dhakane-Lad, J.; Dhumal, S.; Changan, S.; Senapathy, M.; Berwal, M.K.; Sampathrajan, V.; Sayed, A.A.S.; et al. Plant-based proteins and their multifaceted industrial applications. LWT 2022, 154, 112620. [Google Scholar] [CrossRef]
- Niroula, A.; Alshamsi, R.; Sobti, B.; Nazir, A. Optimization of pea protein isolate-stabilized oil-in-water ultra-nanoemulsions by response surface methodology and the effect of electrolytes on optimized nanoemulsions. Colloids Interfaces 2022, 6, 47. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Principles and applications of miniaturized near-infrared (NIR) spectrometers. Chem. Eur. J. 2021, 27, 1514–1532. [Google Scholar] [CrossRef]
- Grabska, J.; Beć, K.B.; Ozaki, Y.; Huck, C.W. Anharmonic DFT study of near-infrared spectra of caffeine: Vibrational analysis of the second overtones and ternary combinations. Molecules 2021, 26, 5212. [Google Scholar] [CrossRef]
- Gál, L.; Oravec, M.; Gemeiner, P.; Čeppan, M. Principal component analysis for the forensic discrimination of black inkjet inks based on the Vis-NIR fibre optics reflection spectra. Forensic Sci. Int. 2015, 257, 285–292. [Google Scholar] [CrossRef]
- Toscano, G.; Rinnan, Å.; Pizzi, A.; Mancini, M. The use of near-infrared (NIR) spectroscopy and principal component analysis (PCA) to discriminate bark and wood of the most common species of the pellet sector. Energy Fuels 2017, 31, 2814–2821. [Google Scholar] [CrossRef]
- Roger, J.-M.; Mallet, A.; Marini, F. preprocessing NIR spectra for aquaphotomics. Molecules 2022, 27, 6795. [Google Scholar] [CrossRef]
- Bampi, M.; Scheer, A.D.P.; De Castilhos, F. Application of near infrared spectroscopy to predict the average droplet size and water content in biodiesel emulsions. Fuel 2013, 113, 546–552. [Google Scholar] [CrossRef]
- Bi, Y.; Yuan, K.; Xiao, W.; Wu, J.; Shi, C.; Xia, J.; Chu, G.; Zhang, G.; Zhou, G. A local pre-processing method for near-infrared spectra, combined with spectral segmentation and standard normal variate transformation. Anal. Chim. Acta 2016, 909, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Grisanti, E.; Totska, M.; Huber, S.; Calderon, C.K.; Hohmann, M.; Lingenfelser, D.; Otto, M. Dynamic localized SNV, Peak SNV, and partial peak SNV: Novel standardization methods for preprocessing of spectroscopic data used in predictive modeling. J. Spectrosc. 2018, 2018, 5037575. [Google Scholar] [CrossRef]
- Heil, K.; Schmidhalter, U. An Evaluation of different NIR-spectral pre-treatments to derive the soil parameters C and N of a humus-clay-rich soil. Sensors 2021, 21, 1423. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Q.; Tang, G.; Feng, Q.; Lin, L. The combined optimization of Savitzky-Golay smoothing and multiplicative scatter correction for ft-nir pls models. ISRN Spectrosc. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Joe, A.F.; Gopal, A. A Study on various preprocessing algorithms used for NIR spectra. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 2752–2757. [Google Scholar]
