Colorimetric Detection and Killing of Bacteria by Enzyme-Instructed Self-Aggregation of Peptide-Modified Gold Nanoparticles
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials and Instruments
2.2. Preparation and Characterization of the 13 nm AuNPs
2.3. Preparation and Characterization of the AuNPs-CF4KYP
2.4. Transmission Electron Microscopy Characterization of AuNPs/AuNPs-CF4KYP
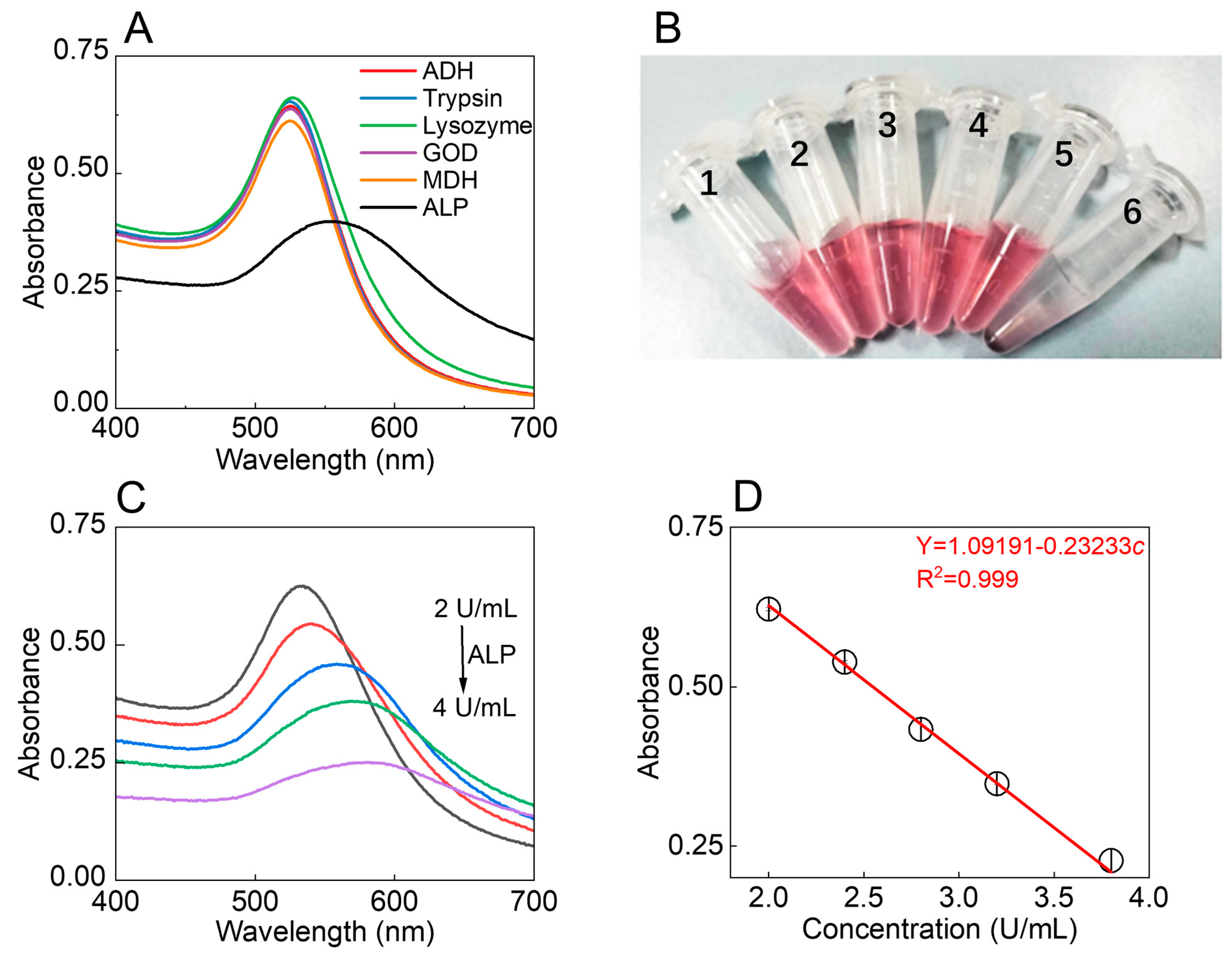
2.5. Selectivity and Sensitivity of AuNPs-CF4KYP for ALP
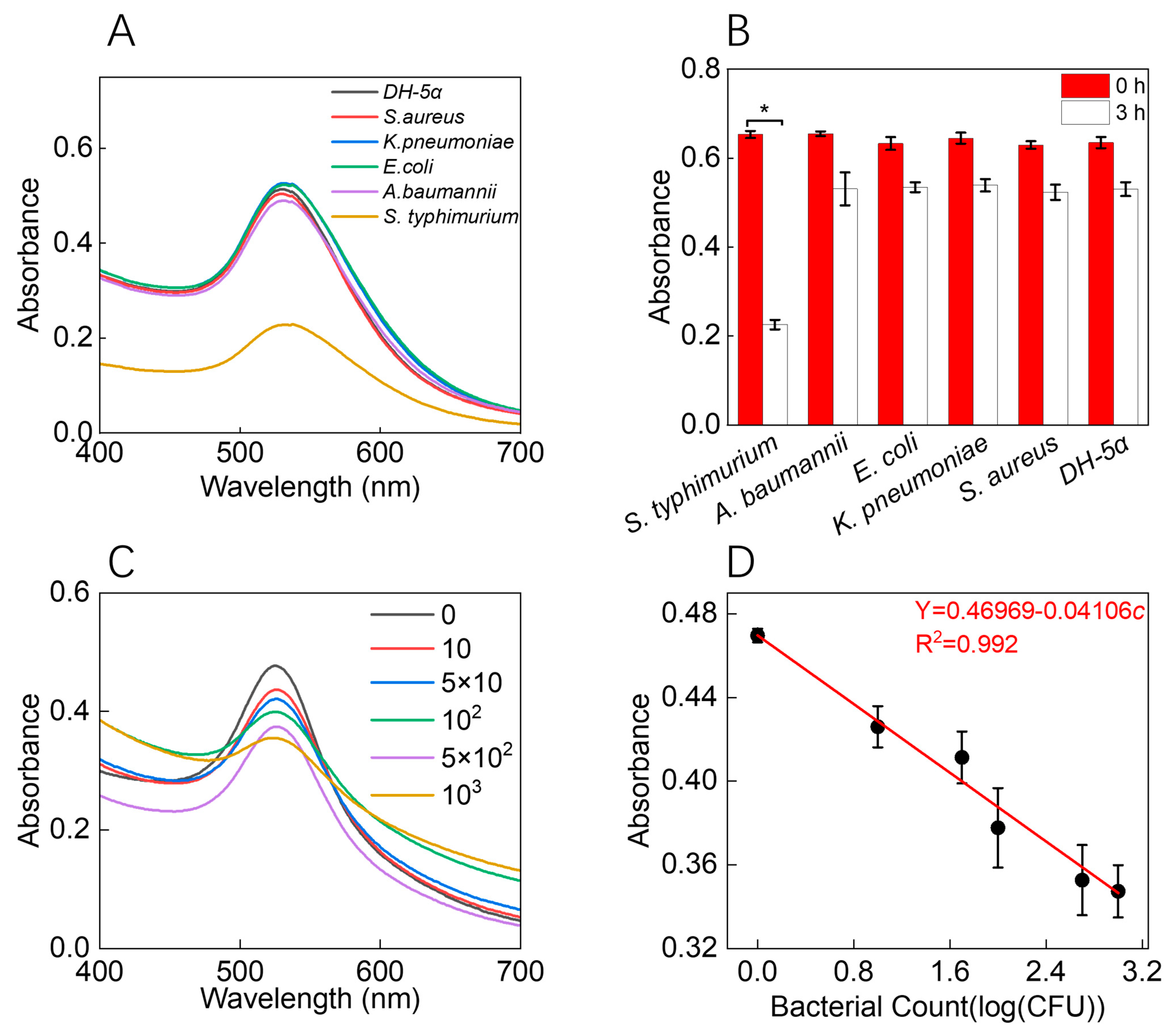
2.6. Selectivity and Sensitivity of AuNPs-CF4KYP for Bacterial ALP
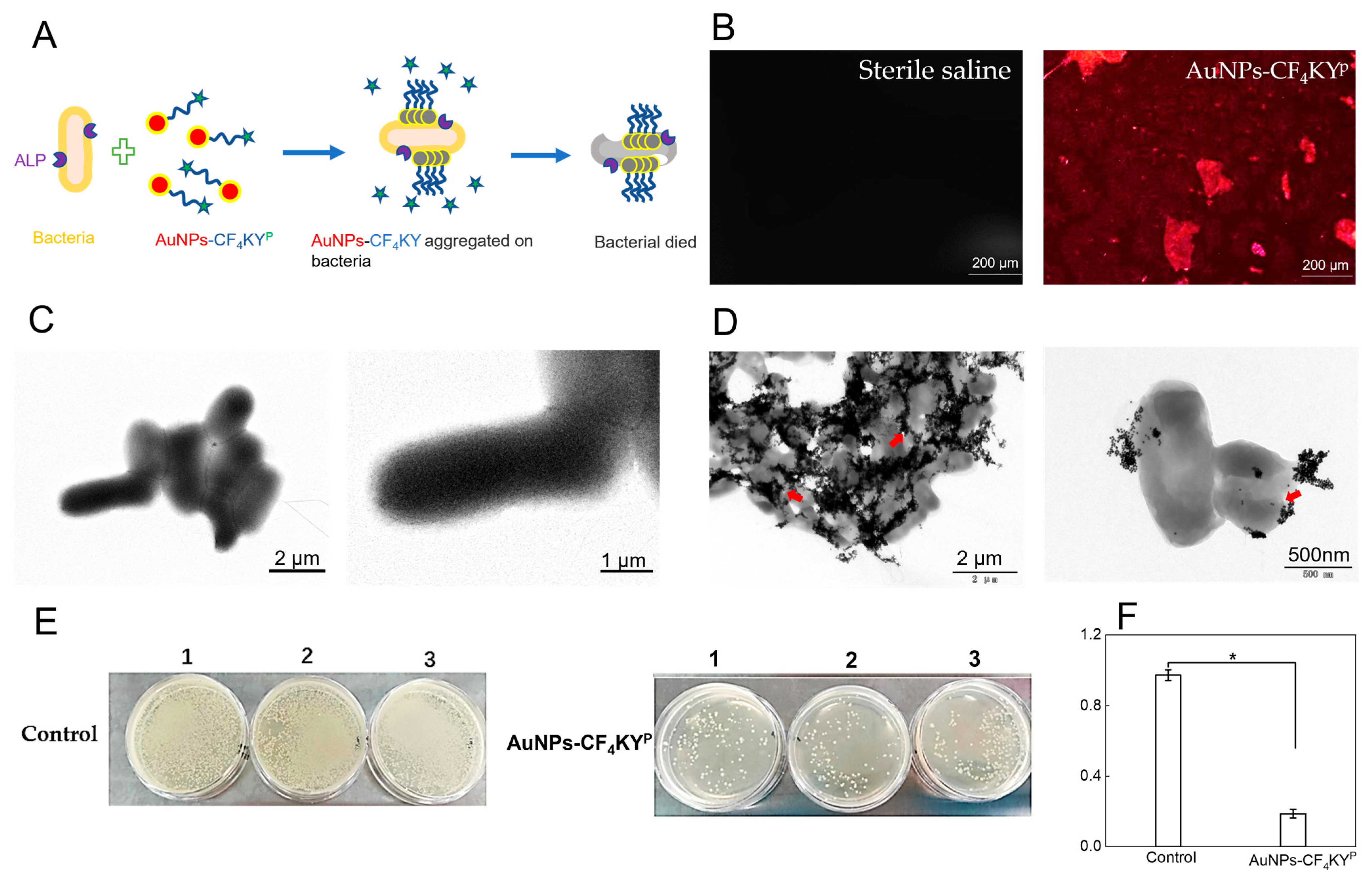
2.7. Antimicrobial Performance of AuNPs-CF4KYP
2.8. TEM Characterization of S. typhimurium
2.9. Statistical Analysis
3. Results and Discussion
3.1. Preparation and Characterization of the 13 nm AuNPs and AuNPs-CF4KYP
3.2. Response Ability of the AuNPs-CF4KYP to ALP
3.3. Response Ability of the AuNPs-CF4KYP to Bacterial ALP
3.4. Antimicrobial Performance of AuNPs-CF4KYP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xie, M.; Chen, T.; Xin, X.; Cai, Z.; Dong, C.; Lei, B. Multiplex detection of foodborne pathogens by real-time loop-mediated isothermal amplification on a digital microfluidic chip. Food Control 2022, 136, 108824. [Google Scholar] [CrossRef]
- Wang, B.; Liu, S.; Sui, Z.; Wang, J.; Wang, Y.; Gu, S. Rapid Flow Cytometric Detection of Single Viable Salmonella Cells in Milk Powder. Foodborne Pathog. Dis. 2020, 17, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Xue, L.; Qi, W.; Cai, G.; Liu, Y.; Lin, J. An ultrasensitive impedance biosensor for Salmonella detection based on rotating high gradient magnetic separation and cascade reaction signal amplification. Biosens. Bioelectron. 2021, 176, 112921. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Teixeira, P.; Oliveira, R.; Azeredo, J. Adhesion to and viability of Listeria monocytogenes on food contact surfaces. J. Food Prot. 2008, 71, 1379–1385. [Google Scholar] [CrossRef]
