Linkage Pathways of DNA–Nanoparticle Conjugates and Biological Applications
Abstract
1. Introduction
- (1)
- Searching for diversely functional DNA sequences. The chemical activity of DNA grows into new areas outside of storing and transferring genetic information in the field of functional DNA nanotechnology. The two main representative categories of functional DNAs that are generated by in vitro selection with particular binding affinities and catalytic capabilities are aptamers and DNAzymes [13,14]. Aptamers are chosen by a procedure called systematic evolution of ligands by exponential enrichment (SELEX), and the selection process has evolved from in vitro to in vivo. Aptamers are widely employed in the assembly of sensitive biosensors, the construction of bioimaging agents, and targeted therapeutics [15]. An additional category of valuable DNA molecules with catalytic activity is DNAzymes. RNA-cleaving DNAzymes are of particular fascination due to their quick reaction time and simplicity in application in life [16]. DNAzymes act by binding to specific metal ions as catalytic cofactors and are ideal for functionalized nanoparticle surfaces [17].
- (2)
- Development of inorganic nanoparticles with facile modification of DNA on the surface. By adding DNA nanotechnology into nanoparticle research, precise geometric construction of nanoparticles and change in surface properties have been described [18,19]. The best candidates for carrying smart DNA walkers/motors, activatable aptamers, and DNAzyme-based systems that respond to stimuli for biosensing, bioimaging, and biomedical applications are nanoparticles with a substantial surface area, favorable biocompatibility, outstanding stability, and beneficial physical and chemical properties, such as gold nanoparticles (AuNPs), upconversion nanomaterials (UCNPs) and so on [20,21,22].
- (3)
- Exploring the way DNA connects to nanoparticles. The facile modification properties of DNA encourage the interface engineering of nanoparticles based on several theories. Mirkin was the first to suggest that sulfurizing DNA and attaching it to AuNPs would result in the development of AuNPs-linked DNA [23,24]. The coordinated binding between DNA base structure and rare earth elements can also connect DNA to UCNPs [25,26,27,28,29]. The interface between DNA molecules and nanoparticles also involves additional covalent and noncovalent connecting techniques, such as the biotin-Streptavidin interaction and click chemistry [30,31]. Thereafter, electrostatic contact is another often employed technique for affixing DNA molecules to positively charged nanoparticles on the surface due to the negative charge of the phosphate backbone on the surface of the DNA structure [32]. Electrostatic interactions exist in various forms, such as hydrogen bonding, hydrophobic contacts, van der Waals forces, and ionic bonding, DNA–nanoparticle conjugates assembled with the above forms also demonstrate excellent