DNAzymes-Embedded Framework Nucleic Acids (FNAzymes) for Metal Ions Imaging in Living Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
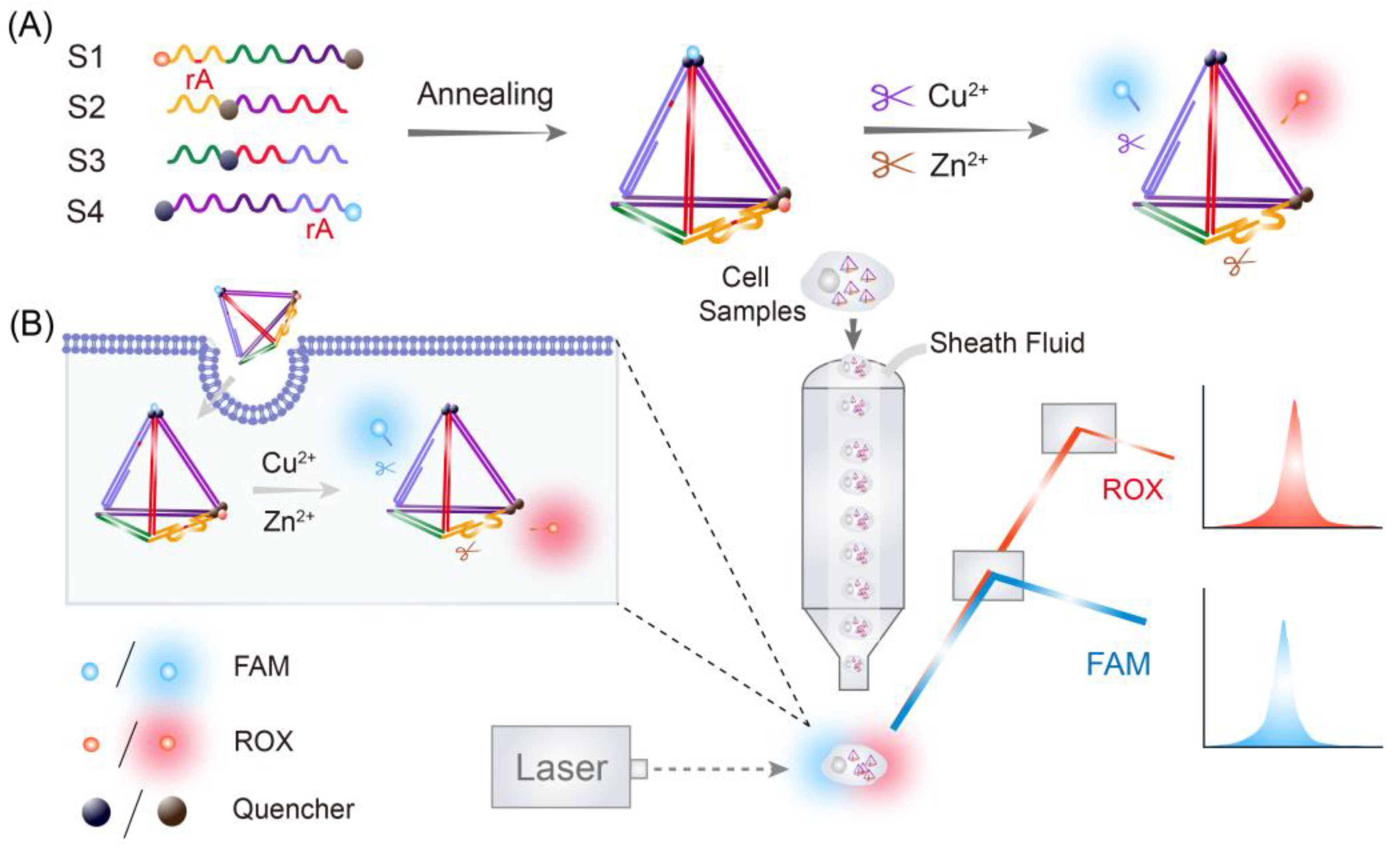
2.2. Assembly of Multifunctional FNAzymes
2.3. AFM Imaging
2.4. Simultaneous Detection of Cu2+ and Zn2+ In Vitro
2.5. Stability of FNAzymes
2.6. Cell Viability
2.7. Confocal Fluorescence Imaging
2.8. Flow Cytometric Assay
2.9. Inductively Coupled Plasma-Mass Spectrometer (ICP-MS) Test
3. Results and Discussion
3.1. Simultaneous Detection of Cu2+ and Zn2+ In Vitro
3.2. Stability and Cytotoxicity of the FNAzymes
3.3. Intracellular Imaging of Cu2+ and Zn2+ with FNAzymes
3.4. Semi-Quantitative Detection of Cu2+ and Zn2+ in Living Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bolognin, S.; Messori, L.; Zatta, P. Metal ion physiopathology in neurodegenerative disorders. Neuromol. Med. 2009, 11, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Krezel, A.; Maret, W. The functions of metamorphic metallothioneins in zinc and copper metabolism. Int. J. Mol. Sci. 2017, 18, 1237. [Google Scholar] [CrossRef] [Green Version]
- Winge, D.R.; Jensen, L.T.; Srinivasan, C. Metal-ion regulation of gene expression in yeast. Curr. Opin. Chem. Biol. 1998, 2, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Stelmashook, E.V.; Isaev, N.K.; Genrikhs, E.E.; Amelkina, G.A.; Khaspekov, L.G.; Skrebitsky, V.G.; Illarioshkin, S.N. Role of zinc and copper ions in the pathogenetic mechanisms of alzheimer’s and parkinson’s diseases. Biochemistry 2014, 79, 391–396. [Google Scholar] [CrossRef]
- Krueger, W.H.H.; Gonye, G.E.; Madison, D.L.; Murray, K.E.; Kumar, M.; Spoerel, N.; Pfeiffer, S.E. Tpo1, a member of a novel protein family, is developmentally regulated in cultured oligodendrocytes. J. Neurochem. 1997, 69, 1343–1355. [Google Scholar] [CrossRef]
- Islamoglu, Y.; Evliyaoglu, O.; Tekbas, E.; Cil, H.; Elbey, M.A.; Atilgan, Z.; Kaya, H.; Bilik, Z.; Akyuz, A.; Alan, S. The relationship between serum levels of Zn and Cu and severity of coronary atherosclerosis. Biol. Trace Elem. Res. 2011, 144, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Milne, A.; Landing, W.; Bizimis, M.; Morton, P. Determination of Mn, Fe, Co, Ni, Cu, Zn, Cd and Pb in seawater using high resolution magnetic sector inductively coupled mass spectrometry (HR-ICP-MS). Anal. Chim. Acta 2010, 665, 200–207. [Google Scholar] [CrossRef]