- Liu, Y.; Liu, Y.; Chen, Y.; Zhang, Y.; Shi, T.; Wang, J.; Hong, Y.; Fei, T.; Zhang, Y. The influence of spectral pretreatment on the selection of representative calibration samples for soil organic matter estimation using Vis-NIR reflectance spectroscopy. Remote Sens. 2019, 11, 450. [Google Scholar] [CrossRef]
- Basile, T.; Marsico, A.D.; Perniola, R. Use of artificial neural networks and NIR spectroscopy for non-destructive grape texture prediction. Foods 2022, 11, 281. [Google Scholar] [CrossRef]
- Gojun, M.; Valinger, D.; Šalić, A.; Zelić, B. Development of NIR-Based ANN models for on-line monitoring of glycerol concentration during biodiesel production in a microreactor. Micromachines 2022, 13, 1590. [Google Scholar] [CrossRef]
- Marsalek, R. Particle size and zeta potential of ZnO. APCBEE Procedia 2014, 9, 13–17. [Google Scholar] [CrossRef]
- Marić, L.; Malešić, E.; Jurinjak Tušek, A.; Benković, M.; Valinger, D.; Jurina, T.; Gajdoš Kljusurić, J. Effects of drying on physical and chemical properties of root vegetables: Artificial neural network modelling. Food Bioprod. Process. 2020, 119, 148–160. [Google Scholar] [CrossRef]
- Jurinjak Tušek, A.; Benković, M.; Malešić, E.; Marić, L.; Jurina, T.; Gajdoš Kljusurić, J.; Valinger, D. Rapid quantification of dissolved solids and bioactives in dried root vegetable extracts using near infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120074. [Google Scholar] [CrossRef] [PubMed]
- Schoot, M.; Kapper, C.; van Kollenburg, G.H.; Postma, G.J.; van Kessel, G.; Buydens, L.M.C.; Jansen, J.J. Investigating the need for preprocessing of near-infrared spectroscopic data as a function of sample size. Chemom. Intell. Lab. Syst. 2020, 204, 104105. [Google Scholar] [CrossRef]



| MLP | R2training/ RMSEtraining | R2test/ RMSEtest | R2validation/ RMSEvalidation | Hidden Activation | Output Activation | ||
|---|---|---|---|---|---|---|---|
| Raw spectra | P | MLP 4-4-2 | 0.8935 49.5475 | 0.8304 54.7184 | 0.7985 60.0452 | Exponential | Tanh |
| C | MLP 4-5-3 | 0.9865 6.0391 | 0.9812 7.7728 | 0.9674 11.3507 | Tanh | Exponential | |
| P + C | MLP 4-6-5 | 0.8745 179.7719 | 0.8558 208.2232 | 0.7830 264.5242 | Exponential | Tanh | |
| SG | P | MLP 4-4-2 | 0.8792 67.5971 | 0.7667 90.3055 | 0.7394 95.8990 | Exponential | Exponential |
| C | MLP 4-5-3 | 0.9803 43.3076 | 0.9749 55.0740 | 0.9612 63.6157 | Tanh | Identity | |
| P + C | MLP 4-4-5 | 0.8833 104.4711 | 0.8227 142.6231 | 0.7954 176.8579 | Exponential | Tanh | |
| SNV | P | MLP 4-3-2 | 0.7891 57.5630 | 0.7532 93.7165 | 0.7184 134.1752 | Logistic | Logistic |