stability and extremely promising applications. For instance, the mechanism of DNA’s denaturation and rehybridization is employed to create hydrogen bonds between base pairs that are complementary on adjacent DNA strands, resulting in the formation of interconnected structures. Then, silicate nanodiscs are utilized to create extra network points by luring electrostatic interactions with the DNA backbone [33]. This improves the mechanical elasticity of the hydrogel formulation and achieves the release of the loaded drug dexamethasone, realizing the conjugate’s ability to treat osteoporosis disease.
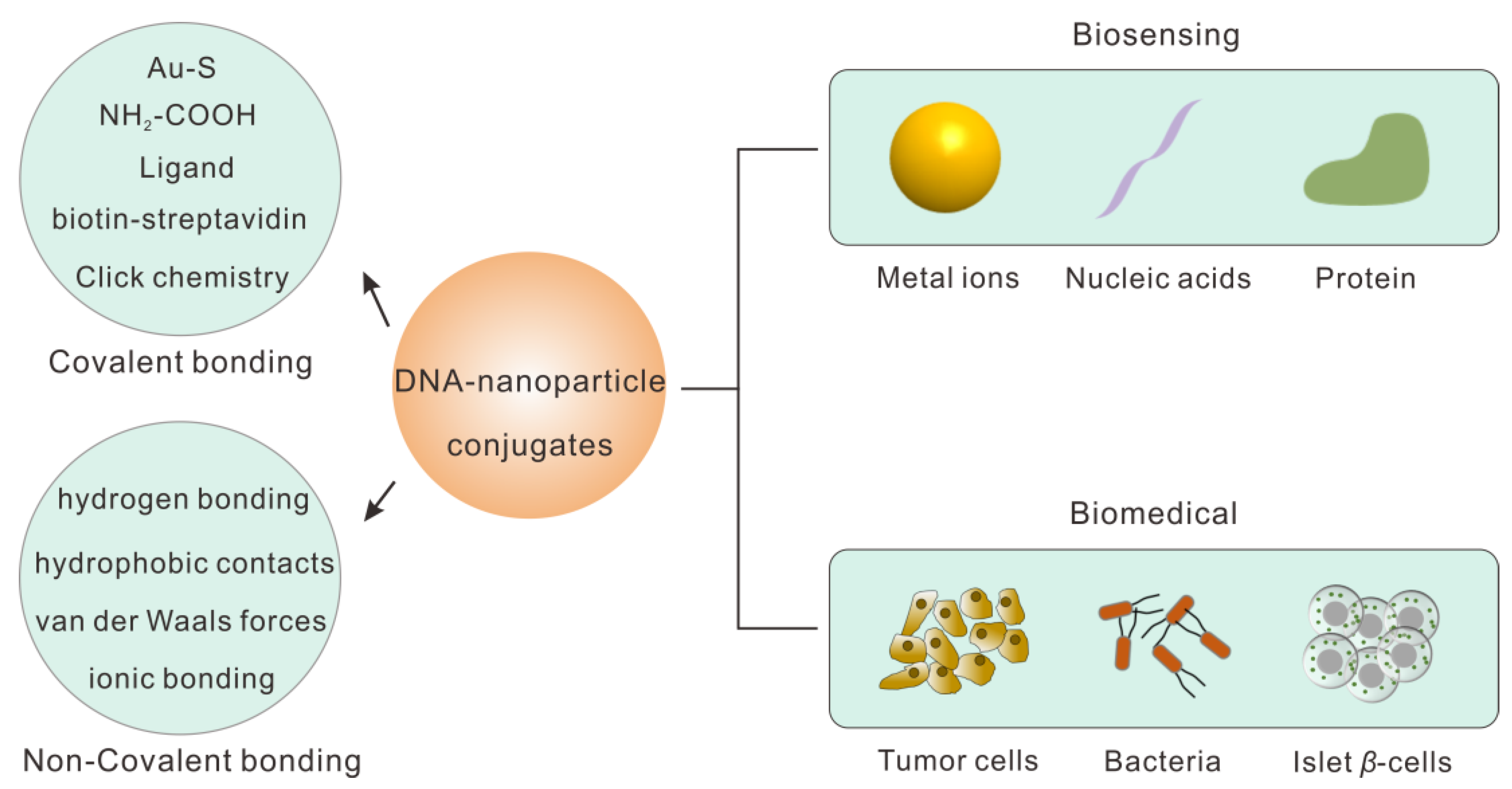
2. DNA–Nanoparticle Conjugates Design
2.1. Covalent Bonding

2.2. Non-Covalent Bonding
3. Biological Applications
3.1. Biosensing
3.2. Biomedical
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashworth, C. DNA nanotechnology: Building big with DNA bricks. Nat. Rev. Mater. 2018, 3, 17092. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef] [PubMed]
- Rothemund PW, K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Knappe, G.A.; Wamhoff, E.-C.; Bathe, M. Functionalizing DNA origami to investigate and interact with biological systems. Nat. Rev. Mater. 2023, 8, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Saliba, D.; Luo, X.; Rizzuto, F.J.; Sleiman, H.F. Programming rigidity into size-defined wireframe DNA nanotubes. Nanoscale 2023, 15, 5403–5413. [Google Scholar] [CrossRef]
- Yutong Huang, F.; Kumar Lat, P.; Sen, D. Unusual Paradigm for DNA–DNA Recognition and Binding: “Socket-Plug” Complementarity. J. Am. Chem. Soc. 2023, 145, 3146–3157. [Google Scholar] [CrossRef]
- Zhang, P.; Ouyang, Y.; Zhuo, Y.; Chai, Y.; Yuan, R. Recent Advances in DNA Nanostructures Applied in Sensing Interfaces and Cellular Imaging. Anal. Chem. 2023, 95, 407–419. [Google Scholar] [CrossRef]
- Wang, W.; Yu, S.; Huang, S.; Bi, S.; Han, H.; Zhang, J.R.; Lu, Y.; Zhu, J.J. Bioapplications of DNA nanotechnology at the solid-liquid interface. Chem. Soc. Rev. 2019, 48, 4892–4920. [Google Scholar] [CrossRef]
- Ahmadi-Sangachin, E.; Bazzi, F.; Xu, G.; Hosseini, M. Biosensing using DNA-based structures integrated with nanosheets. Microchem. J. 2023, 191, 108779. [Google Scholar] [CrossRef]
- Shaikh, S.; Younis, M.; Yuan, L. Functionalized DNA nanostructures for bioimaging. Coord. Chem. Rev. 2022, 469, 214648. [Google Scholar] [CrossRef]
- Chandler, M.; Jain, S.; Halman, J.; Hong, E.; Dobrovolskaia, M.A.; Zakharov, A.V.; Afonin, K.A. Artificial Immune Cell, AI-cell, a New Tool to Predict Interferon Production by Peripheral Blood Monocytes in Response to Nucleic Acid Nanoparticles. Small 2022, 18, 2204941. [Google Scholar] [CrossRef] [PubMed]