- Silver, L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011, 24, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, M.J.; Fromm, K.M.; Ashkarran, A.A.; Jimenez de Aberasturi, D.; de Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef]
- Manzetti, S.; Ghisi, R. The environmental release and fate of antibiotics. Mar. Pollut. Bull. 2014, 79, 7–15. [Google Scholar] [CrossRef]
- Mannoor, M.S.; Tao, H.; Clayton, J.D.; Sengupta, A.; Kaplan, D.L.; Naik, R.R.; Verma, N.; Omenetto, F.G.; McAlpine, M.C. Graphene-based wireless bacteria detection on tooth enamel. Nat. Commun. 2012, 3, 763. [Google Scholar] [CrossRef]
- Disney, M.D.; Zheng, J.; Swager, T.M.; Seeberger, P.H. Detection of bacteria with carbohydrate-functionalized fluorescent polymers. J. Am. Chem. Soc. 2004, 126, 13343–13346. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef]
- Jangampalli Adi, P.; Naidu, J.R.; Matcha, B. Multiplex quantification of Escherichia coli, Salmonella typhi and Vibrio cholera with three DNA targets in single reaction assay. Microb. Pathog. 2017, 110, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Zhang, Y.; Li, L.; Wang, H.; Li, Q.; Tao, X.; Song, Y.; Song, E. Intracellular Pathogen Detection Based on Dual-Recognition Units Constructed Fluorescence Resonance Energy Transfer Nanoprobe. Anal. Chem. 2020, 92, 11462–11468. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Hu, Y.; Liu, F.; Wei, S.; Fang, D.; Shuhendler, A.J.; Liu, H.; Chen, H.-Y.; Ye, D. Activatable NIR Fluorescence/MRI Bimodal Probes for in Vivo Imaging by Enzyme-Mediated Fluorogenic Reaction and Self-Assembly. J. Am. Chem. Soc. 2019, 141, 10331–10341. [Google Scholar] [CrossRef] [PubMed]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef]
- Appaturi, J.N.; Pulingam, T.; Thong, K.L.; Muniandy, S.; Ahmad, N.; Leo, B.F. Rapid and sensitive detection of Salmonella with reduced graphene oxide-carbon nanotube based electrochemical aptasensor. Anal. Biochem. 2020, 589, 113489. [Google Scholar] [CrossRef]