| C | MLP 4-5-3 | 0.9831 19.0224 | 0.9795 27.2635 | 0.9595 28.1258 | Exponential | Exponential | |
| P + C | MLP 4-5-5 | 0.9276 36.0402 | 0.9197 65.9584 | 0.8775 76.8929 | Tanh | Exponential | |
| MSC | P | MLP 4-3-2 | 0.8528 74.246 | 0.8184 79.5826 | 0.7596 80.1605 | Logistic | Identity |
| C | MLP 4-4-3 | 0.9852 11.1603 | 0.9844 12.9489 | 0.9691 20.0684 | Tanh | Logistic | |
| P + C | MLP 4-5-5 | 0.9432 72.3190 | 0.9244 85.4091 | 0.9015 108.2861 | Exponential | Exponential | |
| MLP | R2training/ RMSEtraining | R2test/ RMSEtest | R2validation/ RMSEvalidation | Hidden Activation | Output Activation | ||
|---|---|---|---|---|---|---|---|
| Raw spectra | P | MLP 4-3-2 | 0.8207 34.2804 | 0.8085 62.7199 | 0.7967 75.7252 | Exponential | Tanh |
| C | MLP 4-4-3 | 0.9456 10.3888 | 0.9225 11.9938 | 0.8989 20.0926 | Tanh | Identity | |
| P + C | MLP 4-6-5 | 0.8889 42.5165 | 0.8133 57.3472 | 0.7999 117.9804 | Logistic | Logistic | |
| SG | P | MLP 4-4-2 | 0.8236 33.9466 | 0.7518 52.6949 | 0.7503 76.5493 | Tanh | Exponential |
| C | MLP 4-4-3 | 0.8784 17.9444 | 0.8697 28.7077 | 0.8541 42.7062 | Logistic | Tanh | |
| P + C | MLP 4-5-5 | 0.8611 69.1007 | 0.8493 78.5401 | 0.8387 115.7696 | Logistic | Logistic | |
| SNV | P | MLP 4-5-2 | 0.8745 39.4502 | 0.8485 51.0628 | 0.7282 52.2450 | Tanh | Tanh |
| C | MLP 4-4-3 | 0.9423 13.5427 | 0.9324 30.3847 | 0.8642 43.6561 | Logistic | Logistic | |
| P + C | MLP 4-5-5 | 0.8718 57.9012 | 0.8577 69.7663 | 0.8434 86.8053 | Exponential | Identity | |
| MSC | P | MLP 4-5-2 | 0.8336 24.7871 | 0.8314 47.0239 | 0.7329 5.8123 | Exponential | Exponential |
| C | MLP 4-4-3 | 0.9069 36.5873 | 0.9088 48.8569 | 0.8998 57.8222 | Exponential | Exponential | |
| P + C | MLP 4-5-5 | 0.8724 58.9583 | 0.8267 68.0744 | 0.8245 101.7221 | Logistic | Logistic | |
| Output Variable | R2training/ RMSEtraining | R2test/ RMSEtest | R2validation/ RMSEvalidation | ||
|---|---|---|---|---|---|
| Raw spectra | P | Zeta potential | 0.8966 1.9274 | 0.8963 2.2835 | 0.8666 2.9133 |
| Feret diameter | 0.8907 2.8565 | 0.7941 6.4834 | 0.7005 8.9043 | ||
| C | TPC | 0.9982 3.4746 | 0.9953 3.9415 | 0.9894 6.5336 | |
| DPPH | 0.9899 0.0276 | 0.9695 0.0340 | 0.9630 0.0443 | ||
| FRAP | 0.9756 0.0652 | 0.9742 0.0883 | 0.9499 0.1022 | ||
| P + C | Zeta potential | 0.9612 1.9824 | 0.9083 2.2806 | 0.8418 3.3636 | |
| Feret diameter | 0.7576 11.1085 | 0.7539 14.3959 | 0.6931 17.9466 | ||
| TPC | 0.9282 11.8724 | 0.8918 16.9653 | 0.8741 28.3374 | ||
| DPPH | 0.8627 0.0997 | 0.7991 0.1024 | 0.7636 0.1468 | ||
| FRAP | 0.9623 0.0958 | 0.9256 0.1220 | 0.8433 0.1575 | ||
| SG | P | Zeta potential | 0.8389 2.6495 | 0.8303 3.0937 | 0.7893 6.1019 |
| Feret diameter | 0.9281 11.3214 | 0.7445 11.9740 | 0.7398 14.2207 | ||
| C | TPC | 0.9816 9.3064 | 0.9652 10.4949 | 0.9467 14.3953 | |
| DPPH | 0.9871 0.0293 | 0.9709 0.0466 | 0.9590 0.0594 | ||