- Cui, Y.; Chang, X.; Zhai, Y.; Zhu, X.; Zheng, H.; Lian, N. ICP-AES determination of trace elements after preconcentrated with p-dimethylaminobenzaldehyde-modified nanometer SiO2 from sample solution. Microchem. J. 2006, 83, 35–41. [Google Scholar] [CrossRef]
- Lin, P.H.; Danadurai, K.S.K.; Huang, S.D. Simultaneous determination of cobalt, nickel and copper in seawater with a multi-element electrothermal atomic absorption spectrometer and microcolumn preconcentration. J. Anal. Atom. Spectrom. 2001, 16, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Sheth, V.; Chen, X.; Mettenbrink, E.M.; Yang, W.; Jones, M.A.; M’Saad, O.; Thomas, A.G.; Newport, R.S.; Francek, E.; Wang, L.; et al. Quantifying intracellular nanoparticle distributions with three-dimensional super-resolution microscopy. ACS Nano 2023, 17, 8376–8392. [Google Scholar] [CrossRef]
- Wang, S.; Deng, S.; Cai, X.; Hou, S.; Li, J.; Gao, Z.; Li, J.; Wang, L.; Fan, C. Superresolution imaging of telomeres with continuous wave stimulated emission depletion (STED) microscope. Sci. China Chem. 2016, 59, 1519–1524. [Google Scholar] [CrossRef]
- Bao, L.; Ding, L.; Hui, J.; Ju, H. A light-up imaging protocol for neutral ph-enhanced fluorescence detection of lysosomal neuraminidase activity in living cells. Chem. Commun. 2016, 52, 12897–12900. [Google Scholar] [CrossRef]
- Li, B.; Kou, J.; Mei, H.; Gu, X.; Wang, M.; Xie, X.; Xu, K. A hemicyanine-based “turn-on” fluorescent probe for the selective detection of Cu2+ions and imaging in living cells. Anal. Methods-UK 2020, 12, 4181–4184. [Google Scholar] [CrossRef]
- Zhang, X.; Song, Z.; Chao, Q.; Li, Q.; Kong, R.; Fan, G.; Luo, X. A DNAzyme-based normalized fluorescence strategy for direct quantification of endogenous zinc in living cells. Chem. Commun. 2022, 58, 577–580. [Google Scholar] [CrossRef]
- Patil, M.; Keshav, K.; Kumawat, M.K.; Bothra, S.; Sahoo, S.K.; Srivastava, R.; Rajput, J.; Bendre, R.; Kuwar, A. Monoterpenoid derivative based ratiometric fluorescent chemosensor for bioimaging and intracellular detection of Zn2+and Mg2+ ions. J. Photoch. Photobio. A 2018, 364, 758–763. [Google Scholar] [CrossRef]
- Chen, Y.; Qiu, D.; Zhang, X.; Liu, Y.; Cheng, M.; Lei, J.; Mergny, J.; Ju, H.; Zhou, J. Highly sensitive biosensing applications of a magnetically immobilizable covalent G-quadruplex-hemin DNAzyme catalytic system. Anal. Chem. 2022, 94, 2212–2219. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, J.; Cheng, M.; Monchaud, D.; Zhou, J.; Ju, H. A thermophilic tetramolecular G-quadruplex/hemin DNAzyme. Angew. Chem. Int. Ed. 2017, 56, 16636–16640. [Google Scholar] [CrossRef] [PubMed]