- Zhou, S.; Lu, C.; Li, Y.; Xue, L.; Zhao, C.; Tian, G.; Bao, Y.; Tang, L.; Lin, J.; Zheng, J. Gold Nanobones Enhanced Ultrasensitive Surface-Enhanced Raman Scattering Aptasensor for Detecting Escherichia coli O157:H7. ACS Sens. 2020, 5, 588–596. [Google Scholar] [CrossRef]
- Gao, X.; Yin, Y.; Wu, H.; Hao, Z.; Li, J.; Wang, S.; Liu, Y. Integrated SERS Platform for Reliable Detection and Photothermal Elimination of Bacteria in Whole Blood Samples. Anal. Chem. 2021, 93, 1569–1577. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, T.; Wu, Z.; Zhang, F.; Wang, Y.; Wang, X.; Zhang, Z.; Li, C.; Lv, X.; Chen, D.; et al. Universal Method for Label-Free Detection of Pathogens and Biomolecules by Surface-Enhanced Raman Spectroscopy Based on Gold Nanoparticles. Anal. Chem. 2023, 95, 4050–4058. [Google Scholar] [CrossRef]
- Hu, J.; Tang, F.; Jiang, Y.Z.; Liu, C. Rapid screening and quantitative detection of Salmonella using a quantum dot nanobead-based biosensor. Analyst 2020, 145, 2184–2190. [Google Scholar] [CrossRef]
- Li, Z.; Askim, J.R.; Suslick, K.S. The Optoelectronic Nose: Colorimetric and Fluorometric Sensor Arrays. Chem. Rev. 2019, 119, 231–292. [Google Scholar] [CrossRef]
- Ma, X.; Song, L.; Zhou, N.; Xia, Y.; Wang, Z. A novel aptasensor for the colorimetric detection of S. typhimurium based on gold nanoparticles. Int. J. Food Microbiol. 2017, 245, 1–5. [Google Scholar] [CrossRef]
- Wu, S.; Duan, N.; Qiu, Y.; Li, J.; Wang, Z. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int. J. Food Microbiol. 2017, 261, 42–48. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, X.; Gong, F.; Liu, T.; Liu, Z. 2D Nanomaterials for Cancer Theranostic Applications. Adv. Mater. 2020, 32, 1902333. [Google Scholar] [CrossRef]
- Xu, J.; Hu, Y.; Wang, S.; Ma, X.; Guo, J. Nanomaterials in electrochemical cytosensors. Analyst 2020, 145, 2058–2069. [Google Scholar] [CrossRef]
- Pirsa, S.; Alizadeh, N. Nanoporous Conducting Polypyrrole Gas Sensor Coupled to a Gas Chromatograph for Determination of Aromatic Hydrocarbons Using Dispersive Liquid-Liquid Microextraction Method. IEEE Sens. J. 2011, 11, 3400–3405. [Google Scholar] [CrossRef]