| FRAP | 0.9883 0.0536 | 0.9776 0.0650 | 0.9724 0.0715 | ||
| P + C | Zeta potential | 0.7125 3.8091 | 0.6882 4.6766 | 0.6613 5.6044 | |
| Feret diameter | 0.9177 9.0139 | 0.8272 10.3678 | 0.7259 11.8604 | ||
| TPC | 0.9776 9.8112 | 0.9288 12.4844 | 0.8968 19.9628 | ||
| DPPH | 0.9011 0.0831 | 0.8805 0.0848 | 0.8009 0.1066 | ||
| FRAP | 0.9526 0.1085 | 0.9406 0.1297 | 0.8949 0.1301 | ||
| SNV | P | Zeta potential | 0.8544 2.6495 | 0.7481 3.0937 | 0.7192 6.1019 |
| Feret diameter | 0.7583 11.3214 | 0.7098 11.9740 | 0.7025 14.2207 | ||
| C | TPC | 0.9884 9.3064 | 0.9694 10.4948 | 0.9504 14.3953 | |
| DPPH | 0.9905 0.0293 | 0.9863 0.0466 | 0.9763 0.0594 | ||
| FRAP | 0.9784 0.0536 | 0.9746 0.0650 | 0.9516 0.0715 | ||
| P + C | Zeta potential | 0.9391 3.8091 | 0.8566 4.6766 | 0.8021 5.6044 | |
| Feret diameter | 0.9722 9.0139 | 0.8686 10.3678 | 0.8372 11.8604 | ||
| TPC | 0.9946 9.8112 | 0.9889 12.4844 | 0.9804 19.9628 | ||
| DPPH | 0.9597 0.0831 | 0.8639 0.0848 | 0.8541 0.1066 | ||
| FRAP | 0.9782 0.1085 | 0.9642 0.1297 | 0.9634 0.1301 | ||
| MSC | P | Zeta potential | 0.9051 2.3980 | 0.8718 2.7542 | 0.7820 3.9036 |
| Feret diameter | 0.9236 11.8704 | 0.7649 12.0450 | 0.7142 12.3861 | ||
| C | TPC | 0.9983 4.7244 | 0.9935 5.0862 | 0.9863 6.3345 | |
| DPPH | 0.9863 0.0304 | 0.9847 0.0370 | 0.9527 0.0636 | ||
| FRAP | 0.9757 0.0718 | 0.9733 0.0797 | 0.9701 0.1649 | ||
| P + C | Zeta potential | 0.9107 2.1143 | 0.8438 3.3898 | 0.7974 3.5012 | |
| Feret diameter | 0.9547 8.1953 | 0.8004 11.3799 | 0.7973 12.1886 | ||
| TPC | 0.9879 5.4605 | 0.9869 7.9706 | 0.9831 8.0748 | ||
| DPPH | 0.9628 0.0526 | 0.9479 0.0529 | 0.9372 0.0857 | ||
| FRAP | 0.9925 0.0701 | 0.9797 0.0874 | 0.9740 0.0881 | ||
| Output Variable | R2training/ RMSEtraining | R2test/ RMSEtest | R2validation/ RMSEvalidation | ||
|---|---|---|---|---|---|
| Raw spectra | P | Zeta potential | 0.8510 4.1106 | 0.8497 4.7355 | 0.7922 5.5010 |
| Feret diameter | 0.7916 6.4024 | 0.7654 10.3573 | 0.7342 11.0086 | ||
| C | TPC | 0.9718 4.5579 | 0.9469 4.8426 | 0.8519 7.9168 | |
| DPPH | 0.9493 0.0316 | 0.9436 0.0410 | 0.8942 0.0434 | ||
| FRAP | 0.9719 0.0458 | 0.9158 0.0488 | 0.8556 0.0523 | ||
| P + C | Zeta potential | 0.9577 2.1112 | 0.8748 4.1546 | 0.8706 4.9363 | |
| Feret diameter | 0.8614 8.6155 | 0.8475 9.7587 | 0.8142 14.3121 | ||
| TPC | 0.9877 3.7226 | 0.9756 3.9809 | 0.9638 4.1216 | ||
| DPPH | 0.9124 0.0370 | 0.9025 0.0478 | 0.8896 0.0494 | ||
| FRAP | 0.9441 0.0429 | 0.9355 0.0550 | 0.9056 0.0883 | ||
| SG | P | Zeta potential | 0.8425 4.0238 | 0.8415 4.6788 | 0.8063 5.6473 |
| Feret diameter | 0.8046 6.5721 | 0.7726 9.3411 | 0.7265 11.0094 | ||
| C | TPC | 0.9161 5.9254 | 0.9112 9.2414 | 0.8752 12.2198 | |
| DPPH | 0.8509 0.0440 | 0.7916 0.0714 | 0.7771 0.0649 | ||