- De Fazio, A.F.; Misatziou, D.; Baker, Y.R.; Muskens, O.L.; Brown, T.; Kanaras, A.G. Chemically modified nucleic acids and DNA intercalators as tools for nanoparticle assembly. Chem. Soc. Rev. 2021, 50, 13410–13440. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.M.; Zhang, X.B.; Chen, Z.; Tan, W. Aptamer-assembled nanomaterials for biosensing and biomedical applications. Small 2011, 7, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Fu, X.; Peng, F.; Lee, J.; Yang, Q.; Zhang, F.; Xiong, M.; Kong, G.; Meng, H.M.; Ke, G.; Zhang, X.B. Aptamer-Functionalized DNA Nanostructures for Biological Applications. Top. Curr. Chem. 2020, 378, 21. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, J.; Zhou, Q. Functional DNA sensors integrated with nucleic acid signal amplification strategies for non-nucleic acid targets detection. Biosens. Bioelectron. 2023, 230, 115282. [Google Scholar] [CrossRef]
- Su, J.; Sun, C.; Du, J.; Xing, X.; Wang, F.; Dong, H. RNA-Cleaving DNAzyme-Based Amplification Strategies for Biosensing and Therapy. Adv. Healthcare Mater. 2023, 2300367. [Google Scholar] [CrossRef]
- Zhao, Y.; Shi, L.; Kuang, H.; Xu, C. DNA-Driven Nanoparticle Assemblies for Biosensing and Bioimaging. Top. Curr. Chem. 2020, 378, 18. [Google Scholar] [CrossRef]
- Hu, Q.; Wu, J.; Chen, L.; Lou, X.; Xia, F. Recent Development of DNA-modified AIEgen Probes for Biomedical Application. Chem. Res. Chin. Univ. 2021, 37, 66–72. [Google Scholar] [CrossRef]
- Ohta, S.; Glancy, D.; Chan, W.C.W. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 2016, 351, 841–845. [Google Scholar] [CrossRef]
- Jia, R.; Wang, Y.; Ma, W.; Huang, J.; Sun, H.; Chen, B.; Cheng, H.; He, X.; Wang, K. Activatable Dual Cancer-Related RNA Imaging and Combined Gene-Chemotherapy through the Target-Induced Intracellular Disassembly of Functionalized DNA Tetrahedron. Anal. Chem. 2022, 94, 5937–5945. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Song, Y.; He, Z.; Zhang, J.-R.; Zhu, J.-J. Self-assembled nanomaterials for biosensing and therapeutics: Recent advances and challenges. Analyst 2021, 146, 2807–2817. [Google Scholar] [CrossRef] [PubMed]
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical nucleic acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef]
- Callmann, C.E.; Vasher, M.K.; Das, A.; Kusmierz, C.D.; Mirkin, C.A. In Vivo Behavior of Ultrasmall Spherical Nucleic Acids. Small 2023, 19, 2300097. [Google Scholar] [CrossRef]
- Li, L.L.; Wu, P.; Hwang, K.; Lu, Y. An exceptionally simple strategy for DNA-functionalized up-conversion nanoparticles as biocompatible agents for nanoassembly, DNA delivery, and imaging. J. Am. Chem. Soc. 2013, 135, 2411–2414. [Google Scholar] [CrossRef]
- Huang, L.J.; Yu, R.Q.; Chu, X. DNA-functionalized upconversion nanoparticles as biosensors for rapid, sensitive, and selective detection of Hg(2+) in complex matrices. Analyst 2015, 140, 4987–4990. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Wang, D.; Pan, Y.; Guo, Y.; Li, H.; Zhang, F.; Zhu, X.; Li, Y.; Zhang, C.; Huang, L. Sequence-Dependent DNA Functionalization of Upconversion Nanoparticles and Their Programmable Assemblies. Angew. Chem. Int. Ed. 2020, 59, 8133–8137. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Song, G.; He, Y.; Zhang, X.; Liu, Y.; Ju, H. A DNA-Azobenzene Nanopump Fueled by Upconversion Luminescence for Controllable Intracellular Drug Release. Angew. Chem. Int. Ed. 2019, 58, 18207–18211. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Zhang, Y.; Zhang, X.; Liu, Y.; Ju, H. A Near-Infrared Photo-Switched MicroRNA Amplifier for Precise Photodynamic Therapy of Early-Stage Cancers. Angew. Chem. Int. Ed. 2020, 59, 21454–21459. [Google Scholar] [CrossRef]
- Lu, J.; Chen, Y.; Liu, D.; Ren, W.; Lu, Y.; Shi, Y.; Piper, J.; Paulsen, I.; Jin, D. One-Step Protein Conjugation to Upconversion Nanoparticles. Anal. Chem. 2015, 87, 10406–10413. [Google Scholar] [CrossRef]
- Wu, D.; Yang, K.; Zhang, Z.; Feng, Y.; Rao, L.; Chen, X.; Yu, G. Metal-free bioorthogonal click chemistry in cancer theranostics. Chem. Soc. Rev. 2022, 51, 1336–1376. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.; Chu, H.; Mi, Y.; Zhou, Z.; Di, Z.; Zhao, M.; Li, L. Light-Activated Nanoprobes for Biosensing and Imaging. Adv. Mater. 2019, 31, 1804745. [Google Scholar] [CrossRef]
- Basu, S.; Pacelli, S.; Feng, Y.; Lu, Q.; Wang, J.; Paul, A. Harnessing the Noncovalent Interactions of DNA Backbone with 2D Silicate Nanodisks To Fabricate Injectable Therapeutic Hydrogels. ACS Nano 2018, 12, 9866–9880. [Google Scholar] [CrossRef]
- Baig, N.; Kammakakam, I.; Falath, W. Nanomaterials: A review of synthesis methods, properties, recent progress, and challenges. Mater. Adv. 2021, 2, 1821–1871. [Google Scholar] [CrossRef]
- Zhao, H.; Lee, Y.; Han, M.; Sharma, B.K.; Chen, X.; Ahn, J.-H.; Rogers, J.A. Nanofabrication approaches for functional three-dimensional architectures. Nano Today 2020, 30, 100825. [Google Scholar] [CrossRef]
- Liddle, J.A.; Gallatin, G.M. Nanomanufacturing: A Perspective. ACS Nano 2016, 10, 2995–3014. [Google Scholar] [CrossRef] [PubMed]
- Carnerero, J.M.; Jimenez-Ruiz, A.; Castillo, P.M.; Prado-Gotor, R. Covalent and Non-Covalent DNA–Gold-Nanoparticle Interactions: New Avenues of Research. ChemPhysChem 2017, 18, 17–33. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, C.; Li, Y.; Ma, Y.; Deng, J.; Li, L.; Sun, J. Thermophoretic Detection of Exosomal microRNAs by Nanoflares. J. Am. Chem. Soc. 2020, 142, 4996–5001. [Google Scholar] [CrossRef]
- Wang, W.-J.; Li, J.-J.; Rui, K.; Gai, P.-P.; Zhang, J.-R.; Zhu, J.-J. Sensitive Electrochemical Detection of Telomerase Activity Using Spherical Nucleic Acids Gold Nanoparticles Triggered Mimic-Hybridization Chain Reaction Enzyme-Free Dual Signal Amplification. Anal. Chem. 2015, 87, 3019–3026. [Google Scholar] [CrossRef]
- Zhu, D.; Li, J.; Wang, L.; Li, Q.; Wang, L.; Song, B.; Zhou, R.; Fan, C. Hydrophobic collapse-driven nanoparticle coating with poly-adenine adhesives. Chem. Commun. 2021, 57, 3801–3804. [Google Scholar] [CrossRef]