- Lin, S.Y.; Wu, S.H.; Chen, C.H. A simple strategy for prompt visual sensing by gold nanoparticles: General applications of interparticle hydrogen bonds. Angew. Chem. Int. Ed. 2006, 45, 4948–4951. [Google Scholar] [CrossRef] [PubMed]
- Li, R.S.; Zhang, H.Z.; Ling, J.; Huang, C.Z.; Wang, J. Plasmonic platforms for colorimetric sensing of cysteine. Appl. Spectrosc. Rev. 2016, 51, 129–147. [Google Scholar] [CrossRef]
- Yang, S.; Yao, D.; Wang, Y.; Yang, W.; Zhang, B.; Wang, D. Enzyme-triggered self-assembly of gold nanoparticles for enhanced retention effects and photothermal therapy of prostate cancer. Chem. Commun. 2018, 54, 9841–9844. [Google Scholar] [CrossRef]
- Zhao, W.; Chiuman, W.; Brook, M.A.; Li, Y. Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. ChemBioChem Eur. J. Chem. Biol. 2007, 8, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M.; Grzybowski, B. Self-assembly at all scales. Science 2002, 295, 2418–2421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Emerging biological materials through molecular self-assembly. Biotechnol. Adv. 2002, 20, 321–339. [Google Scholar] [CrossRef]
- Song, S.; Qin, Y.; He, Y.; Huang, Q.; Fan, C.; Chen, H.Y. Functional nanoprobes for ultrasensitive detection of biomolecules. Chem. Soc. Rev. 2010, 39, 4234–4243. [Google Scholar] [CrossRef]
- Li, H.; Huang, H.; Feng, J.J.; Luo, X.; Fang, K.M.; Wang, Z.G.; Wang, A.J. A polypeptide-mediated synthesis of green fluorescent gold nanoclusters for Fe3+ sensing and bioimaging. J. Colloid Interface Sci. 2017, 506, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Li, R.S.; Liu, J.; Shi, H.; Hu, P.P.; Wang, Y.; Gao, P.F.; Wang, J.; Jia, M.; Li, H.; Li, Y.F.; et al. Transformable Helical Self-Assembly for Cancerous Golgi Apparatus Disruption. Nano Lett. 2021, 21, 8455–8465. [Google Scholar] [CrossRef]
- Lim, E.K.; Keem, J.O.; Yun, H.S.; Jung, J.; Chung, B.H. Smart nanoprobes for the detection of alkaline phosphatase activity during osteoblast differentiation. Chem. Commun. 2015, 51, 3270–3272. [Google Scholar] [CrossRef]