| FRAP | 0.9568 0.0485 | 0.8731 0.0548 | 0.8546 0.0583 | ||
| P + C | Zeta potential | 0.9066 3.1347 | 0.8598 4.369 | 0.8446 6.4960 | |
| Feret diameter | 0.8466 5.7399 | 0.8263 9.2201 | 0.7410 11.9014 | ||
| TPC | 0.9155 6.6264 | 0.8957 8.1915 | 0.8877 8.4050 | ||
| DPPH | 0.8332 0.0465 | 0.7877 0.0720 | 0.7612 0.0725 | ||
| FRAP | 0.9855 0.0330 | 0.9513 0.0398 | 0.9212 0.0938 | ||
| SNV | P | Zeta potential | 0.9672 1.8772 | 0.9337 2.7578 | 0.8781 4.5652 |
| Feret diameter | 0.8153 7.5254 | 0.7268 8.8445 | 0.6782 9.8430 | ||
| C | TPC | 0.9398 7.7047 | 0.8780 9.3440 | 0.8075 10.9179 | |
| DPPH | 0.9681 0.0251 | 0.9638 0.0277 | 0.9364 0.0336 | ||
| FRAP | 0.9509 0.0388 | 0.9486 0.0391 | 0.9233 0.0506 | ||
| P + C | Zeta potential | 0.9392 2.5100 | 0.8824 3.5757 | 0.8328 4.9122 | |
| Feret diameter | 0.8485 3.5498 | 0.6723 9.9379 | 0.5764 10.8354 | ||
| TPC | 0.9434 5.9875 | 0.9184 6.5890 | 0.9096 7.7269 | ||
| DPPH | 0.9073 0.0382 | 0.9073 0.0585 | 0.8777 0.0617 | ||
| FRAP | 0.9819 0.0318 | 0.9479 0.0349 | 0.9149 0.0941 | ||
| MSC | P | Zeta potential | 0.9642 3.1605 | 0.8858 9.6204 | 0.7970 10.1486 |
| Feret diameter | 0.8703 3.9175 | 0.6987 8.778 | 0.5881 12.6877 | ||
| C | TPC | 0.8539 8.4527 | 0.8323 9.8816 | 0.8088 10.7537 | |
| DPPH | 0.9721 0.0527 | 0.9522 0.0735 | 0.9224 0.0881 | ||
| FRAP | 0.9876 0.0343 | 0.9653 0.0355 | 0.9232 0.0534 | ||
| P + C | Zeta potential | 0.9633 2.7911 | 0.9109 3.0450 | 0.8837 4.2572 | |
| Feret diameter | 0.6794 9.4931 | 0.5664 9.6395 | 0.5166 9.4931 | ||
| TPC | 0.9722 4.6885 | 0.9529 5.3897 | 0.91277 7.1686 | ||
| DPPH | 0.9639 0.0234 | 0.8939 0.0445 | 0.8632 0.0679 | ||
| FRAP | 0.9679 0.0394 | 0.9250 0.0404 | 0.9084 0.0496 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirovec, S.; Benković, M.; Valinger, D.; Sokač Cvetnić, T.; Gajdoš Kljusurić, J.; Jurinjak Tušek, A.; Jurina, T. Development of ANN Models for Prediction of Physical and Chemical Characteristics of Oil-in-Aqueous Plant Extract Emulsions Using Near-Infrared Spectroscopy. Chemosensors 2023, 11, 278. https://doi.org/10.3390/chemosensors11050278
Sirovec S, Benković M, Valinger D, Sokač Cvetnić T, Gajdoš Kljusurić J, Jurinjak Tušek A, Jurina T. Development of ANN Models for Prediction of Physical and Chemical Characteristics of Oil-in-Aqueous Plant Extract Emulsions Using Near-Infrared Spectroscopy. Chemosensors. 2023; 11(5):278. https://doi.org/10.3390/chemosensors11050278
Chicago/Turabian StyleSirovec, Sara, Maja Benković, Davor Valinger, Tea Sokač Cvetnić, Jasenka Gajdoš Kljusurić, Ana Jurinjak Tušek, and Tamara Jurina. 2023. "Development of ANN Models for Prediction of Physical and Chemical Characteristics of Oil-in-Aqueous Plant Extract Emulsions Using Near-Infrared Spectroscopy" Chemosensors 11, no. 5: 278. https://doi.org/10.3390/chemosensors11050278