- Ye, Y.; Hou, S.; Wu, X.; Cheng, X.; He, S. Freeze-Driven Adsorption of Poly-A DNA on Gold Nanoparticles: From a Stable Biointerface to Plasmonic Dimers. Langmuir 2022, 38, 4625–4632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Peng, R.; Liu, W.; Donovan, M.J.; Wang, L.; Ismail, I.; Li, J.; Li, J.; Qu, F.; Tan, W. Engineering DNA on the Surface of Upconversion Nanoparticles for Bioanalysis and Therapeutics. ACS Nano 2021, 15, 17257–17274. [Google Scholar] [CrossRef] [PubMed]
- Francés-Soriano, L.; Estebanez, N.; Pérez-Prieto, J.; Hildebrandt, N. DNA-Coated Upconversion Nanoparticles for Sensitive Nucleic Acid FRET Biosensing. Adv. Funct. Mater. 2022, 32, 2201541. [Google Scholar] [CrossRef]
- Yang, Z.; Loh, K.Y.; Chu, Y.-T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Nakamata Huynh, S.M.; Hwang, K.; Li, L.; Xing, H.; et al. Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef] [PubMed]
- Vizzini, P.; Manzano, M.; Farre, C.; Meylheuc, T.; Chaix, C.; Ramarao, N.; Vidic, J. Highly sensitive detection of Campylobacter spp. In chicken meat using a silica nanoparticle enhanced dot blot DNA biosensor. Biosens. Bioelectron. 2021, 171, 112689. [Google Scholar] [CrossRef]
- D’Agata, R.; Palladino, P.; Spoto, G. Streptavidin-coated gold nanoparticles: Critical role of oligonucleotides on stability and fractal aggregation. Beilstein J. Nanotechnol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Li, M.; Zhang, T.; Zhang, Y. Ultrasensitive electrochemical sensing platform for miRNA-21 detection based on manganese dioxide-gold nanoparticle nanoconjugates coupled with hybridization chain reaction and horseradish peroxidase signal amplification. Analyst 2023, 148, 2180–2188. [Google Scholar] [CrossRef]
- Chen, M.; Song, Z.; Yang, X.; Song, Z.; Luo, X. Antifouling peptides combined with recognizing DNA probes for ultralow fouling electrochemical detection of cancer biomarkers in human bodily fluids. Biosens. Bioelectron. 2022, 206, 114162. [Google Scholar] [CrossRef]
- van der Meer, S.B.; Loza, K.; Wey, K.; Heggen, M.; Beuck, C.; Bayer, P.; Epple, M. Click Chemistry on the Surface of Ultrasmall Gold Nanoparticles (2 nm) for Covalent Ligand Attachment Followed by NMR Spectroscopy. Langmuir 2019, 35, 7191–7204. [Google Scholar] [CrossRef]
- Yan, W.; Zhong, Z.; Ma, J.; Rujiralai, T. Highly sensitive colorimetric sensing of copper(ii) ions based on “CLICK-17” DNAzyme-catalyzed azide modified gold nanoparticles and alkyne capped dsDNA cycloaddition. RSC Adv. 2021, 11, 24196–24205. [Google Scholar] [CrossRef]
- Hu, L.; Takezawa, Y.; Shionoya, M. CuII-mediated DNA base pairing of a triazole-4-carboxylate nucleoside prepared by click chemistry. Chem. Commun. 2023, 59, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ge, Y.; Liu, M.; Pei, Y.; He, P.; Song, W.; Zhang, S. DNA-Au Janus Nanoparticles for In Situ SERS Detection and Targeted Chemo-photodynamic Synergistic Therapy. Anal. Chem. 2022, 94, 7823–7832. [Google Scholar] [CrossRef]
- Kyriazi, M.-E.; Giust, D.; El-Sagheer, A.H.; Lackie, P.M.; Muskens, O.L.; Brown, T.; Kanaras, A.G. Multiplexed mRNA Sensing and Combinatorial-Targeted Drug Delivery Using DNA-Gold Nanoparticle Dimers. ACS Nano 2018, 12, 3333–3340. [Google Scholar] [CrossRef]