- Kaliannan, K.; Hamarneh, S.R.; Economopoulos, K.P.; Nasrin Alam, S.; Moaven, O.; Patel, P.; Malo, N.S.; Ray, M.; Abtahi, S.M.; Muhammad, N.; et al. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 7003–7008. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.R.; Awan, F.R.; Najam, S.S.; Islam, M.; Siddique, T.; Zain, M. Elevated serum level of human alkaline phosphatase in obesity. J. Pak. Med. Assoc. 2015, 65, 1182–1185. [Google Scholar]
- Ooi, K.; Shiraki, K.; Morishita, Y.; Nobori, T. High-molecular intestinal alkaline phosphatase in chronic liver diseases. J. Clin. Lab. Anal. 2007, 21, 133–139. [Google Scholar] [CrossRef]
- Zhou, J.; Du, X.; Berciu, C.; He, H.; Shi, J.; Nicastro, D.; Xu, B. Enzyme-Instructed Self-Assembly for Spatiotemporal Profiling of the Activities of Alkaline Phosphatases on Live Cells. Chem 2016, 1, 246–263. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Zhang, Q.; Wang, J.; Yi, M.; He, H.; Xu, B. Enzymatic Assemblies of Thiophosphopeptides Instantly Target Golgi Apparatus and Selectively Kill Cancer Cells. Angew. Chem. Int. Ed. 2021, 60, 12796–12801. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zheng, D.; Xie, L.; Zhang, X.; Zhang, Z.; Wang, L.; Hu, Z.W.; Yang, Z. Enzyme-Instructed Peptide Assembly Favored by Preorganization for Cancer Cell Membrane Engineering. J. Am. Chem. Soc. 2023, 145, 4366–4371. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Q.; Shy, A.N.; Yi, M.; He, H.; Lu, S.; Xu, B. Enzymatically Forming Intranuclear Peptide Assemblies for Selectively Killing Human Induced Pluripotent Stem Cells. J. Am. Chem. Soc. 2021, 143, 15852–15862. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, Y.; Zhang, C.; Ye, C.; Bian, Q.; Cheng, X.; Xia, H.; Zheng, J.; Liu, H. A novel colorimetric method for H2O2 sensing and its application: Fe2+-catalyzed H2O2 prevents aggregation of AuNPs by oxidizing cysteine (FeHOAuC). Anal. Chim. Acta 2022, 1207, 339840. [Google Scholar] [CrossRef]