- McGhee, C.E.; Loh, K.Y.; Lu, Y. DNAzyme sensors for detection of metal ions in the environment and imaging them in living cells. Curr. Opin. Biotech. 2017, 45, 191–201. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.; He, Y.; Xiong, B.; Li, Y.; Wang, F. Chemical and structural modification of RNA-cleaving DNAzymes for efficient biosensing and biomedical applications. Trac-Trend Anal. Chem. 2023, 159, 116910. [Google Scholar] [CrossRef]
- Hu, L.; Fu, X.; Kong, G.; Yin, Y.; Meng, H.-M.; Ke, G.; Zhang, X.-B. DNAzyme-gold nanoparticle-based probes for biosensing and bioimaging. J. Mater. Chem. B 2020, 8, 9449–9465. [Google Scholar] [CrossRef]
- Khan, S.; Burciu, B.; Filipe, C.D.M.; Li, Y.; Dellinger, K.; Didar, T.F. DNAzyme-based biosensors: Immobilization strategies, applications, and future prospective. ACS Nano 2021, 15, 13943–13969. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerber, P.P.; Donde, M.J.; Matheson, N.J.; Taylor, A.I. XNAzymes targeting the SARS-CoV-2 genome inhibit viral infection. Nat. Commun. 2022, 13, 6716. [Google Scholar] [CrossRef]
- Ge, Z.; Li, Q.; Fan, C. Framework nucleic acids for cell imaging and therapy. Chem. Res. Chin. Univ. 2020, 36, 1–9. [Google Scholar] [CrossRef]
- Ge, Z.; Gu, H.; Li, Q.; Fan, C. Concept and development of framework nucleic acids. J. Am. Chem. Soc. 2018, 140, 17808–17819. [Google Scholar] [CrossRef]
- Liu, Q.; Ge, Z.; Mao, X.; Zhou, G.; Zuo, X.; Shen, J.; Shi, J.; Li, J.; Wang, L.; Chen, X.; et al. Valency-controlled framework nucleic acid signal amplifiers. Angew. Chem. Int. Ed. 2018, 57, 7131–7135. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, M.; Duan, X.; Guo, B.; Cheng, W.; Ding, S.; Ju, H. Collapse of DNA tetrahedron nanostructure for “off-on” fluorescence detection of DNA methyltransferase activity. ACS Appl. Mater. Interfaces 2017, 9, 40087–40093. [Google Scholar] [CrossRef] [PubMed]
- Qing, Z.H.; Hu, J.L.; Xu, J.Y.; Zou, Z.; Lei, Y.L.; Qing, T.P.; Yang, R.H. An intramolecular catalytic hairpin assembly on a DNA tetrahedron for mRNA imaging in living cells: Improving reaction kinetics and signal stability. Chem. Sci. 2020, 11, 1985–1990. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Li, J.; Li, Q.; Huang, Q.; Shi, J.; Yan, H.; Fan, C. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. 2014, 53, 7745–7750. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. A DNAzyme catalytic beacon sensor for paramagnetic Cu2+ ions in aqueous solution with high sensitivity and selectivity. J. Am. Chem. Soc. 2007, 129, 9838–9839. [Google Scholar] [CrossRef]