- Damavandi, F.; Wang, W.; Shen, W.-Z.; Cetinel, S.; Jordan, T.; Jovel, J.; Montemagno, C.; Wong, G.K.-S. Enrichment of low abundance DNA/RNA by oligonucleotide-clicked iron oxide nanoparticles. Sci. Rep. 2021, 11, 13053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fischer, A.; Ouyang, Y.; Wang, J.; Sohn, Y.S.; Nechushtai, R.; Pikarsky, E.; Fan, C.; Willner, I. Aptamer-modified DNA tetrahedra-gated metal–organic framework nanoparticle carriers for enhanced chemotherapy or photodynamic therapy. Chem. Sci. 2021, 12, 14473–14483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gao, J.; Xue, W.; Di, Z.; Xing, H.; Lu, Y.; Li, L. Upconversion Luminescence-Activated DNA Nanodevice for ATP Sensing in Living Cells. J. Am. Chem. Soc. 2018, 140, 578–581. [Google Scholar] [CrossRef]
- Morzy, D.; Tekin, C.; Caroprese, V.; Rubio-Sánchez, R.; Di Michele, L.; Bastings, M.M.C. Interplay of the mechanical and structural properties of DNA nanostructures determines their electrostatic interactions with lipid membranes. Nanoscale 2023, 15, 2849–2859. [Google Scholar] [CrossRef]
- Hao, Y.; Li, Y.; Song, L.; Deng, Z. Flash Synthesis of Spherical Nucleic Acids with Record DNA Density. J. Am. Chem. Soc. 2021, 143, 3065–3069. [Google Scholar] [CrossRef]
- Zhao, X.; Han, Q.; Na, N.; Ouyang, J. Spatiotemporally Controlled DNA Nanoclamps: Single-Molecule Imaging of Receptor Protein Oligomerization. Anal. Chem. 2021, 93, 14514–14520. [Google Scholar] [CrossRef]
- Chai, X.; Fan, Z.; Yu, M.-M.; Zhao, J.; Li, L. A Redox-Activatable DNA Nanodevice for Spatially-Selective, AND-Gated Imaging of ATP and Glutathione in Mitochondria. Nano Lett. 2021, 21, 10047–10053. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Yu, M.; Gu, Z.; Li, L.; Zhao, Y. Time-Resolved Activation of pH Sensing and Imaging in Vivo by a Remotely Controllable DNA Nanomachine. Nano Lett. 2020, 20, 874–880. [Google Scholar] [CrossRef]
- Chu, H.; Zhao, J.; Mi, Y.; Zhao, Y.; Li, L. Near-Infrared Light-Initiated Hybridization Chain Reaction for Spatially and Temporally Resolved Signal Amplification. Angew. Chem. Int. Ed. 2019, 58, 14877–14881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, L.; Gao, W.; Yu, X.; Gu, W.; Fu, W.; Luo, Y. Spatiotemporally Controllable MicroRNA Imaging in Living Cells via a Near-Infrared Light-Activated Nanoprobe. ACS Appl. Mater. Interfaces 2020, 12, 35958–35966. [Google Scholar] [CrossRef]
- O’Hagan, M.P.; Duan, Z.; Huang, F.; Laps, S.; Dong, J.; Xia, F.; Willner, I. Photocleavable Ortho-Nitrobenzyl-Protected DNA Architectures and Their Applications. Chem. Rev. 2023, 123, 6839–6887. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Kong, Y.; Zhang, C.; Lv, Y.; Cheng, S.; Hou, D.; Xian, Y. Near-Infrared Light Controllable DNA Walker Driven by Endogenous Adenosine Triphosphate for in Situ Spatiotemporal Imaging of Intracellular MicroRNA. ACS Nano 2021, 15, 14253–14262. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Zhao, J.; Chu, H.; Li, Z.; Yu, M.; Li, L. Upconversion Luminescence-Controlled DNA Computation for Spatiotemporally Resolved, Multiplexed Molecular Imaging. Anal. Chem. 2021, 93, 2500–2509. [Google Scholar] [CrossRef]