- Nie, H.-Y.; Romanovskaia, E.; Romanovski, V.; Hedberg, J.; Hedberg, Y.S. Detection of gold cysteine thiolate complexes on gold nanoparticles with time-of-flight secondary ion mass spectrometry. Biointerphases 2021, 16, 021005. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, D.A.; Deng, T.; Li, J.S. Combination of Alkaline Phosphatase/Graphene Oxide Nanoconjugates and D-Glucose-6-Phosphate-Functionalized Gold Nanoparticles for the Rapid Colorimetric Assay of Pathogenic Bacteria. Biosens. Bioelectron. 2022, 216, 114611. [Google Scholar] [CrossRef]
- Roh, S.G.; Robby, A.I.; Phuong, P.T.M.; In, I.; Park, S.Y. Photoluminescence-Tunable Fluorescent Carbon Dots-Deposited Silver Nanoparticle for Detection and Killing of Bacteria. Mater. Sci. Eng. C. 2019, 97, 613–623. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, C.H.; Hu, F.; Gao, Y.; Wang, Z.Y.; Li, H.Q.; Liu, J.F.; Liu, B.; Yang, C.H. Detection of Bacterial Alkaline Phosphatase Activity by Enzymatic in Situ Self-Assembly of the Aiegen-Peptide Conjugate. Anal. Chem. 2020, 92, 5185–5190. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, X.; Hu, H.; Huang, Y.; Yang, X.; Wang, Q.; Chen, X. Improving the Detection Limit of Salmonella Colorimetry Using Long Ssdna of Asymmetric-Pcr and Non-Functionalized Aunps. Anal. Biochem. 2021, 626, 114229. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiao, M.; Lai, W.; Wan, Y.; Li, L.; Pei, H. Stochastic DNA Dual-Walkers for Ultrafast Colorimetric Bacteria Detection. Anal. Chem. 2020, 92, 4990–4995. [Google Scholar] [CrossRef]
- Le, T.N.; Tran, T.D.; Kim, M.I. A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles. Nanomaterials 2020, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.B.; Mazrad, Z.a.I.; Robby, A.I.; In, I.; Park, S.Y. Alkaline Phosphatase-Responsive Fluorescent Polymer Probe Coated Surface for Colorimetric Bacteria Detection. Eur. Polym. J. 2018, 105, 217–225. [Google Scholar] [CrossRef]