- Endo, M.; Takeuchi, Y.; Suzuki, Y.; Emura, T.; Hidaka, K.; Wang, F.; Willner, I.; Sugiyama, H. Single-molecule visualization of the activity of a Zn2+-dependent DNAzyme. Angew. Chem. Int. Ed. 2015, 54, 10550–10554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, P.; Jiang, J.; Li, L.; Wang, S.; Yu, H.; Feng, Y.; Zhu, M.; Zhang, B.; Yin, H.; Guo, Q.; et al. A mitochondria-targeted ratiometric two-photon fluorescent probe for biological zinc ions detection. Biosens. Bioelectron. 2016, 77, 921–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, M.; Liu, C.; Zhang, Y.; Rong, X.; Wang, X.; Li, X.; Wang, K.; Zhu, H.; Zhu, B. Rational design of a water-soluble TICT-AIEE-active fluorescent probe for mercury ion detection. Anal. Chim. Acta 2022, 1230, 340337. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, Y.; Dong, Y.; Chu, Y.; Song, N.; Yang, D. Dynamic assembly of DNA nanostructures in living cells for mitochondrial interference. J. Am. Chem. Soc. 2022, 144, 4667–4677. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Wu, Z.; Yu, R.; Jiang, J. Three-dimensional DNA nanostructures for dual-color microRNA imaging in living cells via hybridization chain reaction. Chem. Commun. 2020, 56, 6668–6671. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, D.; Huang, J.; Xia, Y.; Su, S.; Zuo, X.; Li, Q.; Wang, L. DNAzymes-Embedded Framework Nucleic Acids (FNAzymes) for Metal Ions Imaging in Living Cells. Chemosensors 2023, 11, 358. https://doi.org/10.3390/chemosensors11070358
Zhu D, Huang J, Xia Y, Su S, Zuo X, Li Q, Wang L. DNAzymes-Embedded Framework Nucleic Acids (FNAzymes) for Metal Ions Imaging in Living Cells. Chemosensors. 2023; 11(7):358. https://doi.org/10.3390/chemosensors11070358
Chicago/Turabian StyleZhu, Dan, Jiaxuan Huang, Yanting Xia, Shao Su, Xiaolei Zuo, Qian Li, and Lianhui Wang. 2023. "DNAzymes-Embedded Framework Nucleic Acids (FNAzymes) for Metal Ions Imaging in Living Cells" Chemosensors 11, no. 7: 358. https://doi.org/10.3390/chemosensors11070358
APA StyleZhu, D., Huang, J., Xia, Y., Su, S., Zuo, X., Li, Q., & Wang, L. (2023). DNAzymes-Embedded Framework Nucleic Acids (FNAzymes) for Metal Ions Imaging in Living Cells. Chemosensors, 11(7), 358. https://doi.org/10.3390/chemosensors11070358