- Zhao, T.; Gao, Y.; Wang, J.; Cui, Y.; Niu, S.; Xu, S.; Luo, X. From Passive Signal Output to Intelligent Response: “On-Demand” Precise Imaging Controlled by Near-Infrared Light. Anal. Chem. 2021, 93, 12329–12336. [Google Scholar] [CrossRef]
- Gao, P.; Shen, X.; Liu, X.; Cui, B.; Wang, M.; Wan, X.; Li, N.; Tang, B. Covalent Organic Framework-Derived Carbonous Nanoprobes for Cancer Cell Imaging. ACS Appl. Mater. Interfaces 2021, 13, 41498–41506. [Google Scholar] [CrossRef]
- Wang, L.; Wang, K.; Wang, X.; Niu, R.; Chen, X.; Zhu, Y.; Sun, Z.; Yang, J.; Liu, G.; Luo, Y. Intelligent Dual-Lock Deoxyribonucleic Acid Automatons Boosting Precise Tumor Imaging. ACS Appl. Mater. Interfaces 2023, 15, 3826–3838. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Y.; Wang, W.; Min, Q.; Zhang, J.R.; Zhu, J.J. A Telomerase-Assisted Strategy for Regeneration of DNA Nanomachines in Living Cells. Angew. Chem. Int. Ed. 2023, 62, e202213884. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, Y.; Li, L.; Li, Z.; Li, M. Endogenous Enzyme-Operated Spherical Nucleic Acids for Cell-Selective Protein Capture and Localization Regulation. Angew. Chem. Int. Ed. 2023, 62, e202214958. [Google Scholar] [CrossRef]
- Li, L.; Xing, H.; Zhang, J.; Lu, Y. Functional DNA Molecules Enable Selective and Stimuli-Responsive Nanoparticles for Biomedical Applications. Acc. Chem. Res. 2019, 52, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, B.; Li, M.; Huang, Y.; Li, L. Heterostructures Made of Upconversion Nanoparticles and Metal–Organic Frameworks for Biomedical Applications. Adv. Sci. 2022, 9, 2103911. [Google Scholar] [CrossRef]
- Xu, R.; Li, Y.; Zhu, C.; Liu, D.; Yang, Y.R. Cellular Ingestible DNA Nanostructures for Biomedical Applications. Adv. NanoBiomed Res. 2023, 3, 2200119. [Google Scholar] [CrossRef]
- Tunc, C.U.; Culha, M. Gold nanoparticles conjugated DNA-tile nanomaterials for simultaneous delivery of morpholino antisense oligonucleotides and doxorubicin. J. Drug Delivery Sci. Technol. 2022, 74, 103546. [Google Scholar] [CrossRef]
- Xue, C.; Hu, S.; Gao, Z.-H.; Wang, L.; Luo, M.-X.; Yu, X.; Li, B.-F.; Shen, Z.; Wu, Z.-S. Programmably tiling rigidified DNA brick on gold nanoparticle as multi-functional shell for cancer-targeted delivery of siRNAs. Nat. Commun. 2021, 12, 2928. [Google Scholar] [CrossRef]
- Tebcharani, L.; Akter, N.; Fan, D.; Lieleg, O.; Gibbs, J.M.; Boekhoven, J. Hydrolyzable emulsions as a dual release platform for hydrophobic drugs and DNA. Chem. Commun. 2023, 59, 8099. [Google Scholar] [CrossRef]