- Wang, K.-Y.; Bu, S.-J.; Ju, C.-J.; Li, C.-T.; Li, Z.-Y.; Han, Y.; Ma, C.-Y.; Wang, C.-Y.; Hao, Z.; Liu, W.-S.; et al. Hemin-Incorporated Nanoflowers as Enzyme Mimics for Colorimetric Detection of Foodborne Pathogenic Bacteria. Bioorg. Med. Chem. Lett. 2018, 28, 3802–3807. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, Y.; Li, Z.; Dong, M.; Dou, W.; Xu, X.; He, S. Bridge-DNA Synthesis Triggered by an Allosteric Aptamer for the Colorimetric Detection of Pathogenic Bacteria. Anal. Methods 2023, 15, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, J.; Warden, A.R.; Ding, X. Colorimetric and Photographic Detection of Bacteria in Drinking Water by Using 4-Mercaptophenylboronic Acid Functionalized Aunps. Food Control 2020, 108, 106885. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, C.; Zhao, W.; Zhang, H.; Yao, S.; Shi, Y.; Li, J.; Wang, J. Multi-Functional Mno2-Doped Fe3o4 Nanoparticles as an Artificial Enzyme for the Colorimetric Detection of Bacteria. Anal. Bioanal. Chem. 2020, 412, 3135–3140. [Google Scholar] [CrossRef]
- Wang, C.; Gao, X.; Wang, S.; Liu, Y. A Smartphone-Integrated Paper Sensing System for Fluorescent and Colorimetric Dual-Channel Detection of Foodborne Pathogenic Bacteria. Anal. Bioanal. Chem. 2020, 412, 611–620. [Google Scholar] [CrossRef]
- Chen, Q.M.; Gao, R.; Jia, L. Enhancement of the Peroxidase-Like Activity of Aptamers Modified Gold Nanoclusters by Bacteria for Colorimetric Detection of Salmonella Typhimurium. Talanta 2021, 221, 121476. [Google Scholar] [CrossRef]
- Bahadoran, A.; Jabarabadi, M.K.; Mahmood, Z.H.; Bokov, D.; Janani, B.J.; Fakhri, A. Quick and Sensitive Colorimetric Detection of Amino Acid with Functionalized-Silver/Copper Nanoparticles in the Presence of Cross Linker, and Bacteria Detection by Using DNA-Template Nanoparticles as Peroxidase Activity. Spectrochim. Acta A 2022, 268, 120636. [Google Scholar] [CrossRef]
- Bagheri Pebdeni, A.; Hosseini, M. Fast and Selective Whole Cell Detection of Staphylococcus Aureus Bacteria in Food Samples by Paper Based Colorimetric Nanobiosensor Using Peroxidase-Like Catalytic Activity of DNA-Au/Pt Bimetallic Nanoclusters. Microchem. J. 2020, 159, 105475. [Google Scholar] [CrossRef]
- Sun, J.; Huang, J.; Li, Y.; Lv, J.; Ding, X. A Simple and Rapid Colorimetric Bacteria Detection Method Based on Bacterial Inhibition of Glucose Oxidase-Catalyzed Reaction. Talanta 2019, 197, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Verdoodt, N.; Basso, C.R.; Rossi, B.F.; Pedrosa, V.A. Development of a Rapid and Sensitive Immunosensor for the Detection of Bacteria. Food Chem. 2017, 221, 1792–1796. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, B.; Liu, X.; Li, Z.; Zhu, S.; Liang, Y.; Cui, Z.; Wu, S. Recent Progress in Photocatalytic Antibacterial. ACS Appl. Bio Mater. 2021, 4, 3909–3936. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Wu, J.; Li, M.; Tan, J.; Fu, W.; Tang, H.; Zhang, P. Negatively Charged Sulfur Quantum Dots for Treatment of Drug-Resistant Pathogenic Bacterial Infections. Nano Lett. 2021, 21, 9433–9441. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, D.; Li, X.; Wang, X.; Liu, J.-Z.; She, W.-Z.; Liu, J.; Ling, J.; Li, R.S.; Cao, Q. Colorimetric Detection and Killing of Bacteria by Enzyme-Instructed Self-Aggregation of Peptide-Modified Gold Nanoparticles. Chemosensors 2023, 11, 484. https://doi.org/10.3390/chemosensors11090484
Yin D, Li X, Wang X, Liu J-Z, She W-Z, Liu J, Ling J, Li RS, Cao Q. Colorimetric Detection and Killing of Bacteria by Enzyme-Instructed Self-Aggregation of Peptide-Modified Gold Nanoparticles. Chemosensors. 2023; 11(9):484. https://doi.org/10.3390/chemosensors11090484
Chicago/Turabian StyleYin, Dan, Xiao Li, Xin Wang, Jin-Zhou Liu, Wen-Zhi She, Jiahui Liu, Jian Ling, Rong Sheng Li, and Qiue Cao. 2023. "Colorimetric Detection and Killing of Bacteria by Enzyme-Instructed Self-Aggregation of Peptide-Modified Gold Nanoparticles" Chemosensors 11, no. 9: 484. https://doi.org/10.3390/chemosensors11090484
APA StyleYin, D., Li, X., Wang, X., Liu, J.-Z., She, W.-Z., Liu, J., Ling, J., Li, R. S., & Cao, Q. (2023). Colorimetric Detection and Killing of Bacteria by Enzyme-Instructed Self-Aggregation of Peptide-Modified Gold Nanoparticles. Chemosensors, 11(9), 484. https://doi.org/10.3390/chemosensors11090484