- Pal, S.; Koneru, J.K.; Andreou, C.; Rakshit, T.; Rajasekhar, V.K.; Wlodarczyk, M.; Healey, J.H.; Kircher, M.F.; Mondal, J. DNA-Functionalized Gold Nanorods for Perioperative Optical Imaging and Photothermal Therapy of Triple-Negative Breast Cancer. ACS Appl. Nano Mater. 2022, 5, 9159–9169. [Google Scholar] [CrossRef]
- Shankar, R.; Nayak, S.; Singh, S.; Sen, A.; Kumar, N.; Bhushan, R.; Aggarwal, M.; Das, P. Simultaneous Sustained Drug Delivery, Tracking, and On-Demand Photoactivation of DNA–Hydrogel Formulated from a Biomass-Derived DNA Nanoparticle. ACS Appl. Bio Mater. 2023, 6, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
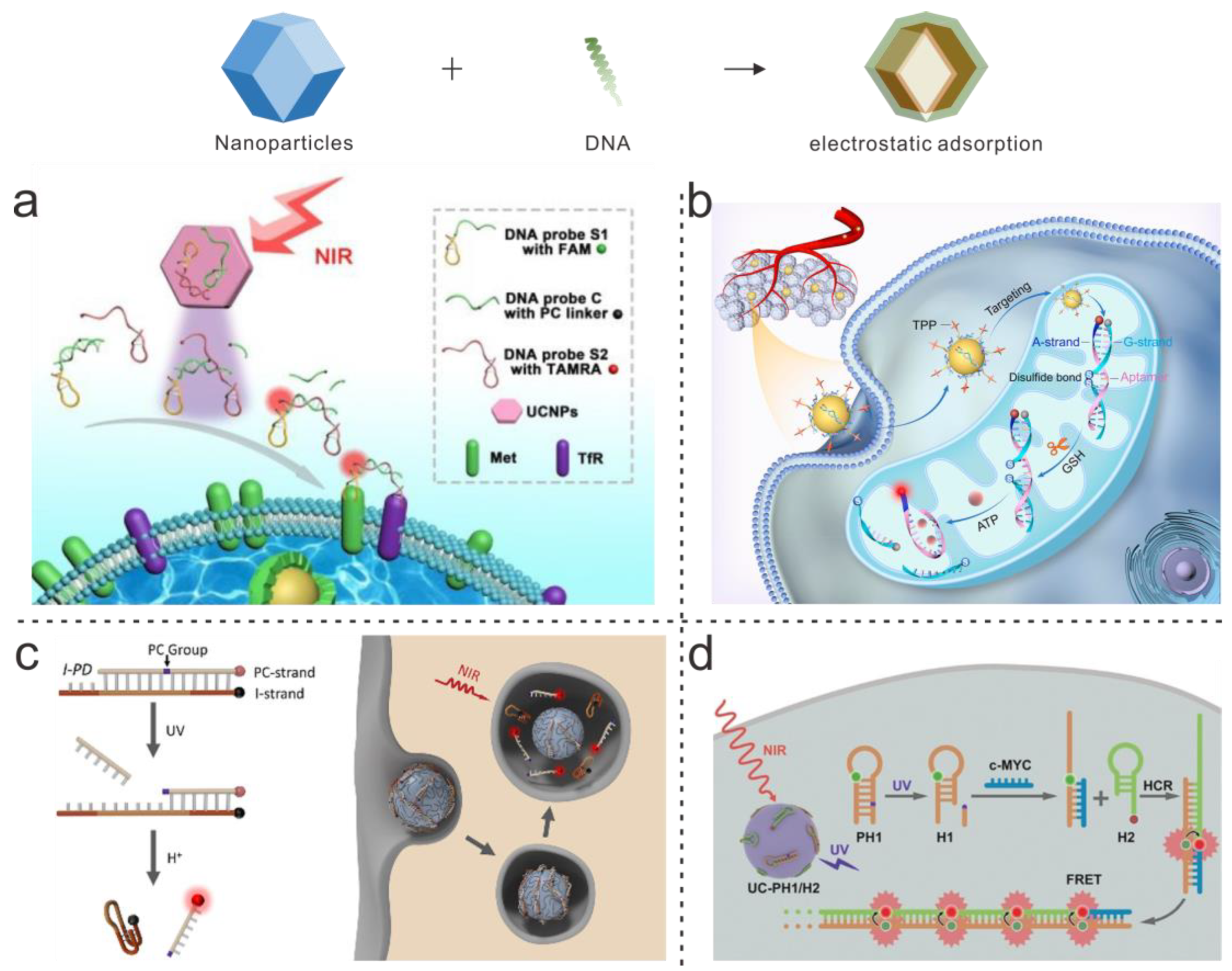
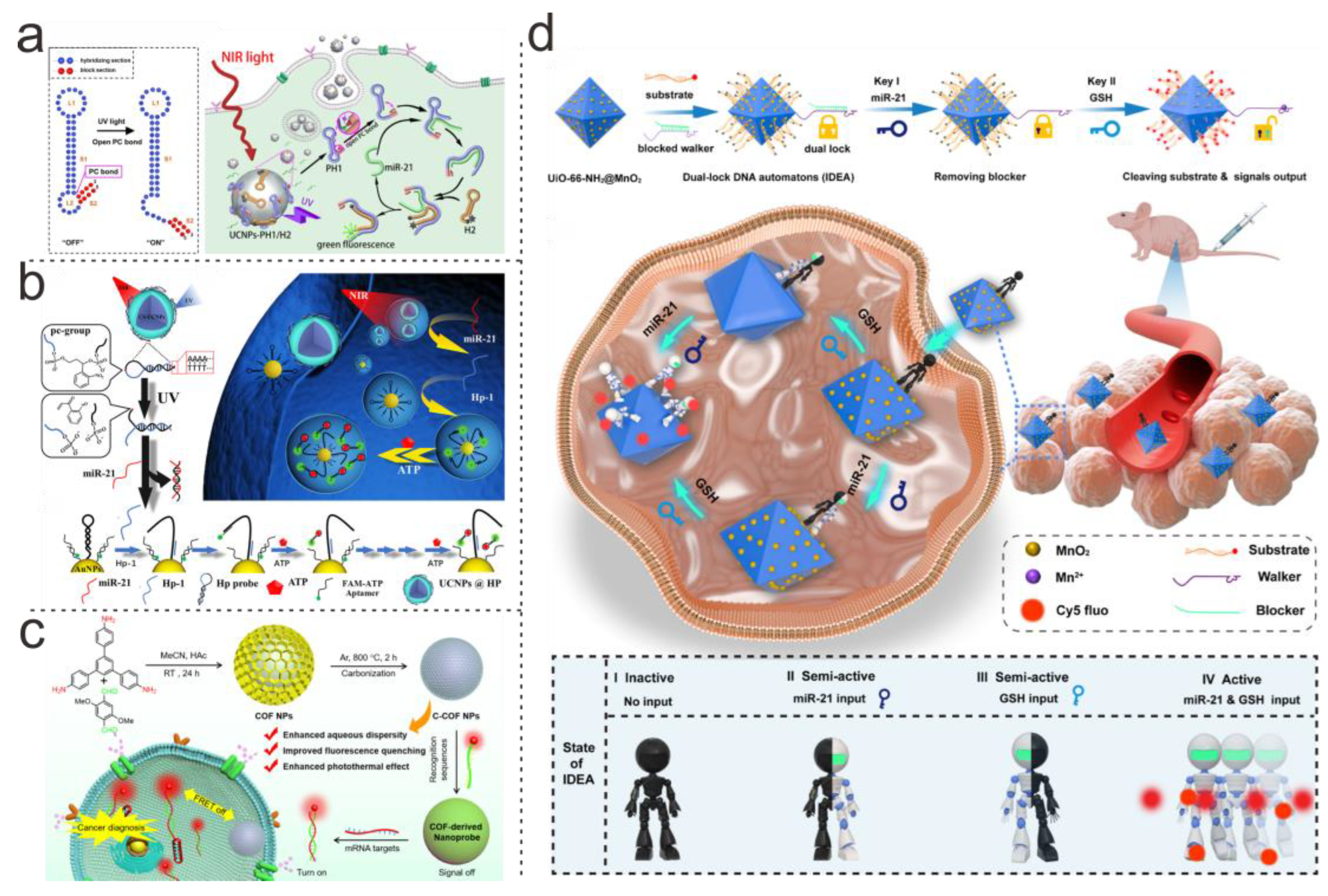
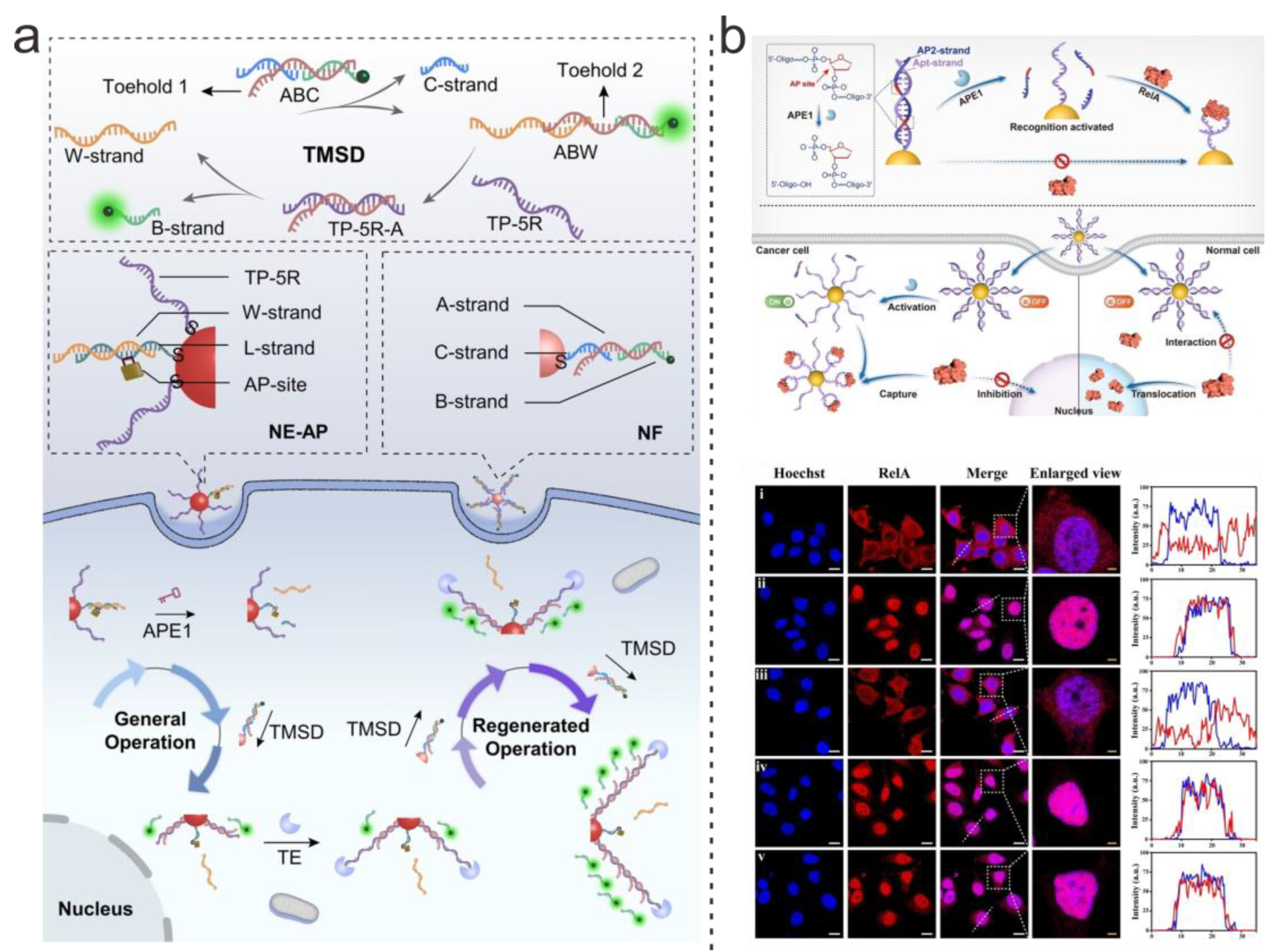

| DNA Structure | Nanoparticle’s Type | Connection Type | Analyte | LOD | Application | Ref. |
|---|---|---|---|---|---|---|
| DNA dumbbell structure | UCNPs | Allotropic bond | miRNA-21 | Single cell | In situ imaging | [62] |
| Hairpin DNA structure | UCNPs | Allotropic bond | miRNA-21 | Single cell | In situ imaging | [64] |
| Double-stranded | UCNPs | Allotropic bond | ATP, miRNA-21 | Single cell | In vivo imaging | [65] |
| Triangle structure | UCNPs | Allotropic bond | miRNA-21, miRNA-373, miRNA-155 | Single cell | In situ imaging | [66] |
| Single strand | COFs | Electrostatic adsorption | mRNA | Single cell | Cancer diagnosis | [67] |
| Double-strands | ZrMOF@MnO2 | Covalent bonding | miRNA-21, GSH | Single cell | In vivo imaging | [68] |
| Double-Strands | AuNPs | Covalent bonding | RelA protein | Single cell | In situ imaging | [69] |
| Double-Strands | AuNPs | Covalent bonding | APE1 enzyme | Single cell | In situ imaging | [70] |
| DNA Structure | Nanoparticle’s Type | Connection Type | Treatment Modalities | Application | Ref. |
|---|---|---|---|---|---|
| Y-shaped backbone-rigidified triangular DNA | AuNPs | Covalent bonding | Gene silencing | Cancer therapy | [75] |
| Cholesterol-conjugated DNA | AuNPs | Conjugate connection | Gene silencing | Drug release calculation | [76] |
| Single DNA | AuNRs | Covalent bonding | Phototherapy | Triple-negative breast cancer therapy | [77] |
| Biomass DNA | DNA dots | Conjugate connection | Photoactivated ROS Generation | Cancer therapy | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Zhu, J.-J. Linkage Pathways of DNA–Nanoparticle Conjugates and Biological Applications. Chemosensors 2023, 11, 444. https://doi.org/10.3390/chemosensors11080444
Huang S, Zhu J-J. Linkage Pathways of DNA–Nanoparticle Conjugates and Biological Applications. Chemosensors. 2023; 11(8):444. https://doi.org/10.3390/chemosensors11080444
Chicago/Turabian StyleHuang, Shan, and Jun-Jie Zhu. 2023. "Linkage Pathways of DNA–Nanoparticle Conjugates and Biological Applications" Chemosensors 11, no. 8: 444. https://doi.org/10.3390/chemosensors11080444
APA StyleHuang, S., & Zhu, J.-J. (2023). Linkage Pathways of DNA–Nanoparticle Conjugates and Biological Applications. Chemosensors, 11(8), 444. https://doi.org/10.3390/chemosensors11080444







